Symmetry in Nucleic-Acid Double Helices
Abstract
:1. Introduction
2. General Structural Principles of Nucleic-Acid Double Helices
2.1. Symmetry of Base Pairs
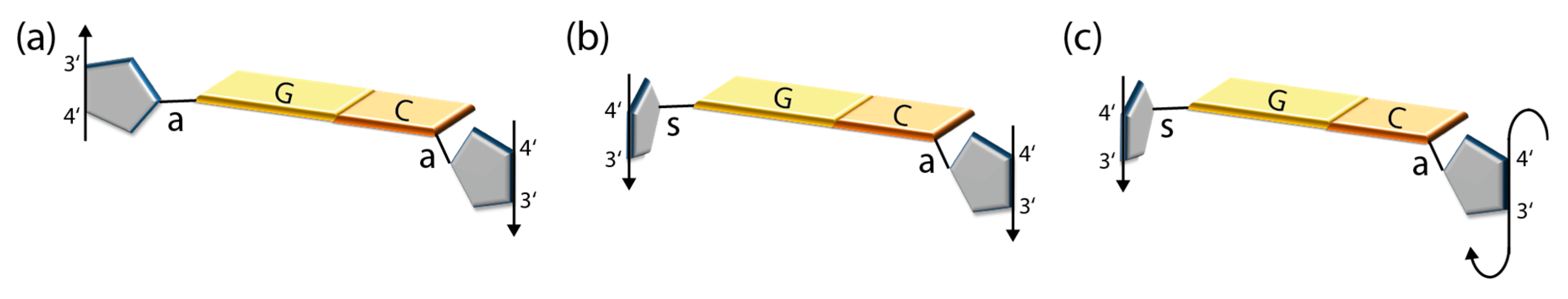
2.2. Symmetry of Nucleoside Pairs
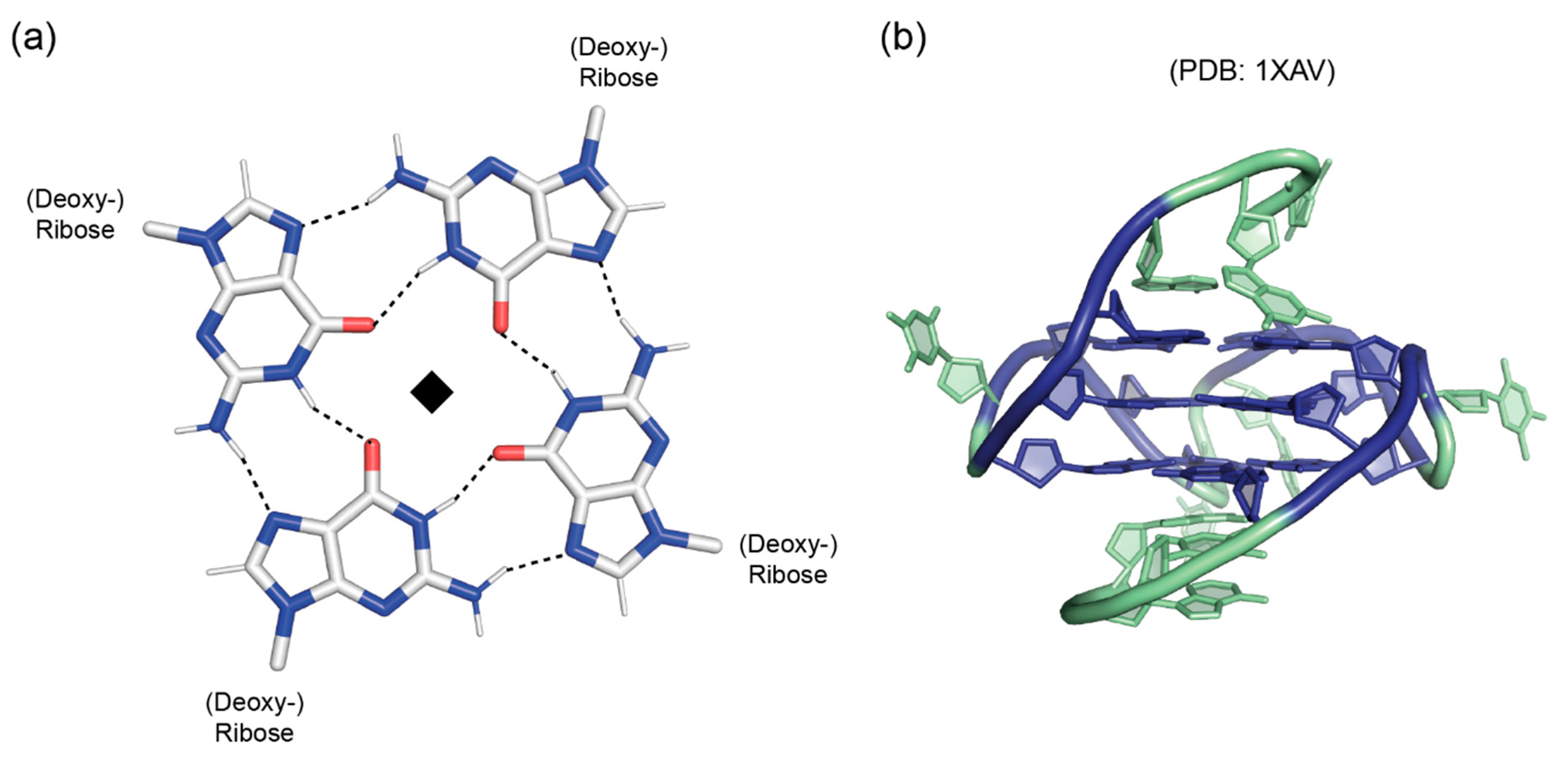
3. Guanine Quadruplexes
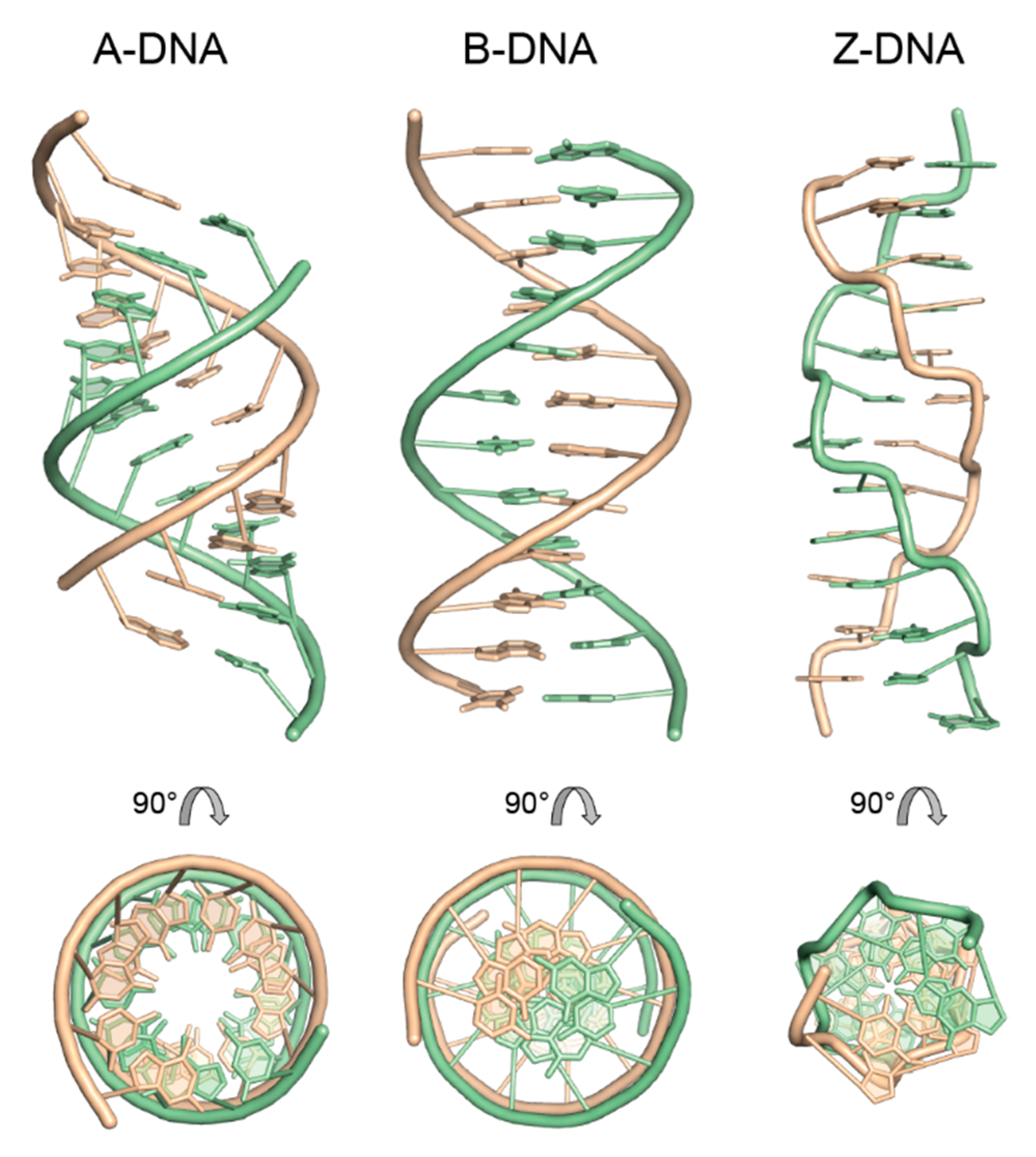
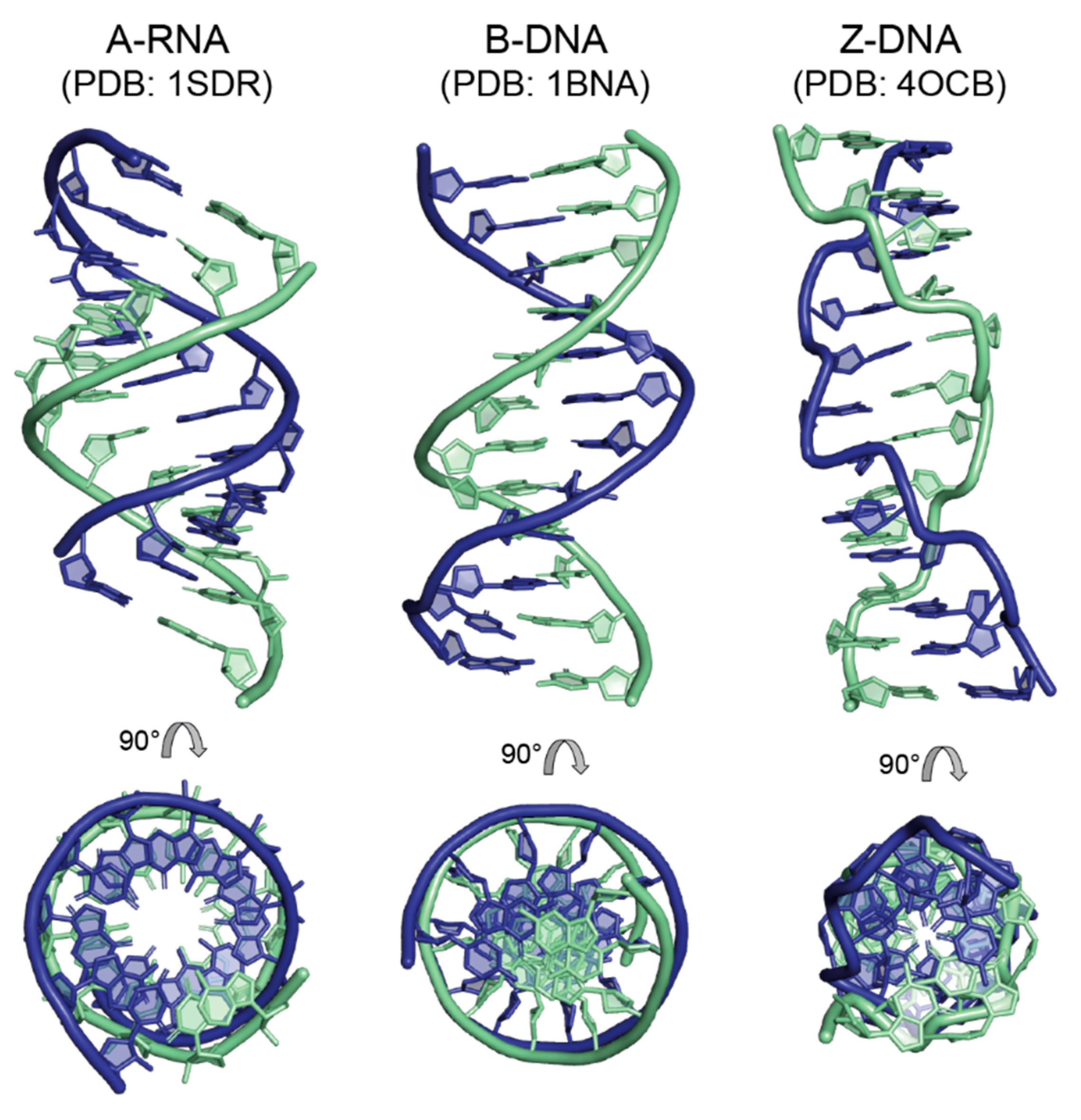
4. Regular Double-Helical Structures as Defined by X-ray Fiber Diffraction Studies
4.1. The A-Form of RNA and DNA
4.2. The B-Form of DNA
4.3. B-Like Forms of DNA
4.4. The Z-Form of DNA
5. Double-Helical Structures as Observed in Single-Crystal X-ray Diffraction Studies
5.1. Sequence-Dependent Helix Modulation Introduced by Base-Pair Stacking Propensities
5.2. Double Helix Structure Modulation by Mis-Pairing and Chemical Modification
5.3. Double Helix Structure Modulation by Ligand Binding
6. The Biology of Double-Helical DNA Structures
Superhelical Structures and Chromatin
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Brodsky, B.; Persikov, A.V. Molecular structure of the collagen triple helix. Adv. Protein. Chem. 2005, 70, 301–339. [Google Scholar]
- Ramachandran, G.N.; Kartha, G. Structure of collagen. Nature 1954, 174, 269–270. [Google Scholar] [CrossRef]
- Chandrasekaran, R. Molecular architecture of polysaccharide helices in oriented fibers. Adv. Carbohydr. Chem. Biochem. 1997, 52, 311–439. [Google Scholar]
