Abstract
In nature and in the test tube, nucleic acids occur in many different forms. Apart from single-stranded, coiled molecules, DNA and RNA prefer to form helical arrangements, in which the bases are stacked to shield their hydrophobic surfaces and expose their polar edges. Focusing on double helices, we describe the crucial role played by symmetry in shaping DNA and RNA structure. The base pairs in nucleic-acid double helices display rotational pseudo-symmetry. In the Watson–Crick base pairs found in naturally occurring DNA and RNA duplexes, the symmetry axis lies in the base-pair plane, giving rise to two different helical grooves. In contrast, anti-Watson–Crick base pairs have a dyad axis perpendicular to the base-pair plane and identical grooves. In combination with the base-pair symmetry, the syn/anti conformation of paired nucleotides determines the parallel or antiparallel strand orientation of double helices. DNA and RNA duplexes in nature are exclusively antiparallel. Watson–Crick base-paired DNA or RNA helices display either right-handed or left-handed helical (pseudo-) symmetry. Genomic DNA is usually in the right-handed B-form, and RNA double helices adopt the right-handed A-conformation. Finally, there is a higher level of helical symmetry in superhelical DNA in which B-form double strands are intertwined in a right- or left-handed sense.
1. Introduction
Nucleic acids are inherently asymmetric molecules, yet in their biologically prevalent, base-paired and helical forms, DNA and RNA display several levels of symmetry. Their constituents, nucleobases, nucleosides and nucleotides, do not have any non-trivial symmetry. However, bases tend to form base pairs, and oligo- and polynucleotides tend to form helices, whose structures are governed by symmetry in a hierarchical order. The rotational symmetry of base pairs determines how they can be arranged in double helices, and the conformation of paired nucleotides determines the parallel or antiparallel strand orientation. Helices have helical symmetry, and DNA double helices can be organized into superhelical structures with higher-order helical symmetry.
The formation of helical structures is a unifying concept in structural and molecular biology. Helices are made of repeating units of similar shape and chemical properties, and they serve the purpose of satisfying hydrogen-bonding and other stereochemical requirements of their building blocks. As early as 1951, Pauling and coworkers proposed a model for helical polypeptides [1]. In addition to the well-known 310-, α- and π-helical conformations of proteins, special helical forms such as the collagen triple helix [2,3] are supported by repetitive polypeptide sequences and/or amino acid modifications. Carbohydrates are also known to form helical structures [4]. The first structural insight into DNA was presented in 1938 by Astbury and Bell [5], and the first detailed models of double-helical DNA [6,7] were famously published in 1953. The review by Goodsell [8] in this volume provided an excellent overview on molecular symmetry in biology.
The symmetries found in nucleic acids are often not exact. Frequently, the execution of a symmetry operation does not lead to an atom-by-atom superposition of molecular structures but to a superposition of shapes or molecular sub-structures. A well-known example of this concept, for which we will use the term “pseudo-symmetry” below, are the Watson–Crick base pairs of double-stranded DNA and RNA, which have similar shapes and whose dyad symmetry axes superimpose their glycosyl bonds after rotation.
Since the 1950s, a vast literature on nucleic acids and their structures has been accumulated. For general principles of nucleic-acid structure, we refer the reader to the classical text book by Saenger [9] and for an introduction into higher-order structure of DNA to Sinden’s book [10]. Basic knowledge covered in these books will be used in this review without any further citation. In this review, we focus on symmetry properties of double-stranded DNA and RNA, an aspect of these structures that has not received much attention so far, and illustrate them with examples from the published literature. We make an attempt to review how our knowledge of the symmetries inherent in DNA and RNA structures has developed over time. Our selection of specific examples is slightly biased towards our own work; we apologize to all authors whose work could not be cited out of considerations of economy and readability.
2. General Structural Principles of Nucleic-Acid Double Helices
2.1. Symmetry of Base Pairs
The four nucleobases A, C, G and T present in DNA and the bases A, C, G and U of RNA are aromatic heterocycles with a propensity for stacking and hydrogen bonding. Base pairing is an efficient way of satisfying the hydrogen bonding potential of the purine and pyrimidine bases. The bases in Figure 1 are shown in their standard non-protonated keto/amino form generally assumed to be predominantly present in nature. However, their hydrogen-bonding and, consequently, their base-pairing patterns can be altered by keto/enol tautomerism or base protonation. The potential for adopting the rare enol/imino forms of bases was noted early on [11] and received renewed interest recently [12,13] because of its possible involvement in genomic mutagenesis. At neutral pH, the bases in DNA and RNA are uncharged. However, at acidic pH or in a spatial environment that changes the microscopic pKa value, protonation at various sites is possible. Bases violating the Watson–Crick pairing scheme (see below) in DNA or RNA double helices as a consequence of base protonation and their effects on duplex structure have been described [14,15,16]. In our following discussion of the symmetry properties of nucleic-acid double helices, we will disregard the unusual enol/imino and protonated forms of the bases and focus entirely on their standard unprotonated keto/amino forms.
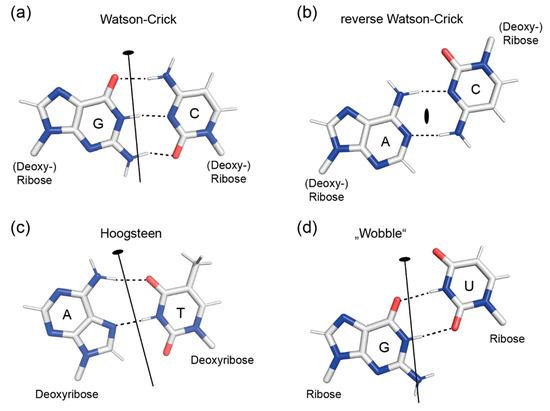
Figure 1.
Rotational (pseudo-) symmetry in base pairs. In the Watson–Crick G:C base pair (a), the two-fold symmetry axis (black line with small oval) is in the base-pair plane. In the reverse Watson–Crick A:C base pair (b), the dyad axis (small oval) is oriented perpendicular to the base-pair plane. In the Hoogsteen A:T (c) and wobble G:U (d) base pairs, the symmetry axes are placed in the base-pair planes. Dashed lines mark hydrogen bonds.
Adenine has three hydrogen bond acceptor atoms (N1, N3 and N7) and one donor atom (N6), guanine has three acceptors (N3, O6 and N7) and two donors (N1 and N2), cytosine has two acceptors (O2 and N3) and one donor (N1), and thymine/uracil have two acceptors (O2 and O4) and one donor (N3). This is the basis for a large number of base pairs that may form between the bases in addition to the standard Watson–Crick (WC) base pairs (Figure 1a), which are commonly, but not exclusively, observed in DNA and RNA double helices in nature. Out of the many alternative pairings, the WC geometry may have been selected in the course of molecular evolution, because only the WC G:C pair is linked with three hydrogen bonds and may thus confer extra stability to helical arrangements in which it occurs. Base-paired arrangements stabilized by two hydrogen bonds are ubiquitous, and the most common of them are depicted in Figure 1.
