Mechanistic Insights into Plant Chiral Growth
Abstract
:1. Introduction
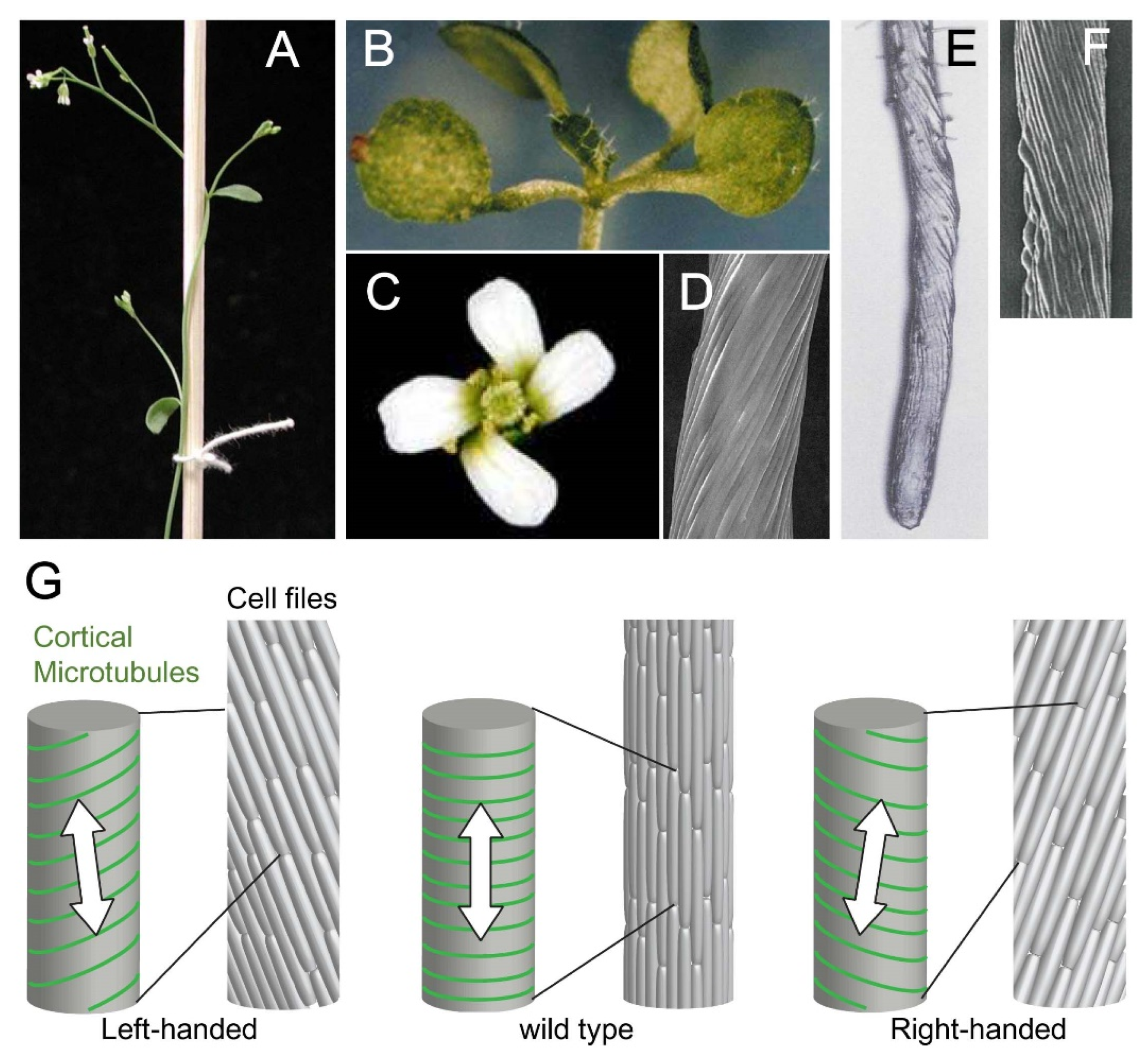
2. Cellular Basis of Organ Twisting
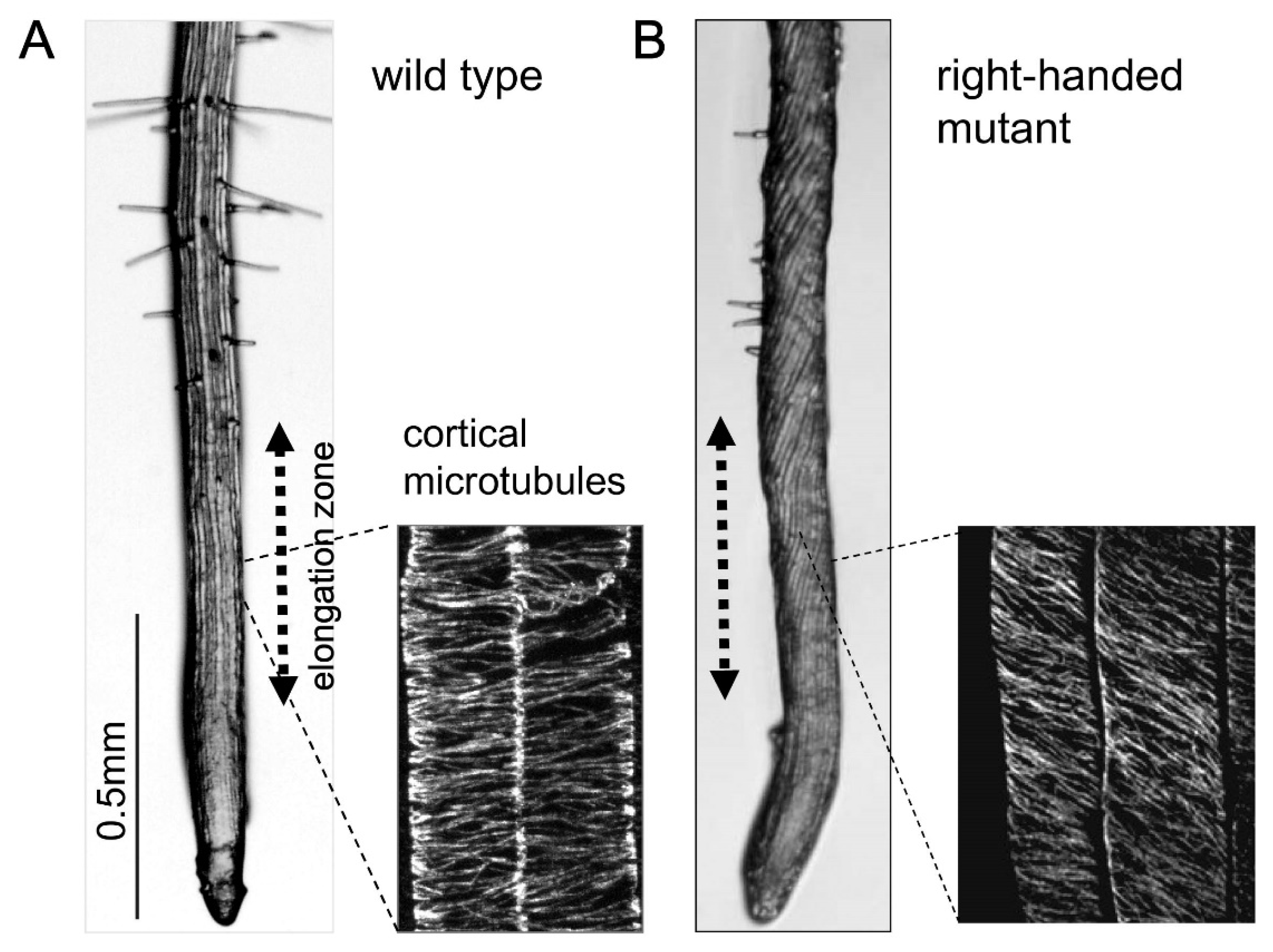
3. Microtubule Defects Cause Helical Growth
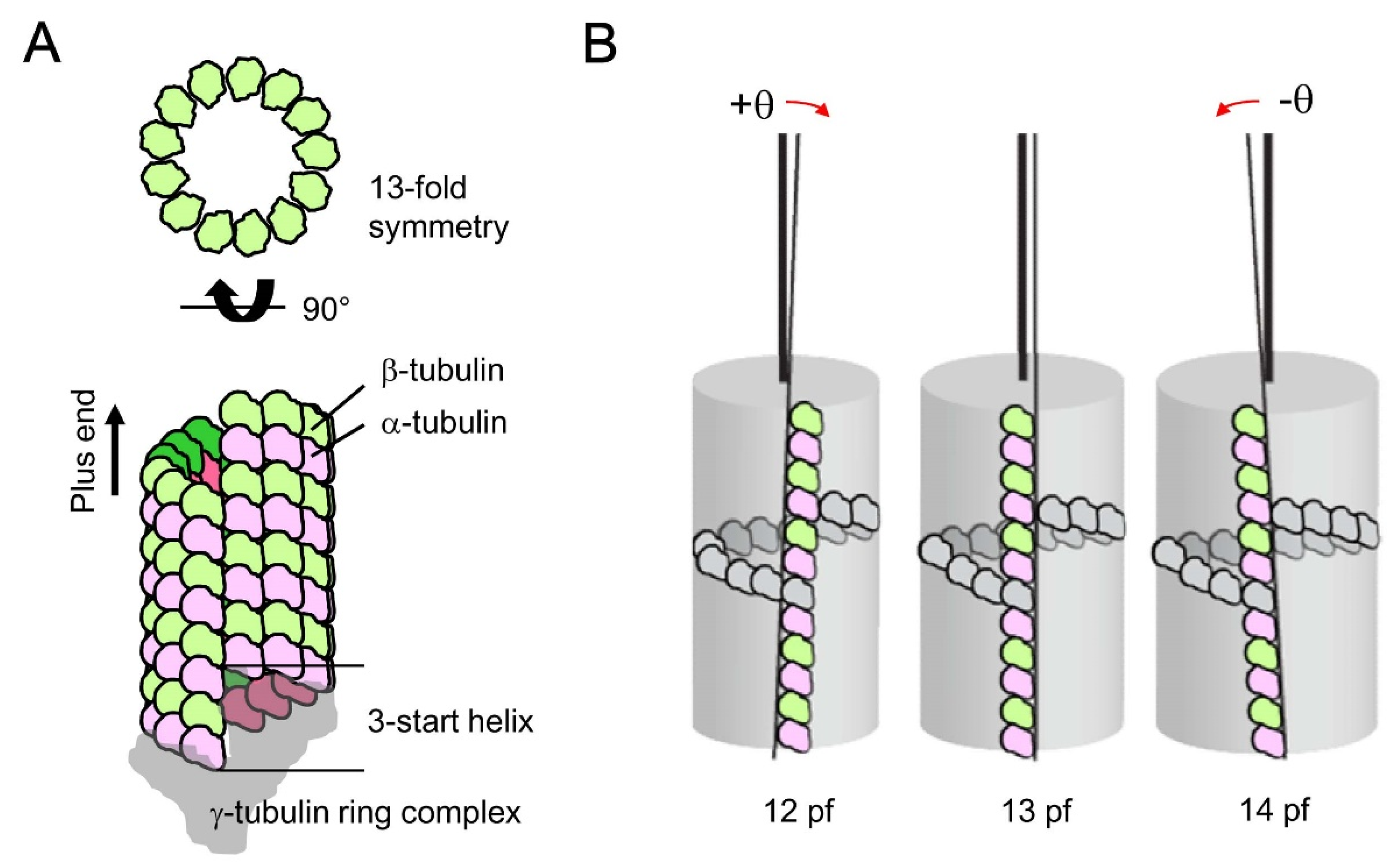
4. Possible Origin of Microtubule Chirality
5. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Darwin, C. On the movements and habits of climbing plants. Bot. J. Linn. Soc. 1865, 9, 1–118. [Google Scholar] [CrossRef]
- Darwin, C. The Power of Movement in Plants; John Murray: London, UK, 2017. [Google Scholar]
- Gianoli, E. The behavioural ecology of climbing plants. AoB Plants 2015, 7, plv013. [Google Scholar] [CrossRef] [Green Version]
- Isnard, S.; Silk, W.K. Moving with climbing plants from Charles Darwin’s time into the 21st century. Am. J. Bot. 2009, 96, 1205–1221. [Google Scholar] [CrossRef] [PubMed]
- Compton, J.A.; Lack, H.W. The discovery, naming and typification of Wisteria floribunda and W. brachybotrys (Fabaveae) with notes on associated names. Willdenowia 2012, 42, 219–240. [Google Scholar] [CrossRef]
- Edwards, W.; Moles, A.T.; Franks, P. The global trend in plant twining direction. Glob. Ecol. Biogeogr. 2007, 16, 795–800. [Google Scholar] [CrossRef]
- Stolarz, M. Circumnutation as a visible plant action and reaction. Plant Signal. Behav. 2009, 4, 380–387. [Google Scholar] [CrossRef]
- Hashimoto, T. Molecular genetic analysis of left-right handedness in plants. Philos. Trans. R. Soc. Biol. Sci. 2002, 357, 799–808. [Google Scholar] [CrossRef]
- Smyth, D.R. Helical growth in plant organs: Mechanisms and significance. Development 2016, 143, 3272–3282. [Google Scholar] [CrossRef] [Green Version]
- Furutani, I.; Watanabe, Y.; Prieto, R.; Masukawa, M.; Suzuki, K.; Naoi, S.; Thitamadee, T.; Shikanai, T. The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 2000, 127, 4443–4453. [Google Scholar] [PubMed]
- Thitamadee, S.; Tuchihara, K.; Hashimoto, T. Microtubule basis for left-handed helical growth in Arabidopsis. Nature 2002, 417, 193–196. [Google Scholar] [CrossRef]