- Astbury, W.T.; Bell, F.O. X-ray study of thymonucleic acid. Nature 1938, 141, 747–748. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Molecular configuration in sodium thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef]
- Goodsell, D.S. Symmetry at the cellular mesoscale. Symmetry 2019, 11, 1170. [Google Scholar] [CrossRef] [Green Version]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: Berlin/Heidelberg, Germany, 1984. [Google Scholar]
- Sinden, R.R. DNA Structure and Function; Academic Press: San Diego, CA, USA, 1994. [Google Scholar]
- Birnbaum, G.I.; Kierdaszuk, B.; Shugar, D. Tautomerism and conformation of the promutagenic analogue N6-methoxy-2′,3′,5′-tri-O-methyladenosine. Nucleic Acids Res. 1984, 12, 2447–2460. [Google Scholar] [CrossRef] [Green Version]
- Kimsey, I.J.; Petzold, K.; Sathyamoorthy, B.; Stein, Z.W.; Al-Hashimi, H.M. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 2015, 519, 315–320. [Google Scholar] [CrossRef] [Green Version]
- Westhof, E. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett. 2014, 588, 2464–2469. [Google Scholar] [CrossRef] [Green Version]
- Garg, A.; Heinemann, U. A novel form of RNA double helix based on G.U and C.A(+) wobble base pairing. RNA 2018, 24, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Hunter, W.N.; Brown, T.; Kennard, O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Res. 1987, 15, 6589–6606. [Google Scholar] [CrossRef] [Green Version]