The standard WC base pairs G:C (depicted in Figure 1a), C:G, A:T and T:A are the core constituents of double-helical DNA in cells. In RNA, of course, A:T is replaced with A:U and T:A with U:A. All WC base pairs are isosteric; superimposing them reveals close shape similarity and accurately places their glycosyl bonds (purine N9 or pyrimidine N1 to sugar C1’) atop each other. This is the basis for the formation of regular double-helical structures of random sequence. Note that the (pseudo-) dyad axis relating a G:C base pair to C:G and A:T to T:A is in the plane of the base pair and intersects the angle between the glycosyl bonds. The important consequence of this symmetry is that the edges of a WC base pair have very different geometries: The edge harboring the purine N7 and N6/O6 and the pyrimidine N4/O4 (at the top in Figure 1a), defined by the >180° angle between the glycosyl bond vectors, differs from the edge defined by the <180° angle between glycosyl bonds (bottom) in terms of shape and hydrogen-bonding potential [17]. In double-stranded DNA or RNA, the base-pair edge with purine N7 and N6/O6 and pyrimidine N4/O4 will define the major groove, the opposite edge with purine N3 and N2 (in guanine) as well as pyrimidine O2 defines the minor groove. We may formulate a general rule: base pairs with (pseudo-) dyad axis in the base-pair plane have different edge properties and distinct grooves when incorporated in double helices.
Reverse Watson–Crick base pairs (Figure 1b) use the same hydrogen bonding atoms as WC pairs, but have a radically different arrangement of the bases and do not obey the Watson–Crick pairing rules [6,18]. Most importantly, their dyad axis is perpendicular to the base-pair plane. This gives rise to an approximately antiparallel orientation of the glycosyl bonds and a similar shape of the two base-pair edges, while their hydrogen-bonding patterns may differ, as seen in the example of the A:C anti-WC base pair.
Two more non-WC base pairs are frequently encountered in DNA or RNA double helices. Hoogsteen base pairs (Figure 1c) are unique in that they involve the purine N7 atoms in hydrogen bonding [19]. A reverse Hoogsteen base pair (not depicted) can be formed by rotating the purine base by 180° against the pyrimidine, facilitating formation of an A (N6) to T (O2) hydrogen bond. The existence of wobble base pairs was proposed by Crick [20] to explain apparently relaxed rules for codon-anticodon pairing during translation. In the G:U wobble base pair (Figure 1d), the pyrimidine base is shifted towards the major groove relative to its position in a WC base pair such that a hydrogen bond between G (N1) and U (O2) is formed. G:U wobble pairs occur frequently in RNA where they may serve regulatory functions [21]. In both Hoogsteen and wobble base pairs, the (pseudo-) dyad axis is in the base-pair plane, creating base-pair edges with distinctly dissimilar features.
2.2. Symmetry of Nucleoside Pairs
Nucleoside conformation adds a further level of complexity to DNA or RNA helix structure, as the glycosyl torsion angles determine the polarity of the sugar–phosphate backbones. As a rule, the nucleosides adopt the anti-conformation of their glycosyl bonds, i.e., the χ torsion angle defined by atoms O4’-C1’-N9-C4 in purine nucleosides and by O4’-C1’-N1-C2 in pyrimidine nucleosides is between 90° and 270° (χ ~195° in A-DNA, ~260° in B-DNA, see below). While pyrimidine nucleosides rarely deviate from anti, in purine nucleosides the rotational barrier is lower, and the syn-conformation is populated in equilibrium [22] and may be adopted in polymeric nucleic acids under certain conditions. Double helices are antiparallel if they are formed by nucleosides in standard anti-conformation (Figure 2a) paired with WC, Hoogsteen or wobble geometry. In the figure, the C4’-C3’ vectors indicating strand polarity point in opposite directions. If helices could be formed by nucleosides in WC, Hoogsteen or wobble base pairs that are all in syn-conformation, these would also be antiparallel. Conversely, double helices formed by base pairs in reverse WC or reverse Hoogsteen geometry with all glycosyl bonds in standard anti-conformation (not shown) are parallel.
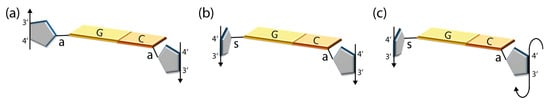
Figure 2.
Symmetry of nucleoside pairs and strand orientation in double helices. The vectors passing through the ribose or deoxyribose C4’ and C3’ atoms mark the conventionally stated 5’-to-3’ strand polarity. (a) Schematic drawing of a Watson–Crick (WC) base pair with glycosyl bonds (a, anti; s, syn) in both nucleosides in anti-conformation: the two strands are in antiparallel orientation. (b) WC base pair with one nucleoside in syn- and one in anti-conformation: the two strands are in parallel orientation. (c) WC G:C base pair with guanosine in syn- and cytidine in anti-conformation with unusual sugar-phosphate backbone as found in Z-DNA: The two strands are in antiparallel orientation.
WC, Hoogsteen and wobble base pairs with the purine nucleoside in syn and the pyrimidine in anti are consistent with parallel strands (Figure 2b), and the related reverse WC and Hoogsteen base pairs with identical glycosyl torsion angles (not shown) also have parallel strands. An interesting deviation from these general principles is found in Z-form DNA double helices (see below). Here, the purine nucleosides are all in syn-conformation and the pyrimidines in anti (Figure 2c), but the strands in Z-DNA are nevertheless antiparallel because of a unique conformation of the sugar–phosphate backbone. This may be taken as an exception from a second general rule that follows from the above considerations, which posits that base-pair symmetry along with the glycosyl torsion angles determines the strand polarity in double-stranded nucleic acids (Table 1).

Table 1.
Strand polarity in double helices is determined by base-pair symmetry and glycosyl torsion angles.
3. Guanine Quadruplexes
As they contain two hydrogen-bond donor (N1 and N2) and three acceptor functions (N3, O6 and N7), guanines are unique among nucleobases in their ability to form quadruplex structures of various topologies in G-repeat nucleic acids [23]. In the planar guanine-tetrad core structure of G-quadruplexes (Figure 3a), the hydrogen-bonding potential of the purine bases is nearly fully saturated by the participation of donor and acceptor functions from both their WC and Hoogsteen edges, leaving only N3 at the minor-groove edge uninvolved. The guanine bases in a G-tetrad are related to each other by a fourfold symmetry axis perpendicular to the tetrad plane. G-tetrads and G-quadruplexes have been observed in both DNA and RNA [24,25,26].
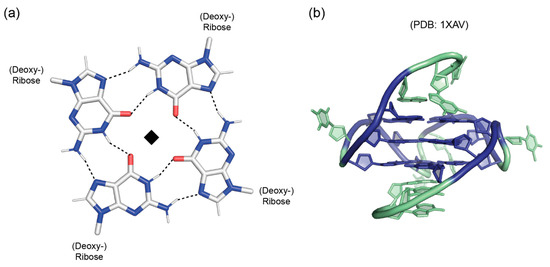
Figure 3.
Guanine quadruplexes. (a) Base pairing scheme in a G-tetrad. The guanine bases are related by a fourfold rotation axis (◆) perpendicular to the tetrad plane. With the exception of N3 and N9, all polar atoms of the bases are engaged in hydrogen bonding. (b) NMR(nuclear magnetic spectroscopy) structure of an intramolecular parallel-stranded G-quadruplex with sequence dTGAGGGTGGGTAGGGTGGGTAA present in the human c-Myc promoter [27]. The guanine nucleotides involved in G-tetrad formation are printed in italic letters and shown in blue in the cartoon drawing. Extra-tetrad nucleotides are colored green. Drawn with PyMOL [28].
