AResU-Net: Attention Residual U-Net for Brain Tumor Segmentation
Abstract
1. Introduction
2. Related Work
3. Method
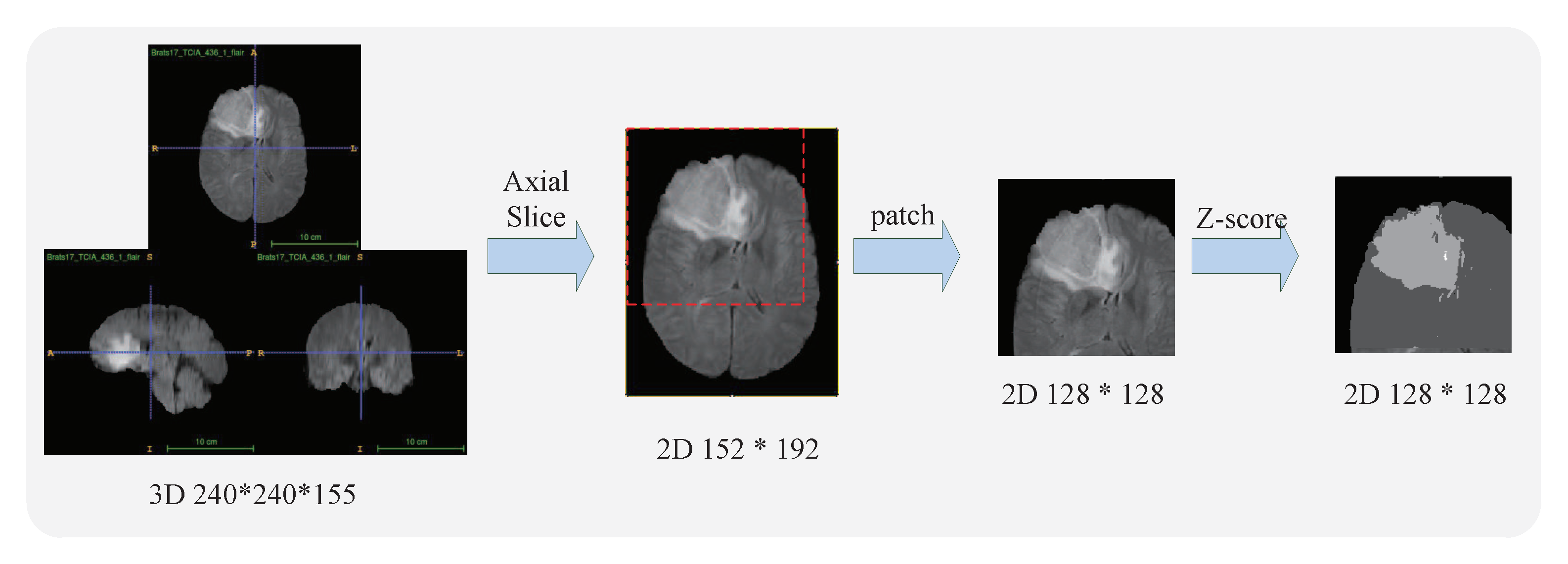
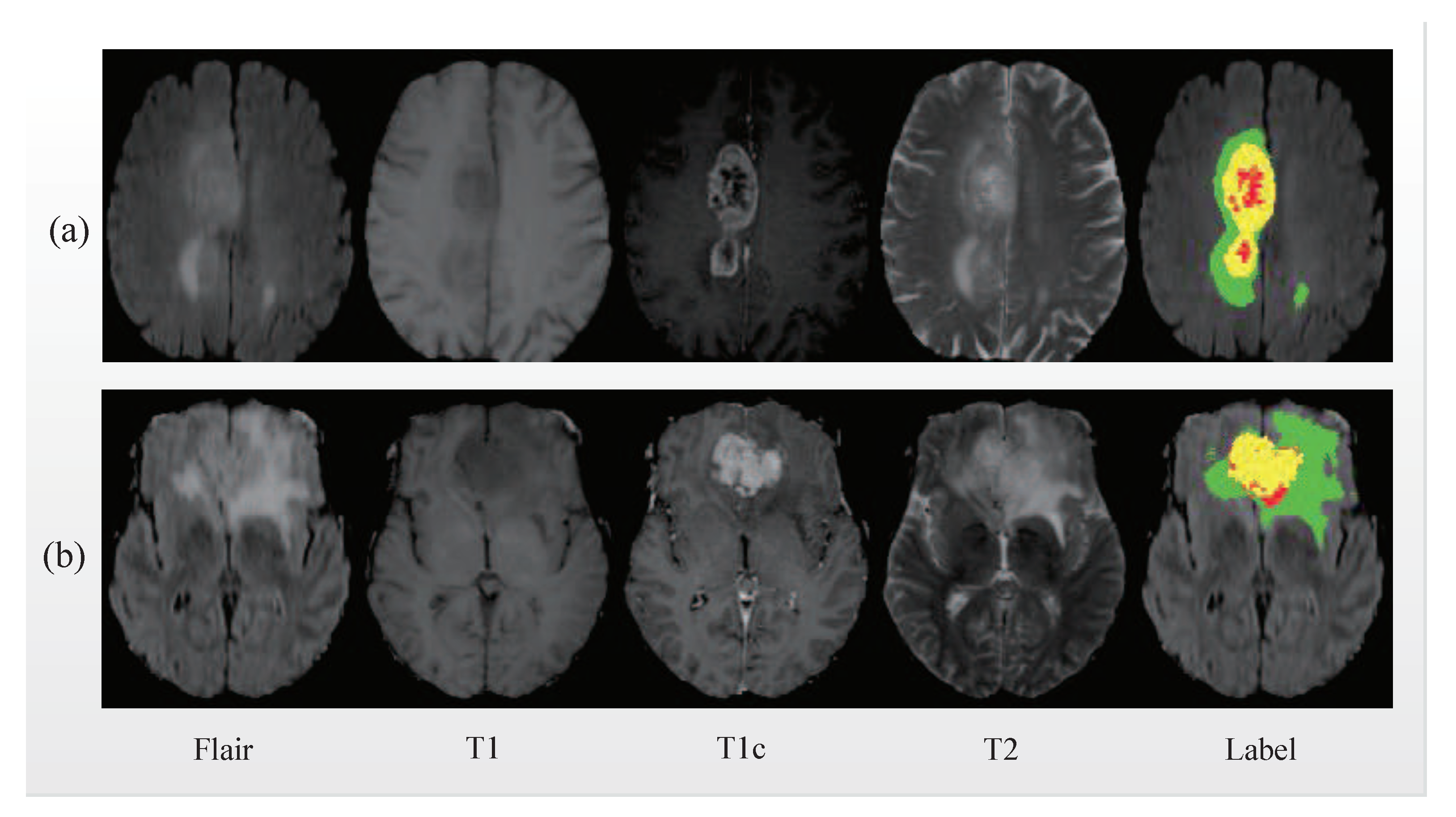
3.1. Data Preprocessing
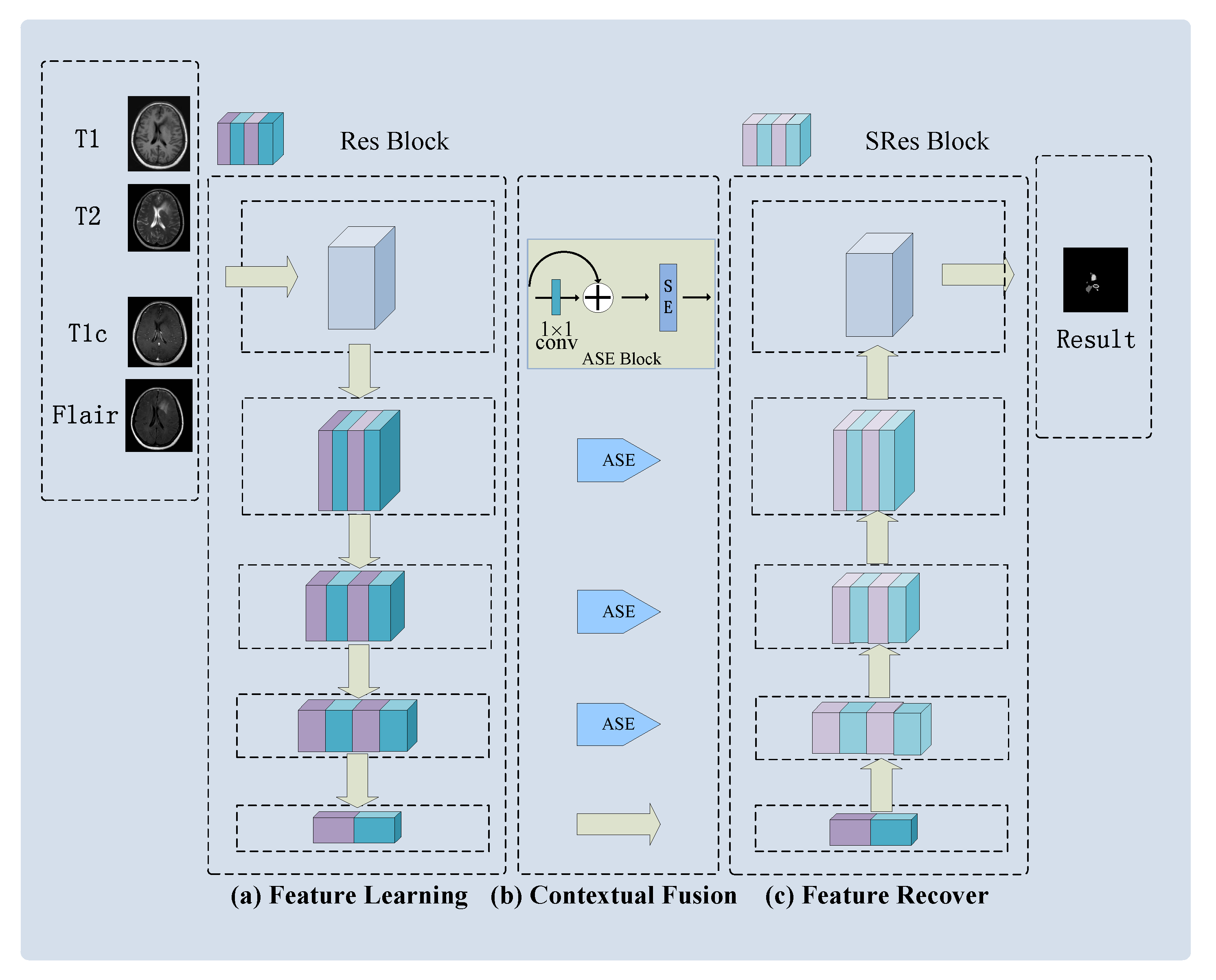
3.2. AResU-Net
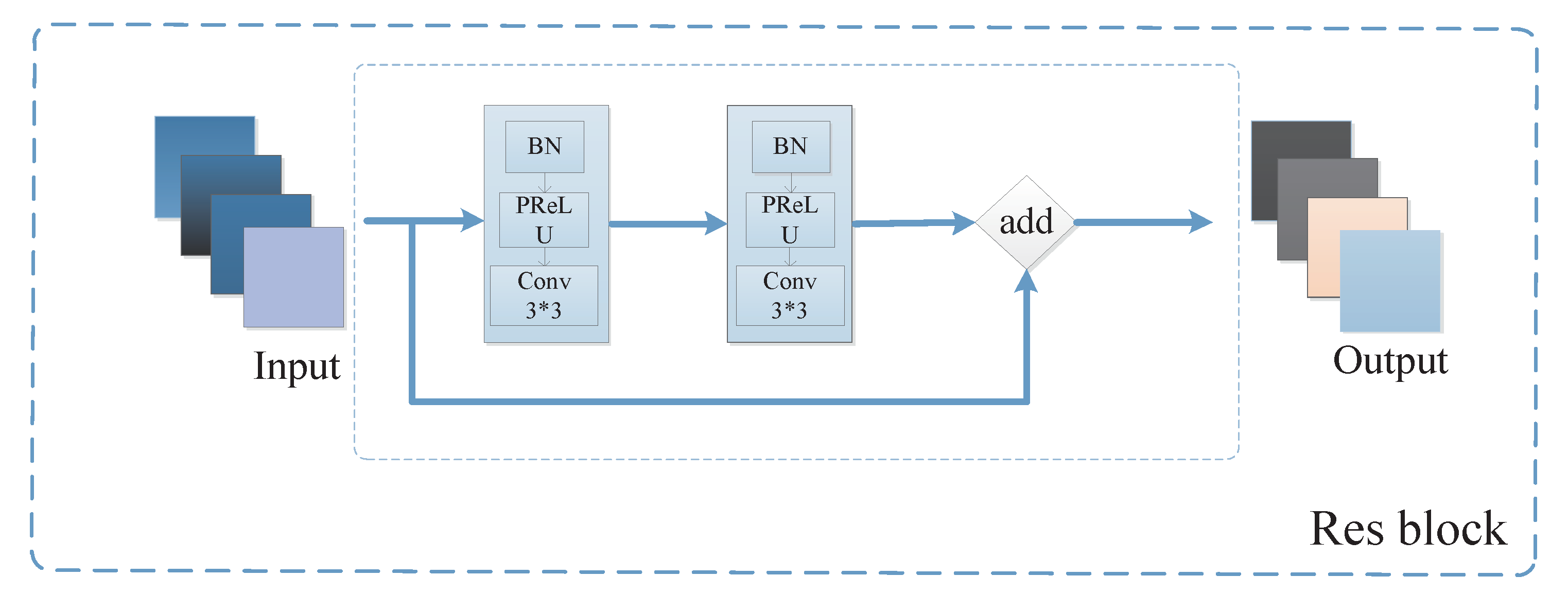
3.2.1. Feature Learning Module
3.2.2. Contextual Fusion Module
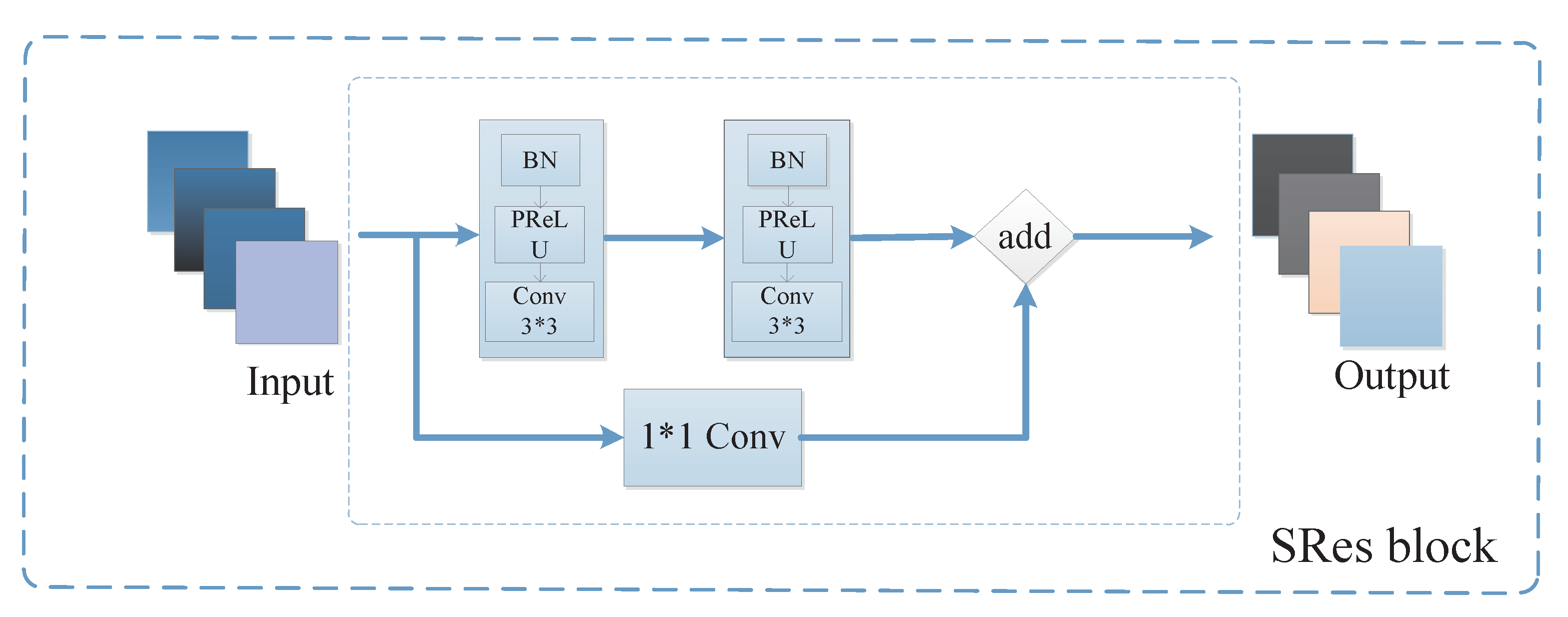
3.2.3. Feature Recovery Module
3.3. Loss Function
4. Experiments and Results
4.1. Datasets
4.2. Experimental Settings
4.3. Evaluation Metric
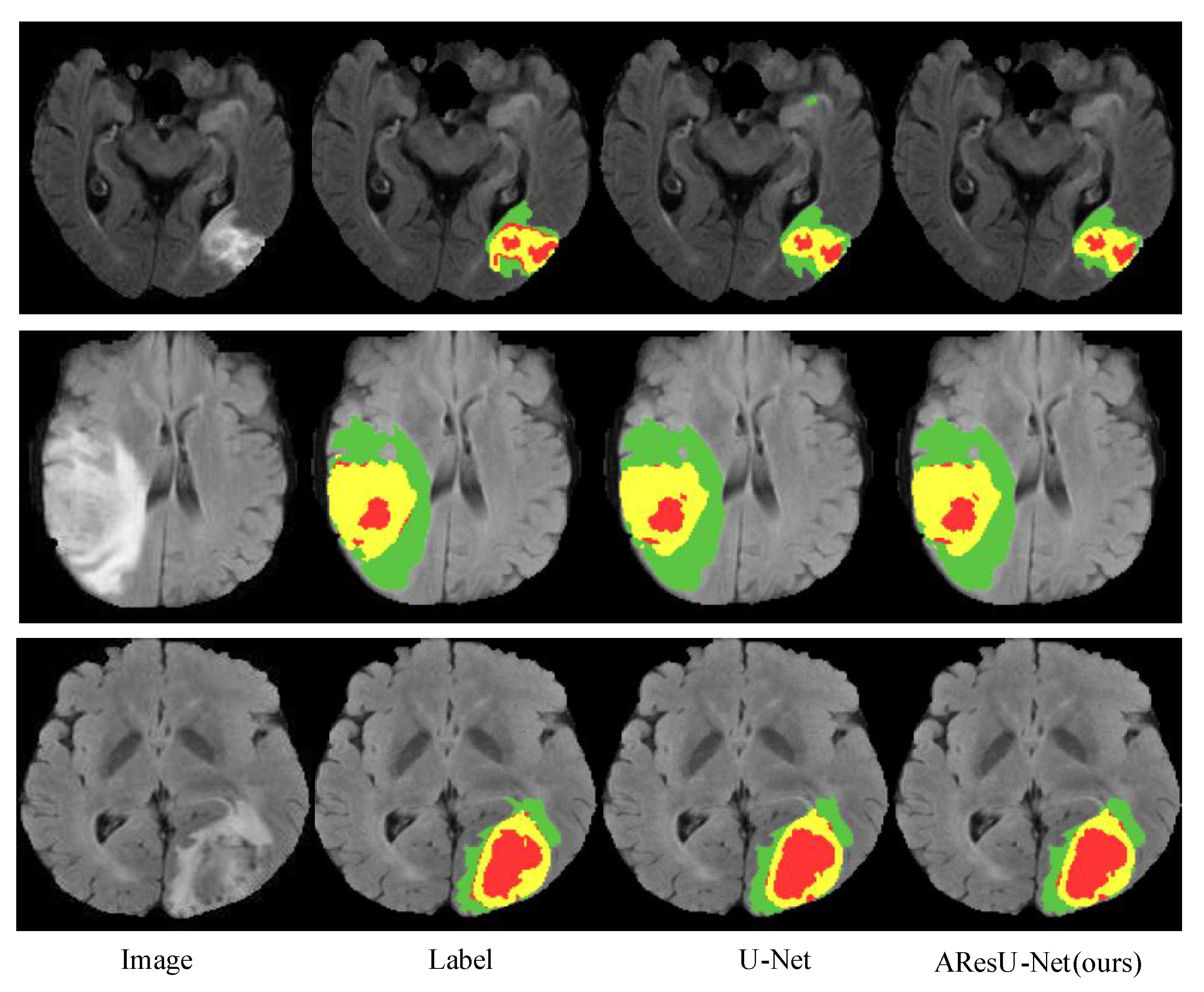
4.4. Experiment Results
4.4.1. Experiment Results on the BraTS 2017 Dataset
4.4.2. Experiment Results on the BraTS 2018 Dataset
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bauer, S.; Wiest, R.; Nolte, L.P.; Reyes, M.A. survey of MRI-based medical image analysis for brain tumor studies. Phys. Med. Biol. 2013, 58, R97. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Mao, L.; Jiang, J.; Liu, C.; Xiong, S. Automatic Semantic Segmentation of Brain Gliomas from MRI Images Using a Deep Cascaded Neural Network. J. Healthc. Eng. 2018, 1, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Işın, A.; Direkoğlu, C.; Şah, M. Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Comput. Sci. 2016, 102, 317–324. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Kayalibay, B.; Jensen, G.; van der Smagt, P. CNN-based segmentation of medical imaging data. arXiv 2017, arXiv:1701.03056. [Google Scholar]
- Abbas, N.; Saba, T.; Mohamad, D.; Rehman, A.; Almazyad, A.S.; Al-Ghamdi, J.S. Machine aided malaria parasitemia detection in Giemsa-stained thin blood smears. Neural Comput. Appl. 2018, 29, 803–818. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. J. Am. Med. Assoc. 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Liu, Y.; Gadepalli, K.; Norouzi, M.; Dahl, G.E.; Kohlberger, T.; Boyko, A.; Venugopalan, S.; Timofeev, A.; Nelson, H.Q.; Corrado, G.S. Detecting cancer metastases on gigapixel pathology images. arXiv 2017, arXiv:1703.02442. [Google Scholar]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H. Deep learning for identifying metastatic breast cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar]
- Chen, H.; Dou, Q.; Yu, L.Q.; Qin, J.; Heng, P.A. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 2017, 170, 446–455. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Kermi, A.; Mahmoudi, I.; Khadir, M.T. Deep Convolutional Neural Networks Using U-Net for Automatic Brain Tumor Segmentation in Multimodal MRI Volumes. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018. [Google Scholar]
- Albiol, A.; Albiol, A.; Albiol, F. Extending 2D Deep Learning Architectures to 3D Image Segmentation Problems. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018. [Google Scholar]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Zhang, H.; Dana, K.; Shi, J.; Zhang, Z.; Wang, X.; Tyagi, A.; Agrawal, A. Context encoding for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Yu, C.; Wang, J.; Peng, C.; Gao, C.; Yu, G.; Sang, N. Learning a discriminative feature network for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Zhao, H.S.; Zhang, Y.; Liu, S.; Jia, J.P.; Loy, C.C.; Lin, D.H.; Jia, J.Y. Psanet: Point-wise spatial attention network for scene parsing. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Zhou, C.H.; Chen, S.C.; Ding, C.X.; Tao, D.C. Learning contextual and attentive information for brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018. [Google Scholar]