- Hunter, W.N.; Brown, T.; Anand, N.N.; Kennard, O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature 1986, 320, 552–555. [Google Scholar] [CrossRef]
- Seeman, N.C.; Rosenberg, J.M.; Rich, A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. USA 1976, 73, 804–808. [Google Scholar] [CrossRef] [Green Version]
- Watson, J.D.; Crick, F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Hoogsteen, K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Cryst. 1959, 12, 822–823. [Google Scholar] [CrossRef]
- Crick, F.H. Codon—Anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Mueller, U.; Schubel, H.; Sprinzl, M.; Heinemann, U. Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution. RNA 1999, 5, 670–677. [Google Scholar] [CrossRef] [Green Version]
- Stolarski, R.; Dudycz, L.; Shugar, D. NMR studies in the syn-anti dynamic equilibrium in purine nucleosides and nucleotides. Eur. J. Biochem. 1980, 108, 111–121. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D. G-Quadruplex DNA and RNA. Methods Mol. Biol. 2019, 2035, 1–24. [Google Scholar] [PubMed]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- PyMOL. The PyMOL Molecular Graphics System, 2.0; Schrödinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Lin, C.; Dickerhoff, J.; Yang, D. NMR studies of G-quadruplex structures and G-quadruplex-interactive compounds. Methods Mol. Biol. 2019, 2035, 157–176. [Google