In G-rich nucleic acids, guanine tetrads can stack atop each other to form guanine quadruplexes [23,24]. These quadruplexes often contain three stacked tetrads stabilized by monovalent cations (Na+ or K+) and may be intermolecular or intramolecular, i.e., formed by several DNA or RNA strands or by a contiguous strand folding back upon itself. The formation of these structures is facilitated by the propensity of guanosine nucleosides to adopt either the trans or the syn conformation of their glycosyl bonds, the former being favored over the latter by just a small difference in energy. As with the base pairs in double-stranded DNA or RNA, the glycosyl bond angles adopted by the nucleosides determine both groove dimensions and strand polarity. If all guanosine glycosyl bonds have identical torsion angles, e.g., all-trans as in most intermolecular G-quadruplexes [24], the globular quadruplex structure will display identical grooves and parallel strands. If adjacent strands have different glycosyl torsion angles, they will be antiparallel, and the quadruplex structure will display grooves of different widths. In intramolecular G-quadruplexes, the combination of different strand polarities with different types of connecting loops gives rise to a bewildering variety of quadruplex topologies [24]. The G-quadruplex based on a DNA sequence found in the human c-Myc promoter (Figure 3b) is an example for an intramolecular parallel-stranded quadruplex with propeller-type loops between strands and all-trans glycosyl-bond conformation [27]. Crystal and NMR structure analyses have revealed a large variety of globular quadruplex topologies [23,24,27,29].
G-quadruplexes readily form under physiological conditions and have been implicated in a variety of cellular processes ranging from maintenance of genomic stability to replication, transcription and translation [24,30]. They can be stabilized, but also destabilized or unfolded by specific G-quadruplex-binding proteins [31,32]. The first well-documented cellular role of G-quadruplexes was in telomeres, at the ends of human and other eukaryotic chromosomes, where they are thought to serve a structurally protective role [30,33]. More recently, guanine quadruplexes have been characterized as transcriptional regulators in the promoter regions of genes [34,35].
4. Regular Double-Helical Structures as Defined by X-ray Fiber Diffraction Studies
In the following, we shall restrict ourselves to antiparallel DNA and RNA double helices, neglecting less common structures such as triple helices or parallel double helices. This does not mean to imply that these unusual structures do not exist in nature or are irrelevant. Triple-helical structures are formed when a third strand is inserted into the major groove of B-form DNA and its bases form Hoogsteen pairs with the purines of the WC base pairs [36]. Intramolecular triplexes (H-DNA) can be formed with various strand arrangements in homopurine tracts of supercoiled DNA [36]. The potential use of triplex-forming oligonucleotides as DNA-modifying agents has been discussed [37]. Finally, parallel double-helix structures have been observed in DNA hairpins with 3’-p-3’ or 5’-p-5’ linkages in the connecting loop [38]. Biochemical and spectroscopic evidence strongly suggests that these molecules contain right-handed parallel duplexes with reverse WC A:T base pairs, which are slightly less stable than their antiparallel counterparts. A parallel double helix mimicking poly (rA) has also been observed at acidic pH in a crystal of rA7 [39].
One more remark may be allowed before we focus our attention on fiber structures of nucleic acids in their canonical conformations. This concerns the handedness of double helices, a property that has been known since the first models [6,7] were presented, but is thoroughly underappreciated by geneticists and biologists in general. Popular science outlets and, regrettably, also part of the scientific literature are full of images of double helices that have all the features of B-DNA, but show a left-handed mirror-image structure. This structure is obviously impossible because of the chiral nature of nucleotides. The prevalence of these wrong images has led to the creation of a gallery that shows hundreds of examples and is at the same time fun and sad to peruse: The Left Handed DNA Hall of Fame [40].
From the pioneering work of Astbury and Bell [5] in the 1930s until the late 1970s, when chemical synthesis of DNA became possible [41], X-ray fiber diffraction was the method of choice for DNA and RNA structure analysis. This work [42] defined the canonical types of nucleic-acid double helices: the A-form typical for RNA and occasionally found in DNA, the B-form of DNA corresponding to the Watson–Crick model [6] and related, over-twisted double helices, and the Z-form [43]. The latter, incidentally, was initially observed in crystals [44] and only later characterized in oriented fibers [45].
Helical symmetry as observed in the A-, B- and Z-forms of double-stranded nucleic acids is generated by screw operators. Fiber models of DNA are constrained to 111 helical symmetry in the A-form or 101 symmetry in the B-form, meaning they have exactly 11 or 10 repeating units per right-handed turn of the helix. An rt screw axis is characterized by a right-handed rotation of (360/r) ° and a translation of t/r of the repeat length, here the pitch of a full helical turn. Hypothetical, but stereochemically impossible left-handed mirror duplexes of A- and B-DNA (see above) would have 1110 or 109 symmetry. Z-DNA obeys 65 symmetry, because it has six repeating units in one left-handed helical turn. All canonical forms of DNA double helices (A, B and Z) have WC base pairs at their center and antiparallel strands. Fiber models are completely regular (Figure 4). There are no sequence-dependent features, even if the underlying sequence is random, such as in calf thymus DNA. Sequence effects have been studied by analyzing structures of homopolymeric nucleic acids or simple repetitive sequences [46,47], and structural variations were introduced by modulating the ionic medium and relative humidity. Local effects on the double-helix structure, such as those induced by covalent modifications of individual bases or backbone sites, including mutagenic lesions, or ligand binding cannot be studied by X-ray fiber diffraction.
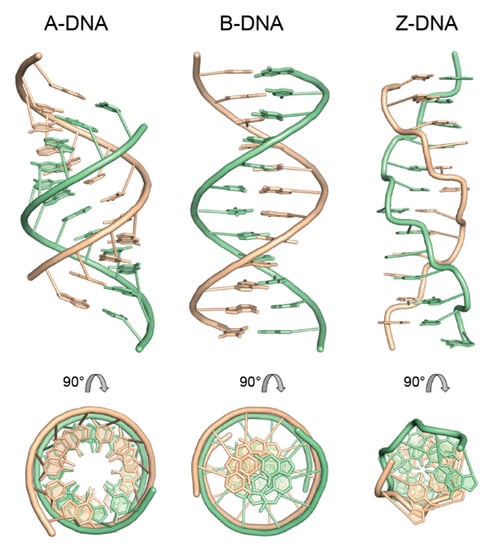
Figure 4.
Canonical double-helical forms of DNA based on X-ray fiber diffraction analysis. Twelve-base-pair fragments of calf thymus A-DNA and B-DNA with arbitrary sequence dATCGATCGATCG and of Z-DNA with alternating sequence dCGCGCGCGCGCG. In the top row, the straight helix axis is vertical, and the view is into the minor groove at the helix center. The bottom row presents the view down the helix axis after 90° rotation. The antiparallel strands are drawn with different colors, and the sugar-phosphate backbone is depicted as smooth tube. Models were generated with Web 3DNA 2.0 [48] based on coordinates derived from X-ray fiber diffraction [46,47] and drawn with PyMOL [28].
4.1. The A-Form of RNA and DNA
RNA duplexes generally adopt the helical A-form (Figure 4, left). Here, a single base pair (nucleotide pair) is the repeating unit of the double helix. The right-handed A-helix is characterized by 11 base pairs per turn (111 symmetry), a rise per base pair along the helix axis of 2.55 Å, an inclination of the base pairs of ~19° against the helix axis, anti-conformation for all glycosyl torsion angles and C3’-endo sugar pucker of all nucleosides. The combination of these parameters gives the helix a stout appearance with a diameter of ~23 Å, deep and narrow major groove and a shallow minor groove. Due to the deep major groove, the base pairs are pushed away from the helix, and the helix appears hollow when looking down its axis. A-RNA helices with 12 base pairs per turn (121 symmetry) have also been modeled based on fiber diffraction data [42].