- Gendreau, E.; Traas, J.; Desnos, T.; Grandjean, O.; Caboche, M.; Hofte, H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997, 114, 295–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weizbauer, R.; Peters, W.S.; Schulz, B. Geometric constraints and the anatomical interpretation of twisted plant organ phenotypes. Front. Plant Sci. 2011, 2, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verger, S.; Liu, M.; Hamant, O. Mechanical conflicts in twisting growth revealed by cell-cell adhesion defects. Front. Plant Sci. 2019, 10, 173. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Kawamura, T.; Hashimoto, T. Role of the SPIRAL gene family in anisotropic growth of Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Savalde-Goldstein, S.; Chory, J. Growth cooridination and the shoot epidermis. Curr. Opin. Plant Biol. 2008, 11, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Vaseva, I.I.; Qudeimat, E.; Potuschak, T.; Du, Y.; Genschik, P.; Vandenbussche, F.; van der Straeten, D. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. USA 2018, 115, E4130–E4139. [Google Scholar] [CrossRef] [Green Version]
- Ishida, T.; Thitamadee, S.; Hashimoto, T. Twisted growth and organization of cortical microtubules. J. Plant Res. 2007, 120, 61–70. [Google Scholar] [CrossRef]
- Wu, G.; Otegui, M.S.; Spalding, E.P. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell 2010, 22, 3295–3304. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, T. Dissecting the cellular functions of plant microtubules using mutant tubulins. Cytoskeleton 2013, 70, 191–200. [Google Scholar] [CrossRef]
- Ishida, T.; Kaneko, Y.; Iwano, M.; Hashimoto, T. Helical microtubule arrays in a collection of tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 8544–8549. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, K.; Furutani, I.; Tachimoto, H.; Matsubara, K.; Hashimoto, T. SPIRAL1 encodes a novel plant-specific microtubule-localized protein required for directional cell control of rapidly expanding Arabidopsis cells. Plant Cell 2004, 16, 1178–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedbrook, J.C.; Ehrhardt, D.W.; Fisher, S.E.; Scheible, W.R.; Somerville, C.R. The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 2004, 16, 1506–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, T.; Narita, N.N.; Hayashi, K.; Asada, J.; Hamada, T.; Sonobe, S.; Nakajima, K.; Hashimoto, T. Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis. Plant Physiol. 2004, 136, 3933–3944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushmann, H.; Fabri, C.O.; Hauptmann, M.; Hutzler, P.; Laux, T.; Lloyd, C.W.; Schäffner, A.R. Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr. Biol. 2004, 14, 1515–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, M.; Wakamatsu, Y.; Itoh, T.J.; Shoji, T.; Hashimoto, T. Arabidopsis SPIRAL2 promotes uninterrupted microtubule growth by suppressing the pause state of microtubule dynamics. J. Cell Sci. 2008, 121, 2372–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, M.; Lideboom, J.J.; Saltini, M.; Mulder, B.M.; Ehrhardt, D.W. SPR2 protects minus ends to promote severing and reorientation of plant cortical microtubule arrays. J. Cell Biol. 2018, 217, 915–927. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Burkart, G.M.; Dixit, R. The Arabidopsis SPIRAL2 protein targets and stabilizes microtubule minus ends. Curr. Biol. 2018, 28, 987–994. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Hashimoto, T. A mutation in the Arabidopsis γ-tubulin-containing complex subunit causes helical growth and abnormal microtubule branching. J. Cell Sci. 2009, 122, 2208–2217. [Google Scholar] [CrossRef] [Green Version]
- Naoi, K.; Hashimoto, T. A semi-dominant mutation in an Arabidopsis mitogen-activated protein kinase phosphatase-like gene compromises cortical microtubule organization. Plant Cell 2004, 16, 1841–1853. [Google Scholar] [CrossRef] [Green Version]
- Fujita, S.; Pytela, J.; Hotta, T.; Kato, T.; Hamada, T.; Akamatsu, R.; Ishida, Y.; Kutsuna, N.; Hasezawa, S.; Nomura, Y.; et al. An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr. Biol. 2013, 23, 1969–1978. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Naoi, K.; Shoji, T.; Hashimoto, T. Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant Cell Physiol. 2004, 45, 1330–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baskin, T.I. On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 2001, 215, 150–171. [Google Scholar] [CrossRef] [PubMed]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purushotham, P.; Ho, R.; Zimmer, J. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 2020, 369, 1089–1094. [Google Scholar] [CrossRef]
- Green, P.B. Mechanism for plant cellular morphogenesis. Science 1962, 138, 1404–1405. [Google Scholar] [CrossRef]
- Hamant, O.; Inoue, D.; Bouchez, D.; Dumais, J.; Mjolsness, E. Are microtubule tension sensors? Nat. Commun. 2019, 10, 2360. [Google Scholar] [CrossRef] [Green Version]