Scholar]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- McRae, E.K.S.; Booy, E.P.; Padilla-Meier, G.P.; McKenna, S.A. On characterizing the interactions between proteins and guanine quadruplex structures of nucleic acids. J. Nucleic Acids 2017, 2017, 9675348. [Google Scholar] [CrossRef] [Green Version]
- Brazda, V.; Haronikova, L.; Liao, J.C.; Fojta, M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef] [Green Version]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I., Jr.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Qin, Y.; Hurley, L.H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 2008, 90, 1149–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Wang, G.; Vasquez, K.M. DNA triple helices: Biological consequences and therapeutic potential. Biochimie 2008, 90, 1117–1130. [Google Scholar] [CrossRef] [Green Version]
- Van de Sande, J.H.; Ramsing, N.B.; Germann, M.W.; Elhorst, W.; Kalisch, B.W.; von Kitzing, E.; Pon, R.T.; Clegg, R.C.; Jovin, T.M. Parallel stranded DNA. Science 1988, 241, 551–557. [Google Scholar] [CrossRef]
- Gleghorn, M.L.; Zhao, J.; Turner, D.H.; Maquat, L.E. Crystal structure of a poly(rA) staggered zipper at acidic pH: Evidence that adenine N1 protonation mediates parallel double helix formation. Nucleic Acids Res. 2016, 44, 8417–8424. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T. The Left Handed DNA Hall of Fame. Available online: http://www.fred.net/tds/leftdna/ (accessed on 20 April 2020).