DNA generally prefers to adopt the B-conformation, which is assumed to be the biologically relevant conformation, but can undergo a transition to the A-form under certain conditions. The conformational shift to the A-form is favored under conditions of low relative humidity (high ionic strength) or if the DNA sequence is G/C-rich. Techniques to control the relative humidity in X-ray fiber diffraction experiments have been known for some time [42,46,47,49] and were recently revisited in a study of hydration forces in A-DNA and B-DNA fibers [50]. The formation of A-DNA is not limited to experimental in vitro situations. It has been reported, for example, that the DNA genome of the SIRV2 virus that infects a hyperthermophilic bacterium is entirely in the A-form [51].
4.2. The B-Form of DNA
The iconic double helical DNA structure presented by Watson and Crick [6] is the first published example of B-DNA and displays many of the features associated with this conformation (Figure 4, center). As in the A-form, a single base pair (nucleotide pair) is the repeating unit of the double helix. The right-handed B-helix is characterized by 10 base pairs per turn (101 symmetry), a rise per base pair along the helix axis of 3.38 Å, base pairs arranged nearly perpendicular to the helix axis, anti-conformation for all glycosyl torsion angles and C2’-endo sugar pucker of all nucleosides. The resulting helix has a diameter of ~20 Å and equally deep grooves, where the major groove is wider than the minor groove. The helix axis passes straight through the base pairs, allowing the helical periodicity to be read out by counting the spokes (the glycosyl bonds) connecting the base pairs to the sugar-phosphate backbone.
4.3. B-Like Forms of DNA
X-ray fiber diffraction studies have identified a family of B-like double helical conformations in DNA, which may be seen as more or less over-wound variants of the former [43,46,47]. C-DNA and D-DNA (not shown) resemble the B-form in having WC base pairs arranged roughly perpendicular to the helix axis, a similar rise per base pair and helix diameter. They differ, however, in their periodicity, with calf thymus C-DNA displaying 28 base pairs per three helical turns (283 symmetry) and a rise of 3.31 Å, while the homopolymeric poly d(AAT):poly d(ATT) in D-conformation has eight base pairs per turn (81 symmetry) and a rise of 3.01 Å. It is unclear whether these B-like conformations occur in nature and what function they may serve.
4.4. The Z-Form of DNA
While the A- and B-forms of DNA as well as A-type RNA helices can accommodate many different nucleotide sequences, Z-DNA is strictly sequence-specific, as it is restricted to alternating purine–pyrimidine sequences. The structure of Z-DNA (Figure 4, right) differs radically from the previously discussed conformations. Here, not a single base pair (nucleotide pair) is the repeating unit of the double helix, but two stacked base pairs. The left-handed Z-helix is characterized by six dinucleotide stacks per turn (65 symmetry), a rise per repeating unit along the helix axis of 7.25 Å and an inclination of the base pairs of ~–9° against the helix axis. A distinctive feature of Z-DNA is the syn-conformation of all purine nucleosides and the usual anti-conformation of pyrimidines. Purines have C3’-endo sugar pucker, while pyrimidines have C2’-endo pucker in Z-DNA. The combination of the dinucleotide stack repeat with the syn-anti alternation along each strand accounts for the wrinkled appearance of the sugar-phosphate backbone in Z-DNA that differs markedly from the smooth trajectory seen in A- and B-helices. The special backbone conformation is required to allow the formation of an antiparallel double helix, although the WC-paired nucleosides have syn and anti glycosyl bonds on their purines and pyrimidines, respectively. The Z-DNA helix appears unusually slender with a diameter of ~18 Å, a shallow major groove and a deep minor groove. Because of the deep minor groove, the base pairs are displaced from the helix axis, and the helix reveals a small central opening when viewed along its axis.
It is noteworthy that a structural transition between right-handed B-DNA and left-handed Z-DNA is a sterically complex event, because the strand polarities in both species are different. Simply unwinding the left-handed Z-DNA helix and twisting it to the opposite handedness will not yield B-DNA, but a right-handed helix with inverted strand polarity, i.e., the lower strand, looking into the minor groove, would run upwards (5’ to 3’) and not downwards, as in B-DNA. The B–Z transition must therefore involve the opening and rejoining of base pairs or the flipping of base pairs around their long axis by 180°. Molecular mechanisms for the B–Z transition have been studied by a variety of biophysical and modeling approaches [52,53].
Since its discovery, the biological significance and function of Z-DNA has been a matter of much debate [54]. It is well established that both covalent DNA modification and superhelical density may trigger the structural transition of oligo-G/C sequences to the Z-form in cells [55]. In addition, proteins were found that associate with Z-DNA in a structure-specific manner [56,57]. Nevertheless, the evidence for a biological function that requires the formation and involvement of Z-DNA is still circumstantial.
To conclude the discussion of DNA and RNA helix types as revealed by X-ray fiber diffraction studies, we may state that the canonical A-, B- and Z-forms are clearly distinct, as they differ in their helical symmetry and several other structural features (Table 2). In these structural models, the helical symmetries are exact and have integral numbers of repeats per turn of the double helix.

Table 2.
Canonical structures of DNA and RNA helices.
5. Double-Helical Structures as Observed in Single-Crystal X-ray Diffraction Studies
Starting in the late 1970s and early 1980s, the crystal structure analysis of short synthetic pieces of DNA and (a short time later) RNA became possible. This work confirmed the general features of A-DNA [58] and B-DNA [59] as established by X-ray fiber diffraction as described above and led to the discovery of a new left-handed form of DNA, the Z-form [44]. Thus, the basic symmetries of the canonical forms of nucleic-acid helices were preserved. In addition, however, the crystal structures (Figure 5) added a new level of structural understanding of nucleic acids, because they revealed sequence-dependent structural variations of the double helix and structural perturbations introduced by base-pair mismatches, chemical modifications of the nucleotides and the binding of small ligands or proteins.
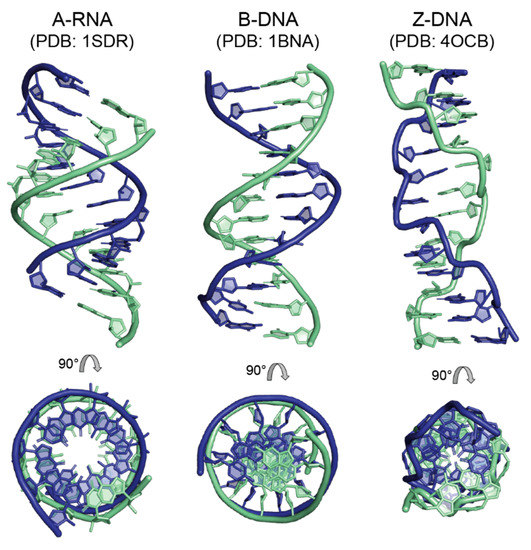
Figure 5.
Examples for the main double-helical forms of nucleic acids based on single-crystal X-ray diffraction analysis of dodecamers. Left, A-RNA with sequence rUAAGGAGGUGAU [70] containing the Shine–Dalgarno sequence [71] important for bacterial and archaeal translation initiation; center, the “Dickerson dodecamer” with sequence dCGCGAATTCGCG [63], the first published B-DNA structure; right, Z-DNA with alternating sequence dCGCGCGCGCGCG [72]. In the top row, the helix axis is vertical and the view is into the minor groove at the helix center. The bottom row presents the view down the helix axis after 90° rotation. PDB entry codes are provided in the figure. Drawn with PyMOL [28].