- Qi, K.H.; Yang, H.; Li, C.; Liu, Z.Y.; Wang, M.Y.; Liu, Q.G.; Wang, S.S. X-Net: Brain Stroke Lesion Segmentation Based on Depthwise Separable Convolution and Long-Range Dependencies. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef]
- Urban, G.; Bendszus, M.; Hamprecht, F. Multi-modal brain tumor segmentation using deep convolutional neural networks. In Proceedings of the MICCAI Multimodal Brain Tumor Segmentation Challenge, Boston, MA, USA, 14–18 September 2014. [Google Scholar]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Zhao, X.M.; Wu, Y.H.; Song, G.D.; Li, Z.Y.; Zhang, Y.Z.; Fan, Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med. Image Anal. 2018, 43, 98–111. [Google Scholar] [CrossRef]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic brain tumor detection and segmentation using u-net based fully convolutional networks. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis, Edinburgh, UK, 11–13 July 2017. [Google Scholar]
- Kong, X.G.; Sun, G.X.; Wu, Q.; Liu, J.; Lin, F.M. Hybrid Pyramid U-Net Model for Brain Tumor Segmentation. In Proceedings of the International Conference on Intelligent Information Processing, Nanning, China, 19–22 October 2018. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017. [Google Scholar]
- Tseng, K.L.; Lin, Y.L.; Hsu, W.; Huang, C.Y. Joint sequence learning and cross-modality convolution for 3d biomedical segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Liu, D.; Zhang, H.; Zhao, M.M.; Yu, X.J.; Yao, S.W.; Zhou, W. Brain Tumor Segmention Based on Dilated Convolution Refine Network. In Proceedings of the International Conference on Software Engineering Research, Management and Applications, Kunming, China, 13–15 June 2018. [Google Scholar]
- Li, H.; Li, A.; Wang, M. A novel end-to-end brain tumor segmentation method using improved fully convolutional networks. Comput. Biol. Med. 2019, 108, 150–160. [Google Scholar] [CrossRef]
- Jin, Q.G.; Meng, Z.P.; Sun, C.M.; Wei, L.Y.; Su, R. RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans. arXiv 2018, arXiv:1811.01328. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.J.; Fang, Z.W.; Lu, H.Q. Dual attention network for scene segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Zhang, H.; Zhang, H.; Wang, C.; Xie, J. Co-occurrent Features in Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Jegou, H.; Douze, M.; Schmid, C.; Perez, P. Aggregating local descriptors into a compact image representation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010. [Google Scholar]