- Buschmann, H.; Hauptmann, M.; Niessing, D.; Lloyd, C.W.; Schäffner, A.R. Helical growth of the Arabidopsis mutant tortifolia2 does not depend on cell division patterns but involves handed twisting of isolated cells. Plant Cell 2009, 21, 2090–2106. [Google Scholar] [CrossRef] [Green Version]
- Murata, T.; Sonobe, S.; Baskin, T.I.; Hyodo, S.; Hasezawa, S.; Nagata, T.; Horio, T.; Hasebe, M. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol. 2005, 7, 961–968. [Google Scholar] [CrossRef]
- Nakamura, M.; Ehrhardt, D.; Hashimoto, T. Microtubule and katanin dependent dynamics of microtubule nucleation complexes in the Arabidopsis cortical array. Nat. Cell Biol. 2010, 12, 1064–1070. [Google Scholar] [CrossRef]
- Shaw, S.; Kamyar, R.; Ehrhardt, D. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 2003, 300, 1715–1718. [Google Scholar] [CrossRef] [Green Version]
- Lindeboom, J.J.; Nakamura, M.; Hibbel, A.; Shundyak, K.; Guierrez, R.; Ketelaar, T.; Emons, A.M.C.; Mulder, B.M.; Kirik, V.; Ehrhardt, D.W. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 2013, 342, 1245533. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Cyr, R. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 2004, 16, 274–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tindermans, S.H.; Deinum, E.E.; Lindeboom, J.J.; Mulder, B.M. Efficient event-driven simulations shed new light on microtubule organization in the plant cortical array. Front. Phys. 2014, 2, 19. [Google Scholar]
- Sedbrook, J.C.; Carroll, K.L.; Hung, K.F.; Masson, P.H.; Somerville, C.R. The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 2002, 14, 1635–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saffer, A.M.; Carpita, N.C.; Irish, V.F. Rhamnose-containing cell wall polymers suppress helical plant growth independently of microtubule orientation. Curr. Biol. 2017, 27, 2248–2259. [Google Scholar] [CrossRef] [PubMed]
- Pyrpassopoulos, S.; Feeser, E.A.; Mazerik, J.N.; Tyska, M.J.; Ostap, E.M. Membrane-bound myo1c powers asymmetric motility of actin filaments. Curr. Biol. 2012, 22, 1688–1692. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.Y.; Uemura, S.; Adachi, K.; Itoh, H.; Kionoshita, K.; Ishiwata, S. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 2002, 9, 464–467. [Google Scholar] [CrossRef]
- Pierson, G.B.; Burton, P.R.; Himes, R.H. Alterations in number of protofilaments in microtubules assembled in vitro. J. Cell Biol. 1978, 76, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Chretien, D.; Wade, R.H. New data on the microtubule surface lattice. Biol. Cell 1991, 71, 161–174. [Google Scholar] [CrossRef]