Some of the distinctive features of A-DNA and B-DNA (see Table 2) became blurred when crystal structures of many different oligonucleotides became available. For example, the clear distinction between the A- and B-form was partly lost, when A-DNA structures with only moderate inclination of base pairs against the helix axis (~7°), a B-DNA-like rise per base pair of 3.2 Å, a wide major groove and the occasional C2’-endo pucker were described [60], suggesting a continuum of conformations between A- and B-DNA. However, since several salient distinctive features such as the groove depth, helix axis position and most of the sugar puckers still differ between helix types, the concept of the A- and B-forms of DNA is still valid today.
5.1. Sequence-Dependent Helix Modulation Introduced by Base-Pair Stacking Propensities
Soon after the first oligonucleotide crystal structures were published, it became clear that the A-DNA [60,61,62] and B-DNA [63,64,65] structures observed at high resolution revealed many subtle sequence-dependent deviations from the fiber models. This becomes apparent, for example, when one compares the B-DNA fiber model (Figure 4, center) with the crystal structure of the “Dickerson dodecamer” (Figure 5, center). The crystal structure appears more irregular with base pairs buckled and propeller-twisted to various degrees, and at its A/T-rich center the minor groove is narrower than in the B-DNA fiber model. This narrow minor groove was linked to the local structure around the A:T base pairs in the center of the helix and to a particular hydration pattern in the minor groove that was made possible by the A/T stretch [66]. This hydration-based concept was validated when G/C-rich and G/C-only B-DNA crystal structures were analyzed [64,65,67,68] that featured wider minor grooves resembling the fiber model of B-DNA and a more irregular hydration in the minor groove.
To comprehend the sequence-dependent features of nucleic-acid helices that were observed in high-resolution crystal structures, universally applicable analytic tools and a new terminology had to be devised. This analysis centers on the geometry and distortions of individual base pairs (Opening, Propeller Twist, Buckle, Stagger, Stretch and Shear), position and orientation of base pairs relative to a helix axis (x-, y-Displacement, Tip and Inclination) and the geometry of base-pair stacking (Twist, Roll, Tilt, Rise, Slide and Shift) [48,69]. Using these analytical tools, the influence of the nucleotide sequence on the helical fine structure could be studied. It became apparent that many of the above parameters of DNA structure vary with rather large amplitudes. The helix Twist, for example, may be as small as 24° and as large as 51° within the same helix [65], but over a full helix turn the Twist values tend to average out at ~36°, the value for the fiber model of B-DNA. Similar observations have been made for other helical parameters. Thus, in spite of significant local structure variations, the overall characteristics of A-, B- and Z-form helices are preserved in crystal structures.
The stacking propensities of the DNA and RNA bases and base pairs are at the origin of the sequence-dependent helical structure modulations. Over the years, many experimental and theoretical approaches have been taken to unravel the energetics of base stacking and the resulting DNA fine structure [73,74]. In spite of all the research effort invested in this field, a simple code relating the DNA sequence to the helical fine structure, which could be exploited by transcription factors and other DNA-binding proteins, has not emerged [75].
5.2. Double Helix Structure Modulation by Mis-Pairing and Chemical Modification
The availability of synthetic DNA and RNA fragments for crystallization opened the door for studying the effects of mismatched bases, chemical modifications of bases or backbone structures or other perturbation of helix conformation. Early crystal structures of mis-incorporated bases in A-DNA [16,76,77] and B-DNA [78,79] duplexes showed that these mismatches had a surprisingly small effect on the underlying helix structure. Even purine-purine mismatches could be incorporated without major structural effect [78,79]. The ability to accommodate mismatched bases into a standard double helix without major structural perturbation is shared by RNA [14,80].
Chemical modifications of bases and the sugar-phosphate backbone were also assessed by structure analysis regarding their effect of DNA or RNA helix structure. Cytosine methylation, known as an important epigenetic modification of chromatin and to stabilize the Z-form of DNA, had very small structural consequences for B-DNA [81,82] or Z-DNA [83]. Similar observations were made with 6-methyladenine incorporated in B-DNA and 5-methylcytosine incorporated in an A-DNA helix [84]. The exchange of deoxyriboses by their ribose analogs is a drastic modification of a DNA sugar-phosphate backbone. Nucleic-acid helices with hybrid DNA/RNA backbones invariably adopt the A-conformation [85,86], because ribonucleotides cannot adopt C2’-endo sugar pucker as required in a B-form helix. Other backbone modifications, such as 3’-methylene phosphonate linkages in A-DNA [87] or chiral phosphorothioate groups in B-DNA [88], were tolerated with minor structural distortions.
5.3. Double Helix Structure Modulation by Ligand Binding
Crystal structure analysis allows researchers to study the influence of ligand binding on DNA helix structure. DNA-binding drug molecules were among the first ligands that were systematically tested for their binding modes. These molecules adopt various strategies to associate with DNA. Here, we will briefly mention two major categories of DNA-binding drug molecules, groove binders and intercalators. Molecules such as netropsin, distamycin and the DNA fluorochrome Hoechst 33258 [89,90] have optimal shapes to bind to the narrow minor groove of A/T-rich DNA without causing much conformational change. The binding of intercalating drugs like triostin A and echinomycin, on the other hand, has very profound consequences for DNA helix structure, as the intercalation is accompanied by a local unwinding and stretching of the double helix and triggers local changes of the base-pairing pattern [91,92]. Other molecules, such as nogalamycin [93], have even more complex binding patterns, acting as intercalators and groove binders simultaneously and causing significant helix distortion.
Hundreds of crystal structures of DNA- or RNA-bound proteins have been determined since the 1980s, and it is beyond the scope of this paper to review even the most basic patterns of protein–nucleic-acid interactions. Regarding the issue of symmetry in double-helical structures, it may be noted that many proteins such as the transcription factors KorB [94], Klf4 [95] and Grhl2 [96] analyzed in our own laboratory bind their target DNA without major helix distortion, preserving the helical symmetry. Of course, there are plenty of examples of proteins that severely bend or distort the helical structure. These include architectural proteins such as the bacterial integration host factor [97] or histone octamers that wrap nuclear DNA into nucleosome structures [98] and transcription factors such as the bacterial catabolite gene activator protein [99] or the eukaryotic TATA-box-binding protein [100]. These proteins achieve DNA helix bending by the intercalation of protein side chains between DNA base pairs and/or asymmetric charge compensation of the DNA backbone, a powerful and well-known mechanism leading to DNA curving [101,102].
To conclude this section, we note that the basic symmetry of DNA and RNA helices is preserved in crystal structures, but it is relaxed such that we no longer find exact 11-, 10- or six-fold helical symmetry, but non-repeating symmetry and non-integral numbers of repeats per helix turn instead. Crystal structure analyses demonstrate that the canonical A-, B- and Z-conformations are remarkably resistant against nucleotide sequence effects, covalent modifications and ligand binding.
6. The Biology of Double-Helical DNA Structures
Until this point, we have limited our discussion of nucleic-acid structure to oriented fibers or short DNA or RNA fragments. Under these conditions, where fibers are fixed in an outstretched shape and the length of synthetic fragments remains well below the persistence length of DNA [103], double helices behave like rigid rods with (mostly) straight axes. Above a length of 100–200 base pairs, DNA helices lose their bending stiffness; the uncooked spaghetti turns into cooked spaghetti. These long DNA molecules no longer have internal symmetry. However, there is one state with defined helical symmetry: the DNA superhelix.