- Li, H.; Xiong, P.; An, J.; Wang, L.X. Pyramid attention network for semantic segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar]
- Kolnogorov, A.V. Gaussian Two-Armed Bandit and Optimization of Batch Data Processing. Probl. Inf. Transm. 2018, 54, 84–100. [Google Scholar] [CrossRef]
- He, K.M.; Zhang, X.; Ren, S.Q.; Sun, J. Deep Residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BraTS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.M.; Chen, Z.F.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M. Tensorflow: A system for large-scale machine learning. In Proceedings of the USENIX Conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016. [Google Scholar]
- Chen, L.; Wu, Y.; DSouza, A.M.; Abidin, A.Z.; Wismüller, A.; Xu, C. MRI tumor segmentation with densely connected 3D CNN. In Proceedings of the International Society for Optics and Photonics, San Diego, CA, USA, 19–23 August 2018. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016. [Google Scholar]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. S3d-unet: Separable 3du-net for brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic brain tumor segmentation using convolutional neural networks with test-time augmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018. [Google Scholar]
- Hu, K.; Gan, Q.; Zhang, Y.; Deng, S.H.; Xiao, F.; Huang, W.; Cao, C.H.; Gao, X.P. Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 2019, 7, 92615–92629. [Google Scholar] [CrossRef]

| Class | Rate % |
|---|---|
| Background | 98.46 |
| edema | 1.02 |
| enhancing tumor | 0.29 |
| necrotic and non-enhancing tumor | 0.23 |
| Methods | Whole | Core | Enhancing |
|---|---|---|---|
| U-Net [29] | 0.831 | 0.801 | 0.750 |
| ResU-Net * [15] | 0.880 | 0.850 | 0.820 |
| FCNN [28] | 0.865 | 0.864 | 0.816 |
| Densely CNN [44] | 0.720 | 0.830 | 0.810 |
| CNN [26] | 0.840 | 0.720 | 0.620 |
| AResU-Net (ours) | 0.892 | 0.853 | 0.825 |
| Methods | Whole | Core | Enhancing |
|---|---|---|---|
| U-Net | 0.870 | 0.762 | 0.700 |
| SegNet [45] | 0.833 | 0.703 | 0.496 |
| PSPNet [34] | 0.809 | 0.701 | 0.554 |
| NovelNet [34] | 0.876 | 0.763 | 0.642 |
| ResU-Net [15] | 0.873 | 0.768 | 0.716 |