- Sui, H.; Downing, K.H. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 2010, 18, 1022–1031. [Google Scholar] [CrossRef] [Green Version]
- Yagi, N.; Matsunaga, S.; Hashimoto, T. Insights into cortical microtubule nucleation and dynamics in Arabidopsis leaf cells. J. Cell Sci. 2018, 131, jcs203778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, T. Microtubule and cell shape determination. In The Plant Cytoskeleton; Liu, B., Ed.; Springer: New York, NY, USA, 2011; Chapter 11; pp. 245–257. [Google Scholar]
- Ledbetter, M.C.; Porter, K.R. Morphology of microtubules of plant cells. Science 1964, 144, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Chaaban, S.; Brouhard, G.J. A microtubule bestiary: Structural diversity in tubulin polymers. Mol. Biol. Cell 2017, 28, 2924–2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalfie, M.; Thomson, J.N. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J. Cell Biol. 1982, 93, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Chaaban, S.; Jariwala, S.; Hsu, C.T.; Redemann, S.; Kollman, J.M.; Müller-Reichert, T.; Sept, D.; Bui, K.H.; Brouhard, G.J. The structure and dynamics of C. elegans tubulin reveals the mechanistic basis of microtubule growth. Dev. Cell 2018, 47, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Wurz, M.; Zupa, E.; Pfeffer, S.; Schiebel, E. Microtubule nucleation: The waltz between γ-tubulin ring complex and associated proteins. Curr. Opin. Cell Biol. 2020, 68, 124–131. [Google Scholar] [CrossRef]
- Chretien, D.; Fuller, S.D. Microtubule switch occasionally into unfavorable configurations during elongation. J. Mol. Biol. 2000, 298, 663–676. [Google Scholar] [CrossRef]
- Vitre, B.; Coquelle, F.M.; Heichette, C.; Garnier, C.; Chretien, D.; Arnal, I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008, 10, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Maurer, S.P.; Fourniol, F.J.; Bohner, G.; Moores, C.A.; Surrey, T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 2012, 149, 371–382. [Google Scholar] [CrossRef] [Green Version]
- Moores, C.A.; Perderiset, M.; Francis, F.; Chelly, J.; Houdusse, A.; Milligan, R.A. Mechanism of microtubule stabilization by doublecortin. Mol. Cell 2004, 14, 833–839. [Google Scholar] [CrossRef]