Superhelical Structures and Chromatin
When a circularly closed plasmid is run on an agarose gel, one usually observes not one but three bands. These bands represent open circles with single-strand breaks, running the slowest, linearized molecules with intermediate speed of migration and covalently closed circles, running the fastest. These fast-running molecules have a compact shape because they are supercoiled, a state that can be directly observed in an electron microscope. DNA supercoiling arises from the activity of DNA topoisomerases [104], a diverse class of enzymes that catalyze the topological remodeling of DNA in cells by introducing and sealing single and double-strand breaks in DNA. Topological remodeling can also be achieved in vitro, e.g., by the combined use of intercalating molecules (see above) and topoisomerases. Depending on the topological change achieved by a topoisomerase in vivo or in vitro, DNA may adopt a positively or negatively supercoiled form (Figure 6).
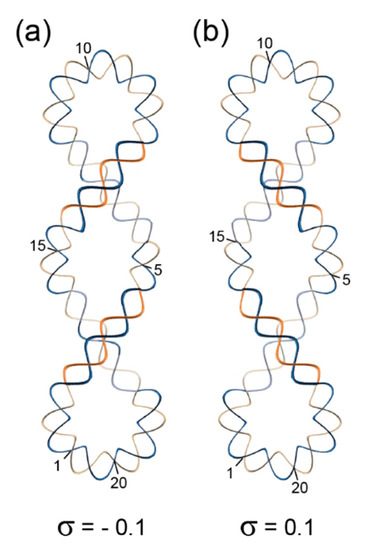
Figure 6.
Superhelical structures formed by a closed circular B-DNA with 210 base pairs and 20 helical turns (10.5 base pairs per turn). (a) Negative superhelical form with two right-handed superhelical turns and a superhelical density σ = −0.1. This form compensates for an initial overwinding of the DNA double strand. (b) Positive superhelical form with two left-handed superhelical turns and a superhelical density σ = 0.1. This form compensates for an initial unwinding of the DNA double strand.
Positively supercoiled DNA displays a left-handed superhelix, negative supercoiling is associated with a right-handed superhelix. Obviously, these superhelices have helical symmetry, much the same as the underlying B-form double helices. If one takes one double-helical turn as the repeating unit of the superhelix, then the right-handed superhelix of the model displayed in Figure 6a can be said to have 101 superhelical symmetry, because it organizes 20 double-helical turns into two superhelical turns. By the same logic, the left-handed superhelix of Figure 6b would have 109 superhelical symmetry. It remains unclear, however, whether this terminology is helpful for understanding superhelical structures. It should be noted in closing that superhelical structures are topologically equivalent with toroidally wrapped DNA helices like those in nucleosomes, i.e., one form can be converted into the other without nicking and re-sealing the DNA and without breaking any covalent bonds.
At higher levels of chromatin organization, the supramolecular entities containing DNA become more and more asymmetric. For that reason, these structures are not covered in this review.
7. Conclusions
We have tried to make the case that symmetry is of central importance for understanding DNA and RNA helical structure. Symmetry determines essential structural features in a hierarchical manner. The rotational (pseudo-) symmetry of base pairs determines if the helices formed by them have grooves with identical shapes or different shapes, such as the major and minor grooves of the A, B- and Z-form duplexes. In combination with the base-pair symmetry, the glycosyl bond conformations determine the parallel or antiparallel orientation of the double-helix strands. The double helices in their different canonical conformations can be described in terms of helical symmetry. This helical symmetry is exact in double-helical models derived from X-ray studies of oriented fibers but relaxed in crystal structures of DNA or RNA duplexes. Crystal structures provide information about sequence-dependent features of helices and about perturbations of the helix structures by covalent modifications or ligand binding. These factors often exert only small effects on the helix conformation, but some ligand binding events lead to major structural rearrangements. Right- or left-handed DNA superhelices represent the highest level of structural organization with clearly recognizable symmetry.
Author Contributions
Writing—original draft preparation and final editing, U.H.; visualization–text review and editing, Y.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The enthusiastic and diligent work of many graduate students and postdoctoral researchers who contributed to studies of nucleic-acid structure and function in the Heinemann laboratory is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef]
- Brodsky, B.; Persikov, A.V. Molecular structure of the collagen triple helix. Adv. Protein. Chem. 2005, 70, 301–339. [Google Scholar]
- Ramachandran, G.N.; Kartha, G. Structure of collagen. Nature 1954, 174, 269–270. [Google Scholar] [CrossRef]
- Chandrasekaran, R. Molecular architecture of polysaccharide helices in oriented fibers. Adv. Carbohydr. Chem. Biochem. 1997, 52, 311–439. [Google Scholar]
- Astbury, W.T.; Bell, F.O. X-ray study of thymonucleic acid. Nature 1938, 141, 747–748. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Franklin, R.E.; Gosling, R.G. Molecular configuration in sodium thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef]
- Goodsell, D.S. Symmetry at the cellular mesoscale. Symmetry 2019, 11, 1170. [Google Scholar] [CrossRef]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: Berlin/Heidelberg, Germany, 1984. [Google Scholar]
- Sinden, R.R. DNA Structure and Function; Academic Press: San Diego, CA, USA, 1994. [Google Scholar]
- Birnbaum, G.I.; Kierdaszuk, B.; Shugar, D. Tautomerism and conformation of the promutagenic analogue N6-methoxy-2′,3′,5′-tri-O-methyladenosine. Nucleic Acids Res. 1984, 12, 2447–2460. [Google Scholar] [CrossRef][Green Version]
- Kimsey, I.J.; Petzold, K.; Sathyamoorthy, B.; Stein, Z.W.; Al-Hashimi, H.M. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 2015, 519, 315–320. [Google Scholar] [CrossRef]
- Westhof, E. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett. 2014, 588, 2464–2469. [Google Scholar] [CrossRef]
- Garg, A.; Heinemann, U. A novel form of RNA double helix based on G.U and C.A(+) wobble base pairing. RNA 2018, 24, 209–218. [Google Scholar] [CrossRef]
- Hunter, W.N.; Brown, T.; Kennard, O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Res. 1987, 15, 6589–6606. [Google Scholar] [CrossRef][Green Version]
- Hunter, W.N.; Brown, T.; Anand, N.N.; Kennard, O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature 1986, 320, 552–555. [Google Scholar] [CrossRef]
- Seeman, N.C.; Rosenberg, J.M.; Rich, A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. USA 1976, 73, 804–808. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Hoogsteen, K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Cryst. 1959, 12, 822–823. [Google Scholar] [CrossRef]
- Crick, F.H. Codon—Anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Mueller, U.; Schubel, H.; Sprinzl, M.; Heinemann, U. Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution. RNA 1999, 5, 670–677. [Google Scholar] [CrossRef]
- Stolarski, R.; Dudycz, L.; Shugar, D. NMR studies in the syn-anti dynamic equilibrium in purine nucleosides and nucleotides. Eur. J. Biochem. 1980, 108, 111–121. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Yang, D. G-Quadruplex DNA and RNA. Methods Mol. Biol. 2019, 2035, 1–24. [Google Scholar] [PubMed]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- PyMOL. The PyMOL Molecular Graphics System, 2.0; Schrödinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Lin, C.; Dickerhoff, J.; Yang, D. NMR studies of G-quadruplex structures and G-quadruplex-interactive compounds. Methods Mol. Biol. 2019, 2035, 157–176. [Google Scholar]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- McRae, E.K.S.; Booy, E.P.; Padilla-Meier, G.P.; McKenna, S.A. On characterizing the interactions between proteins and guanine quadruplex structures of nucleic acids. J. Nucleic Acids 2017, 2017, 9675348. [Google Scholar] [CrossRef]
- Brazda, V.; Haronikova, L.; Liao, J.C.; Fojta, M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I., Jr.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Qin, Y.; Hurley, L.H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 2008, 90, 1149–1171. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Frank-Kamenetskii, M.D.; Mirkin, S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995, 64, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Wang, G.; Vasquez, K.M. DNA triple helices: Biological consequences and therapeutic potential. Biochimie 2008, 90, 1117–1130. [Google Scholar] [CrossRef]
- Van de Sande, J.H.; Ramsing, N.B.; Germann, M.W.; Elhorst, W.; Kalisch, B.W.; von Kitzing, E.; Pon, R.T.; Clegg, R.C.; Jovin, T.M. Parallel stranded DNA. Science 1988, 241, 551–557. [Google Scholar] [CrossRef]
- Gleghorn, M.L.; Zhao, J.; Turner, D.H.; Maquat, L.E. Crystal structure of a poly(rA) staggered zipper at acidic pH: Evidence that adenine N1 protonation mediates parallel double helix formation. Nucleic Acids Res. 2016, 44, 8417–8424. [Google Scholar] [CrossRef]
- Schneider, T. The Left Handed DNA Hall of Fame. Available online: http://www.fred.net/tds/leftdna/ (accessed on 20 April 2020).