| AResU-Net (ours) | 0.881 | 0.780 | 0.719 |
| Methods | Whole | Core | Enhancing |
|---|---|---|---|
| U-Net | 0.860 | 0.790 | 0.767 |
| ResU-Net [15] | 0.867 | 0.803 | 0.768 |
| Ensemble Net [16] | 0.881 | 0.777 | 0.773 |
| 3DU-Net [46] | 0.885 | 0.718 | 0.760 |
| S3DU-Net [47] | 0.894 | 0.831 | 0.749 |
| TTA [48] | 0.873 | 0.783 | 0.754 |
| MCC [49] | 0.882 | 0.748 | 0.718 |
| AResU-Net (ours) | 0.876 | 0.810 | 0.773 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Lv, X.; Zhang, H.; Liu, B. AResU-Net: Attention Residual U-Net for Brain Tumor Segmentation. Symmetry 2020, 12, 721. https://doi.org/10.3390/sym12050721
Zhang J, Lv X, Zhang H, Liu B. AResU-Net: Attention Residual U-Net for Brain Tumor Segmentation. Symmetry. 2020; 12(5):721. https://doi.org/10.3390/sym12050721
Chicago/Turabian StyleZhang, Jianxin, Xiaogang Lv, Hengbo Zhang, and Bin Liu. 2020. "AResU-Net: Attention Residual U-Net for Brain Tumor Segmentation" Symmetry 12, no. 5: 721. https://doi.org/10.3390/sym12050721
APA StyleZhang, J., Lv, X., Zhang, H., & Liu, B. (2020). AResU-Net: Attention Residual U-Net for Brain Tumor Segmentation. Symmetry, 12(5), 721. https://doi.org/10.3390/sym12050721