- Itakura, K.; Riggs, A.D. Chemical DNA synthesis and recombinant DNA studies. Science 1980, 209, 1401–1405. [Google Scholar] [CrossRef]
- Arnott, S. Historical article: DNA polymorphism and the early history of the double helix. Trends Biochem. Sci. 2006, 31, 349–354. [Google Scholar] [CrossRef]
- Ghosh, A.; Bansal, M. A glossary of DNA structures from A to Z. Acta Cryst. D Biol. Cryst. 2003, 59 Pt 4, 620–626. [Google Scholar] [CrossRef]
- Wang, A.H.; Quigley, G.J.; Kolpak, F.J.; Crawford, J.L.; van Boom, J.H.; van der Marel, G.; Rich, A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979, 282, 680–686. [Google Scholar] [CrossRef]
- Arnott, S.; Chandrasekaran, R.; Birdsall, D.L.; Leslie, A.G.; Ratliff, R.L. Left-handed DNA helices. Nature 1980, 283, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Arnott, S. Polynucleotide secondary structres: An historical perspective. In Oxford Handbook of Nucleic Acid Structure; Neidle, S., Ed.; Oxford University Press: Oxford, UK, 1989; pp. 1–38. [Google Scholar]
- Chandrasekaran, R.; Arnott, S. The structures of DNA and RNA helices in oriented fibers. In Nukleinsäuren; Saenger, W., Ed.; Springer: Berlin/Heidelberg, Germany, 1989; Volume 1/II, pp. 31–170. [Google Scholar]
- Li, S.; Olson, W.K.; Lu, X.J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019, 47, W26–W34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrasekaran, R.; Arnott, S. The structure of B-DNA in oriented fibers. J. Biomol. Struct. Dyn. 1996, 13, 1015–1027. [Google Scholar] [CrossRef]
- Case, R.; Schollmeyer, H.; Kohl, P.; Sirota, E.B.; Pynn, R.; Ewert, K.E.; Safinya, C.R.; Li, Y. Hydration forces between aligned DNA helices undergoing B to A conformational change: In-situ X-ray fiber diffraction studies in a humidity and temperature controlled environment. J. Struct. Biol. 2017, 200, 283–292. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, F.; Yu, X.; Rensen, E.; Krupovic, M.; Prangishvili, D.; Egelman, E.H. A virus that infects a hyperthermophile encapsidates a-form DNA. Science 2015, 348, 914–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuertes, M.A.; Cepeda, V.; Alonso, C.; Perez, J.M. Molecular mechanisms for the B-Z transition in the example of poly[d(G-C) x d(G-C)] polymers. A critical review. Chem. Rev. 2006, 106, 2045–2064. [Google Scholar] [CrossRef]
- Moradi, M.; Babin, V.; Roland, C.; Sagui, C. Reaction path ensemble of the B-Z-DNA transition: A comprehensive atomistic study. Nucleic Acids Res. 2013, 41, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Rich, A.; Zhang, S. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003, 4, 566–572. [Google Scholar] [CrossRef]
- Zacharias, W.; Jaworski, A.; Wells, R.D. Cytosine methylation enhances Z-DNA formation in vivo. J. Bacteriol. 1990, 172, 3278–3283. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, T.; Behlke, J.; Lowenhaupt, K.; Heinemann, U.; Rich, A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001, 8, 761–765. [Google Scholar] [CrossRef]
- Schwartz, T.; Rould, M.A.; Lowenhaupt, K.; Herbert, A.; Rich, A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 1999, 284, 1841–1845. [Google Scholar] [CrossRef] [PubMed]
- Shakked, Z.; Rabinovich, D.; Cruse, W.B.; Egert, E.; Kennard, O.; Sala, G.; Salisbury, S.A.; Viswamitra, M.A. Crystalline A-dna: The X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc. R. Soc. Lond B Biol. Sci. 1981, 213, 479–487. [Google Scholar] [PubMed]