- Itakura, K.; Riggs, A.D. Chemical DNA synthesis and recombinant DNA studies. Science 1980, 209, 1401–1405. [Google Scholar] [CrossRef]
- Arnott, S. Historical article: DNA polymorphism and the early history of the double helix. Trends Biochem. Sci. 2006, 31, 349–354. [Google Scholar] [CrossRef]
- Ghosh, A.; Bansal, M. A glossary of DNA structures from A to Z. Acta Cryst. D Biol. Cryst. 2003, 59 Pt 4, 620–626. [Google Scholar] [CrossRef]
- Wang, A.H.; Quigley, G.J.; Kolpak, F.J.; Crawford, J.L.; van Boom, J.H.; van der Marel, G.; Rich, A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979, 282, 680–686. [Google Scholar] [CrossRef]
- Arnott, S.; Chandrasekaran, R.; Birdsall, D.L.; Leslie, A.G.; Ratliff, R.L. Left-handed DNA helices. Nature 1980, 283, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Arnott, S. Polynucleotide secondary structres: An historical perspective. In Oxford Handbook of Nucleic Acid Structure; Neidle, S., Ed.; Oxford University Press: Oxford, UK, 1989; pp. 1–38. [Google Scholar]
- Chandrasekaran, R.; Arnott, S. The structures of DNA and RNA helices in oriented fibers. In Nukleinsäuren; Saenger, W., Ed.; Springer: Berlin/Heidelberg, Germany, 1989; Volume 1/II, pp. 31–170. [Google Scholar]
- Li, S.; Olson, W.K.; Lu, X.J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019, 47, W26–W34. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, R.; Arnott, S. The structure of B-DNA in oriented fibers. J. Biomol. Struct. Dyn. 1996, 13, 1015–1027. [Google Scholar] [CrossRef]
- Case, R.; Schollmeyer, H.; Kohl, P.; Sirota, E.B.; Pynn, R.; Ewert, K.E.; Safinya, C.R.; Li, Y. Hydration forces between aligned DNA helices undergoing B to A conformational change: In-situ X-ray fiber diffraction studies in a humidity and temperature controlled environment. J. Struct. Biol. 2017, 200, 283–292. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, F.; Yu, X.; Rensen, E.; Krupovic, M.; Prangishvili, D.; Egelman, E.H. A virus that infects a hyperthermophile encapsidates a-form DNA. Science 2015, 348, 914–917. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, M.A.; Cepeda, V.; Alonso, C.; Perez, J.M. Molecular mechanisms for the B-Z transition in the example of poly[d(G-C) x d(G-C)] polymers. A critical review. Chem. Rev. 2006, 106, 2045–2064. [Google Scholar] [CrossRef]
- Moradi, M.; Babin, V.; Roland, C.; Sagui, C. Reaction path ensemble of the B-Z-DNA transition: A comprehensive atomistic study. Nucleic Acids Res. 2013, 41, 33–43. [Google Scholar] [CrossRef]
- Rich, A.; Zhang, S. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003, 4, 566–572. [Google Scholar] [CrossRef]
- Zacharias, W.; Jaworski, A.; Wells, R.D. Cytosine methylation enhances Z-DNA formation in vivo. J. Bacteriol. 1990, 172, 3278–3283. [Google Scholar] [CrossRef]
- Schwartz, T.; Behlke, J.; Lowenhaupt, K.; Heinemann, U.; Rich, A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001, 8, 761–765. [Google Scholar] [CrossRef]
- Schwartz, T.; Rould, M.A.; Lowenhaupt, K.; Herbert, A.; Rich, A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 1999, 284, 1841–1845. [Google Scholar] [CrossRef] [PubMed]
- Shakked, Z.; Rabinovich, D.; Cruse, W.B.; Egert, E.; Kennard, O.; Sala, G.; Salisbury, S.A.; Viswamitra, M.A. Crystalline A-dna: The X-ray analysis of the fragment d(G-G-T-A-T-A-C-C). Proc. R. Soc. Lond B Biol. Sci. 1981, 213, 479–487. [Google Scholar] [PubMed]
- Wing, R.; Drew, H.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Crystal structure analysis of a complete turn of B-DNA. Nature 1980, 287, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Lauble, H.; Frank, R.; Blöcker, H. Crystal structure analysis of an A-DNA fragment at 1.8 Å resolution: D(GCCCGGGC). Nucleic Acids Res. 1987, 15, 9531–9550. [Google Scholar] [CrossRef] [PubMed]
- Shakked, Z.; Rabinovich, D.; Kennard, O.; Cruse, W.B.; Salisbury, S.A.; Viswamitra, M.A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J. Mol. Biol. 1983, 166, 183–201. [Google Scholar] [CrossRef]
- Lauble, H.; Frank, R.; Blöcker, H.; Heinemann, U. Three-dimensional structure of d(GGGATCCC) in the crystalline state. Nucleic Acids Res. 1988, 16, 7799–7816. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef]
- Prive, G.G.; Yanagi, K.; Dickerson, R.E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J. Mol. Biol. 1991, 217, 177–199. [Google Scholar] [CrossRef]
- Heinemann, U.; Alings, C. Crystallographic study of one turn of G/C-rich B-DNA. J. Mol. Biol. 1989, 210, 369–381. [Google Scholar] [CrossRef]
- Drew, H.R.; Dickerson, R.E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol. 1981, 151, 535–556. [Google Scholar] [CrossRef]
- Heinemann, U.; Alings, C.; Bansal, M. Double helix conformation, groove dimensions and ligand binding potential of a G/C-stretch in B-DNA. EMBO J. 1992, 11, 1931–1939. [Google Scholar] [CrossRef] [PubMed]
- Chuprina, V.P.; Heinemann, U.; Nurislamov, A.A.; Zielenkiewicz, P.; Dickerson, R.E.; Saenger, W. Molecular dynamics simulation of the hydration shell of a B-DNA decamer reveals two main types of minor groove hydration depending on groove width. Proc. Natl. Acad. Sci. USA 1991, 88, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Olson, W.; Bansal, M.; Burley, S.K.; Dickerson, R.E.; Gerstein, M.; Harvey, S.C.; Heinemann, U.; Neidle, S.; Shakked, Z.; Sklenar, H.; et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001, 313, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, H.; Zhang, M.; Bald, R.; Fürste, J.-P.; Erdmann, V.A.; Heinemann, U. Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 1995, 249, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.; Dalgarno, L. Occurrence of heat-dissociable ribosomal RNA in insects: The presence of three polynucleotide chains in 26 S RNA from cultured Aedes aegypti cells. J. Mol. Biol. 1973, 75, 57–72. [Google Scholar] [CrossRef]
- Luo, Z.; Dauter, M.; Dauter, Z. Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution. Acta Cryst. D Biol. Cryst. 2014, 70 Pt 7, 1790–1800. [Google Scholar] [CrossRef]