- Hamada, T. Microtubule organization and microtubule-associated proteins in plants. In International Review of Cell and Molecular Biology; Academic Press: London, UK, 2014; Volume 312, pp. 1–52. [Google Scholar]
- Ti, S.C.; Alushin, G.M.; Kapoor, T.M. Human β-tubulin isotypes can regulate microtubule protofilament number and stability. Dev. Cell 2018, 47, 175–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumino, Y.; Nagai, K.H.; Shitaka, Y.; Tanaka, D.; Yoshikawa, K.; Chaté, H.; Oiwa, K. Large-scale vortex lattice emerging from collectively moving microtubules. Nature 2012, 483, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Duclos, G.; Adkins, R.; Banerjee, D.; Peterson, M.S.E.; Varghese, M.; Kolvin, I.; Baskaran, A.; Pelcovits, R.A.; Powers, T.R.; Baskaran, A.; et al. Topological structure and dynamics of three-dimensional active nematics. Science 2020, 357, 1120–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokroy, B.; Kang, S.H.; Mahadevan, L.; Aizenberg, J. Self-organization of a mesoscale bristle into ordered, hierarchical helical assemblies. Science 2009, 323, 237–240. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Fallas, J.A.; Lynch, E.; Sheffler, W.; Parry, B.; Jannetty, N.; Decarreau, J.; Wagenbach, M.; Vicente, J.J.; Chen, J.; et al. De novo design of self-assembling helical protein filaments. Science 2018, 362, 705–709. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Furchtgott, L.; Huang, K.C.; Shaevitz, J.W. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc. Natl. Acad. Sci. USA 2012, 109, 3611. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Quint, D.A.; Grason, G.M.; Gopinathan, A.; Huang, K.C. Chiral twisting in a bacterial cytoskeletal polymer affects filament size and orientation. Nat. Commun. 2020, 11, 1408. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, M.; Hashimoto, T. Mechanistic Insights into Plant Chiral Growth. Symmetry 2020, 12, 2056. https://doi.org/10.3390/sym12122056
Nakamura M, Hashimoto T. Mechanistic Insights into Plant Chiral Growth. Symmetry. 2020; 12(12):2056. https://doi.org/10.3390/sym12122056
Chicago/Turabian StyleNakamura, Masayoshi, and Takashi Hashimoto. 2020. "Mechanistic Insights into Plant Chiral Growth" Symmetry 12, no. 12: 2056. https://doi.org/10.3390/sym12122056
APA StyleNakamura, M., & Hashimoto, T. (2020). Mechanistic Insights into Plant Chiral Growth. Symmetry, 12(12), 2056. https://doi.org/10.3390/sym12122056