- Wing, R.; Drew, H.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Crystal structure analysis of a complete turn of B-DNA. Nature 1980, 287, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Lauble, H.; Frank, R.; Blöcker, H. Crystal structure analysis of an A-DNA fragment at 1.8 Å resolution: D(GCCCGGGC). Nucleic Acids Res. 1987, 15, 9531–9550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakked, Z.; Rabinovich, D.; Kennard, O.; Cruse, W.B.; Salisbury, S.A.; Viswamitra, M.A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J. Mol. Biol. 1983, 166, 183–201. [Google Scholar] [CrossRef]
- Lauble, H.; Frank, R.; Blöcker, H.; Heinemann, U. Three-dimensional structure of d(GGGATCCC) in the crystalline state. Nucleic Acids Res. 1988, 16, 7799–7816. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [Green Version]
- Prive, G.G.; Yanagi, K.; Dickerson, R.E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J. Mol. Biol. 1991, 217, 177–199. [Google Scholar] [CrossRef]
- Heinemann, U.; Alings, C. Crystallographic study of one turn of G/C-rich B-DNA. J. Mol. Biol. 1989, 210, 369–381. [Google Scholar] [CrossRef]
- Drew, H.R.; Dickerson, R.E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol. 1981, 151, 535–556. [Google Scholar] [CrossRef]
- Heinemann, U.; Alings, C.; Bansal, M. Double helix conformation, groove dimensions and ligand binding potential of a G/C-stretch in B-DNA. EMBO J. 1992, 11, 1931–1939. [Google Scholar] [CrossRef] [PubMed]
- Chuprina, V.P.; Heinemann, U.; Nurislamov, A.A.; Zielenkiewicz, P.; Dickerson, R.E.; Saenger, W. Molecular dynamics simulation of the hydration shell of a B-DNA decamer reveals two main types of minor groove hydration depending on groove width. Proc. Natl. Acad. Sci. USA 1991, 88, 593–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, W.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Neidle, S.; Shakked, Z.; Sklenar, H.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, H.; Zhang, M.; Bald, R.; Fürste, J.-P.; Erdmann, V.A.; Heinemann, U. Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 1995, 249, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.; Dalgarno, L. Occurrence of heat-dissociable ribosomal RNA in insects: The presence of three polynucleotide chains in 26 S RNA from cultured Aedes aegypti cells. J. Mol. Biol. 1973, 75, 57–72. [Google Scholar] [CrossRef]
- Luo, Z.; Dauter, M.; Dauter, Z. Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution. Acta Cryst. D Biol. Cryst. 2014, 70 Pt 7, 1790–1800. [Google Scholar] [CrossRef] [Green Version]
- Kilchherr, F.; Wachauf, C.; Pelz, B.; Rief, M.; Zacharias, M.; Dietz, H. Single-molecule dissection of stacking forces in DNA. Science 2016, 353, aaf5508. [Google Scholar] [CrossRef]
- Kool, E.T. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.; Zhou, T.; Yang, L.; Dantas Machado, A.C.; Gordan, R.; Rohs, R. Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 2014, 39, 381–399. [Google Scholar] [CrossRef] [Green Version]
- Kneale, G.; Brown, T.; Kennard, O.; Rabinovich, D.G. T base-pairs in a DNA helix: The crystal structure of d(G-G-G-G-T-C-C-C). J. Mol. Biol. 1985, 186, 805–814. [Google Scholar] [CrossRef]
- Brown, T.; Kennard, O.; Kneale, G.; Rabinovich, D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature 1985, 315, 604–606. [Google Scholar] [CrossRef] [PubMed]