- Kilchherr, F.; Wachauf, C.; Pelz, B.; Rief, M.; Zacharias, M.; Dietz, H. Single-molecule dissection of stacking forces in DNA. Science 2016, 353, aaf5508. [Google Scholar] [CrossRef]
- Kool, E.T. Hydrogen bonding, base stacking, and steric effects in dna replication. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef]
- Slattery, M.; Zhou, T.; Yang, L.; Dantas Machado, A.C.; Gordan, R.; Rohs, R. Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 2014, 39, 381–399. [Google Scholar] [CrossRef]
- Kneale, G.; Brown, T.; Kennard, O.; Rabinovich, D.G. T base-pairs in a DNA helix: The crystal structure of d(G-G-G-G-T-C-C-C). J. Mol. Biol. 1985, 186, 805–814. [Google Scholar] [CrossRef]
- Brown, T.; Kennard, O.; Kneale, G.; Rabinovich, D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature 1985, 315, 604–606. [Google Scholar] [CrossRef] [PubMed]
- Webster, G.D.; Sanderson, M.R.; Skelly, J.V.; Neidle, S.; Swann, P.F.; Li, B.F.; Tickle, I.J. Crystal structure and sequence-dependent conformation of the A.G mispaired oligonucleotide d(CGCAAGCTGGCG). Proc. Natl. Acad. Sci. USA 1990, 87, 6693–6697. [Google Scholar] [CrossRef] [PubMed]
- Privé, G.G.; Heinemann, U.; Chandrasegaran, S.; Kan, L.-S.; Kopka, M.L.; Dickerson, R.E. Helix geometry, hydration, and G•A mismatch in a B-DNA decamer. Science 1987, 238, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, S.R.; Cheong, C.; Tinoco, I., Jr.; Kim, S.H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature 1991, 353, 579–581. [Google Scholar] [CrossRef]
- Heinemann, U.; Hahn, M. C-C-A-G-G-C-m5C-T-G-G: Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G. J. Biol. Chem. 1992, 267, 7332–7341. [Google Scholar]
- Heinemann, U.; Alings, C. The conformation of a B-DNA decamer is mainly determined by its sequence and not by crystal environment. EMBO J. 1991, 10, 35–43. [Google Scholar] [CrossRef]
- Fujii, S.; Wang, A.H.; van der Marel, G.; van Boom, J.H.; Rich, A. Molecular structure of (m5 dC-dG)3: The role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA. Nucleic Acids Res. 1982, 10, 7879–7892. [Google Scholar] [CrossRef]
- Frederick, C.A.; Saal, D.; van der Marel, G.A.; van Boom, J.H.; Wang, A.H.; Rich, A. The crystal structure of d(GGm5CCGGCC): The effect of methylation on A-DNA structure and stability. Biopolymers 1987, 26, S145–S160. [Google Scholar] [CrossRef]
- Mueller, U.; Maier, G.; Mochi Onori, A.; Cellai, L.; Heumann, H.; Heinemann, U. Crystal structure of an eight-base pair duplex containing the 3′-DNA-RNA-5′ junction formed during initiation of minus-strand synthesis of HIV replication. Biochemistry 1998, 37, 12005–12011. [Google Scholar] [CrossRef]
- Wang, A.H.; Fujii, S.; van Boom, J.H.; van der Marel, G.A.; van Boeckel, S.A.; Rich, A. Molecular structure of r(GCG)d(TATACGC): A DNA--RNA hybrid helix joined to double helical DNA. Nature 1982, 299, 601–604. [Google Scholar] [CrossRef]
- Heinemann, U.; Rudolph, L.-N.; Alings, C.; Morr, M.; Heikens, W.; Frank, R.; Blöcker, H. Effect of a single 3’-methylene phosphonate linkage on the conformation of an A-DNA octamer double helix. Nucleic Acids Res. 1991, 19, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Cruse, W.B.; Salisbury, S.A.; Brown, T.; Cosstick, R.; Eckstein, F.; Kennard, O. Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C]. J. Mol. Biol. 1986, 192, 891–905. [Google Scholar] [CrossRef]
- Pjura, P.E.; Grzeskowiak, K.; Dickerson, R.E. Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 1987, 197, 257–271. [Google Scholar] [CrossRef]
- Kopka, M.L.; Yoon, C.; Goodsell, D.; Pjura, P.; Dickerson, R.E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl. Acad. Sci. USA 1985, 82, 1376–1380. [Google Scholar] [CrossRef] [PubMed]
- Ughetto, G.; Wang, A.H.; Quigley, G.J.; van der Marel, G.A.; van Boom, J.H.; Rich, A. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985, 13, 2305–2323. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.H.; Ughetto, G.; Quigley, G.J.; Hakoshima, T.; van der Marel, G.A.; van Boom, J.H.; Rich, A. The molecular structure of a DNA-triostin A complex. Science 1984, 225, 1115–1121. [Google Scholar] [CrossRef]
- Liaw, Y.C.; Gao, Y.G.; Robinson, H.; van der Marel, G.A.; van Boom, J.H.; Wang, A.H. Antitumor drug nogalamycin binds DNA in both grooves simultaneously: Molecular structure of nogalamycin-DNA complex. Biochemistry 1989, 28, 9913–9918. [Google Scholar] [CrossRef]
- Khare, D.; Ziegelin, G.; Lanka, E.; Heinemann, U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 2004, 11, 656–663. [Google Scholar] [CrossRef]
- Schuetz, A.; Nana, D.; Rose, C.; Zocher, G.; Milanovic, M.; Koenigsmann, J.; Blasig, R.; Heinemann, U.; Carstanjen, D. The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell Mol. Life Sci. 2011, 68, 3121–3131. [Google Scholar] [CrossRef]
- Ming, Q.; Roske, Y.; Schuetz, A.; Walentin, K.; Ibraimi, I.; Schmidt-Ott, K.M.; Heinemann, U. Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucleic Acids Res. 2018, 46, 2082–2095. [Google Scholar] [CrossRef]
- Rice, P.A.; Yang, S.; Mizuuchi, K.; Nash, H.A. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell 1996, 87, 1295–1306. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Schultz, S.C.; Shields, G.C.; Steitz, T.A. Crystal structure of a CAP-DNA complex: The DNA is bent by 90 degrees. Science 1991, 253, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Geiger, J.H.; Hahn, S.; Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993, 365, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.K.; Maher, L.J., 3rd. DNA bending by asymmetric phosphate neutralization. Science 1994, 266, 1829–1834. [Google Scholar] [CrossRef] [PubMed]
- Mirzabekov, A.D.; Rich, A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc. Natl. Acad. Sci. USA 1979, 76, 1118–1121. [Google Scholar] [CrossRef]
- Bednar, J.; Furrer, P.; Katritch, V.; Stasiak, A.Z.; Dubochet, J.; Stasiak, A. Determination of DNA persistence length by cryo-electron microscopy. Separation of the static and dynamic contributions to the apparent persistence length of DNA. J. Mol. Biol. 1995, 254, 579–594. [Google Scholar] [CrossRef]
- Champoux, J.J. NA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).