- Webster, G.D.; Sanderson, M.R.; Skelly, J.V.; Neidle, S.; Swann, P.F.; Li, B.F.; Tickle, I.J. Crystal structure and sequence-dependent conformation of the A.G mispaired oligonucleotide d(CGCAAGCTGGCG). Proc. Natl. Acad. Sci. USA 1990, 87, 6693–6697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Privé, G.G.; Heinemann, U.; Chandrasegaran, S.; Kan, L.-S.; Kopka, M.L.; Dickerson, R.E. Helix geometry, hydration, and G•A mismatch in a B-DNA decamer. Science 1987, 238, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, S.R.; Cheong, C.; Tinoco, I., Jr.; Kim, S.H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature 1991, 353, 579–581. [Google Scholar] [CrossRef]
- Heinemann, U.; Hahn, M. C-C-A-G-G-C-m5C-T-G-G: Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G. J. Biol. Chem. 1992, 267, 7332–7341. [Google Scholar]
- Heinemann, U.; Alings, C. The conformation of a B-DNA decamer is mainly determined by its sequence and not by crystal environment. EMBO J. 1991, 10, 35–43. [Google Scholar] [CrossRef]
- Fujii, S.; Wang, A.H.; van der Marel, G.; van Boom, J.H.; Rich, A. Molecular structure of (m5 dC-dG)3: The role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA. Nucleic Acids Res. 1982, 10, 7879–7892. [Google Scholar] [CrossRef] [Green Version]
- Frederick, C.A.; Saal, D.; van der Marel, G.A.; van Boom, J.H.; Wang, A.H.; Rich, A. The crystal structure of d(GGm5CCGGCC): The effect of methylation on A-DNA structure and stability. Biopolymers 1987, 26, S145–S160. [Google Scholar] [CrossRef]
- Mueller, U.; Maier, G.; Mochi Onori, A.; Cellai, L.; Heumann, H.; Heinemann, U. Crystal structure of an eight-base pair duplex containing the 3′-DNA-RNA-5′ junction formed during initiation of minus-strand synthesis of HIV replication. Biochemistry 1998, 37, 12005–12011. [Google Scholar] [CrossRef]
- Wang, A.H.; Fujii, S.; van Boom, J.H.; van der Marel, G.A.; van Boeckel, S.A.; Rich, A. Molecular structure of r(GCG)d(TATACGC): A DNA--RNA hybrid helix joined to double helical DNA. Nature 1982, 299, 601–604. [Google Scholar] [CrossRef]
- Heinemann, U.; Rudolph, L.-N.; Alings, C.; Morr, M.; Heikens, W.; Frank, R.; Blöcker, H. Effect of a single 3’-methylene phosphonate linkage on the conformation of an A-DNA octamer double helix. Nucleic Acids Res. 1991, 19, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruse, W.B.; Salisbury, S.A.; Brown, T.; Cosstick, R.; Eckstein, F.; Kennard, O. Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C]. J. Mol. Biol. 1986, 192, 891–905. [Google Scholar] [CrossRef]
- Pjura, P.E.; Grzeskowiak, K.; Dickerson, R.E. Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 1987, 197, 257–271. [Google Scholar] [CrossRef]
- Kopka, M.L.; Yoon, C.; Goodsell, D.; Pjura, P.; Dickerson, R.E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. USA 1985, 82, 1376–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ughetto, G.; Wang, A.H.; Quigley, G.J.; van der Marel, G.A.; van Boom, J.H.; Rich, A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985, 13, 2305–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.H.; Ughetto, G.; Quigley, G.J.; Hakoshima, T.; van der Marel, G.A.; van Boom, J.H.; Rich, A. The molecular structure of a DNA-triostin A complex. Science 1984, 225, 1115–1121. [Google Scholar] [CrossRef]
- Liaw, Y.C.; Gao, Y.G.; Robinson, H.; van der Marel, G.A.; van Boom, J.H.; Wang, A.H. Antitumor drug nogalamycin binds DNA in both grooves simultaneously: Molecular structure of nogalamycin-DNA complex. Biochemistry 1989, 28, 9913–9918. [Google Scholar] [CrossRef]
- Khare, D.; Ziegelin, G.; Lanka, E.; Heinemann, U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 2004, 11, 656–663. [Google Scholar] [CrossRef]
- Schuetz, A.; Nana, D.; Rose, C.; Zocher, G.; Milanovic, M.; Koenigsmann, J.; Blasig, R.; Heinemann, U.; Carstanjen, D. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell Mol. Life Sci. 2011, 68, 3121–3131. [Google Scholar] [CrossRef]
- Ming, Q.; Roske, Y.; Schuetz, A.; Walentin, K.; Ibraimi, I.; Schmidt-Ott, K.M.; Heinemann, U. Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucleic Acids Res. 2018, 46, 2082–2095. [Google Scholar] [CrossRef] [Green Version]
- Rice, P.A.; Yang, S.; Mizuuchi, K.; Nash, H.A. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell 1996, 87, 1295–1306. [Google Scholar] [CrossRef] [Green Version]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Schultz, S.C.; Shields, G.C.; Steitz, T.A. Crystal structure of a CAP-DNA complex: The DNA is bent by 90 degrees. Science 1991, 253, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Geiger, J.H.; Hahn, S.; Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993, 365, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.K.; Maher, L.J., 3rd. DNA bending by asymmetric phosphate neutralization. Science 1994, 266, 1829–1834. [Google Scholar] [CrossRef] [PubMed]
- Mirzabekov, A.D.; Rich, A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc. Natl. Acad. Sci. USA 1979, 76, 1118–1121. [Google Scholar] [CrossRef] [Green Version]
- Bednar, J.; Furrer, P.; Katritch, V.; Stasiak, A.Z.; Dubochet, J.; Stasiak, A. Determination of DNA persistence length by cryo-electron microscopy. Separation of the static and dynamic contributions to the apparent persistence length of DNA. J. Mol. Biol. 1995, 254, 579–594. [Google Scholar] [CrossRef]
- Champoux, J.J. NA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef] [Green Version]
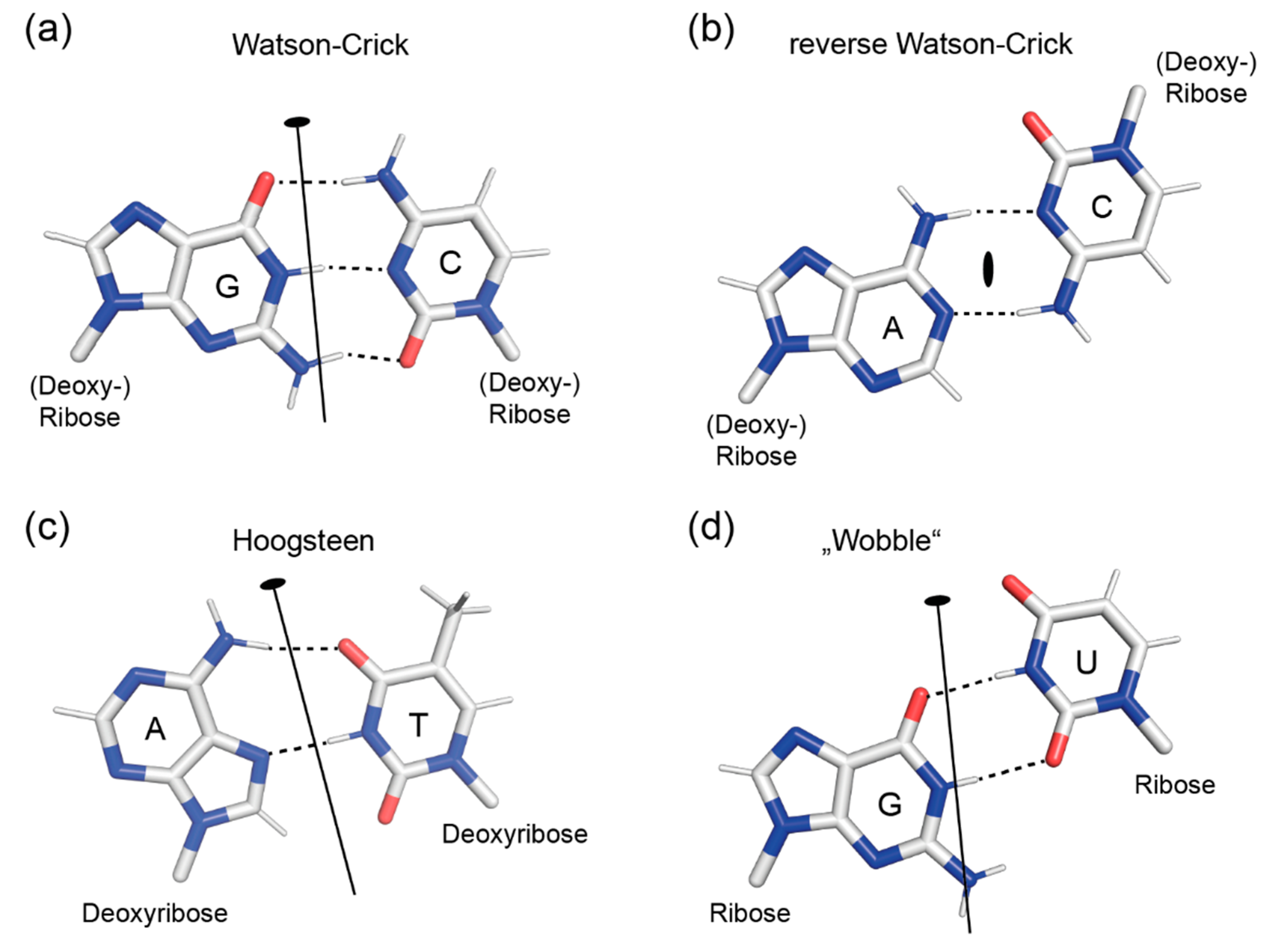
| Base Pairs 1 | |||||
|---|---|---|---|---|---|
| WC | rWC | H | rH | ||
| Glycosyl Bonds 2 | aa | ap 3 | P | ap | P |
| ss | ap | P | ap | p | |
| as | p | ap | p | ap | |
| sa | p | ap | p | ap | |
| A | B | Z | |
|---|---|---|---|
| Helix symmetry | 111 (121) | 101 | 65 |
| Repeat | bp | bp | 2 bp |
| Base-pair inclination (°) | 19 | 0 | −9 |
| Rise/bp (Å) | 2.55 | 3.38 | 3.63 |
| Glycosyl torsion angles (°) | anti | anti | Pu: syn Py: anti |
| Sugar pucker | C3’-endo | C2’-endo | Pu: C3’-endo Py: C2’-endo |
| Position of helix axis | major groove | bp | minor groove |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heinemann, U.; Roske, Y. Symmetry in Nucleic-Acid Double Helices. Symmetry 2020, 12, 737. https://doi.org/10.3390/sym12050737
Heinemann U, Roske Y. Symmetry in Nucleic-Acid Double Helices. Symmetry. 2020; 12(5):737. https://doi.org/10.3390/sym12050737
Chicago/Turabian StyleHeinemann, Udo, and Yvette Roske. 2020. "Symmetry in Nucleic-Acid Double Helices" Symmetry 12, no. 5: 737. https://doi.org/10.3390/sym12050737
APA StyleHeinemann, U., & Roske, Y. (2020). Symmetry in Nucleic-Acid Double Helices. Symmetry, 12(5), 737. https://doi.org/10.3390/sym12050737






