Cells with Broken Left–Right Symmetry: Roles of Intrinsic Cell Chirality in Left–Right Asymmetric Epithelial Morphogenesis
Abstract
1. Chirality Appears as a Hierarchical Structure in Biology
2. Cells Have Chirality
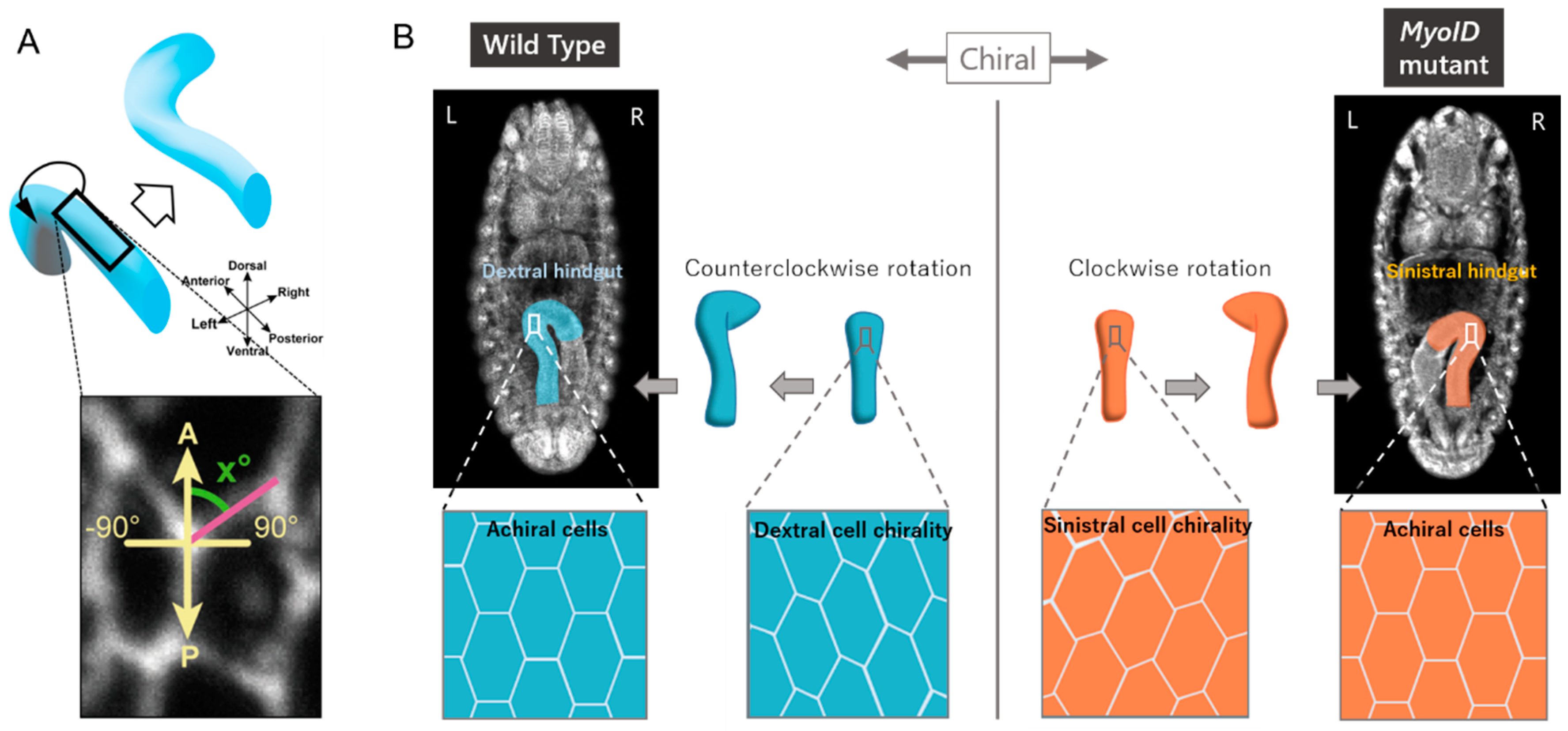
3. Cellular Basis for LR Asymmetric Morphogenesis in Drosophila
4. Cell Chirality Drives LR Asymmetric Development in Drosophila
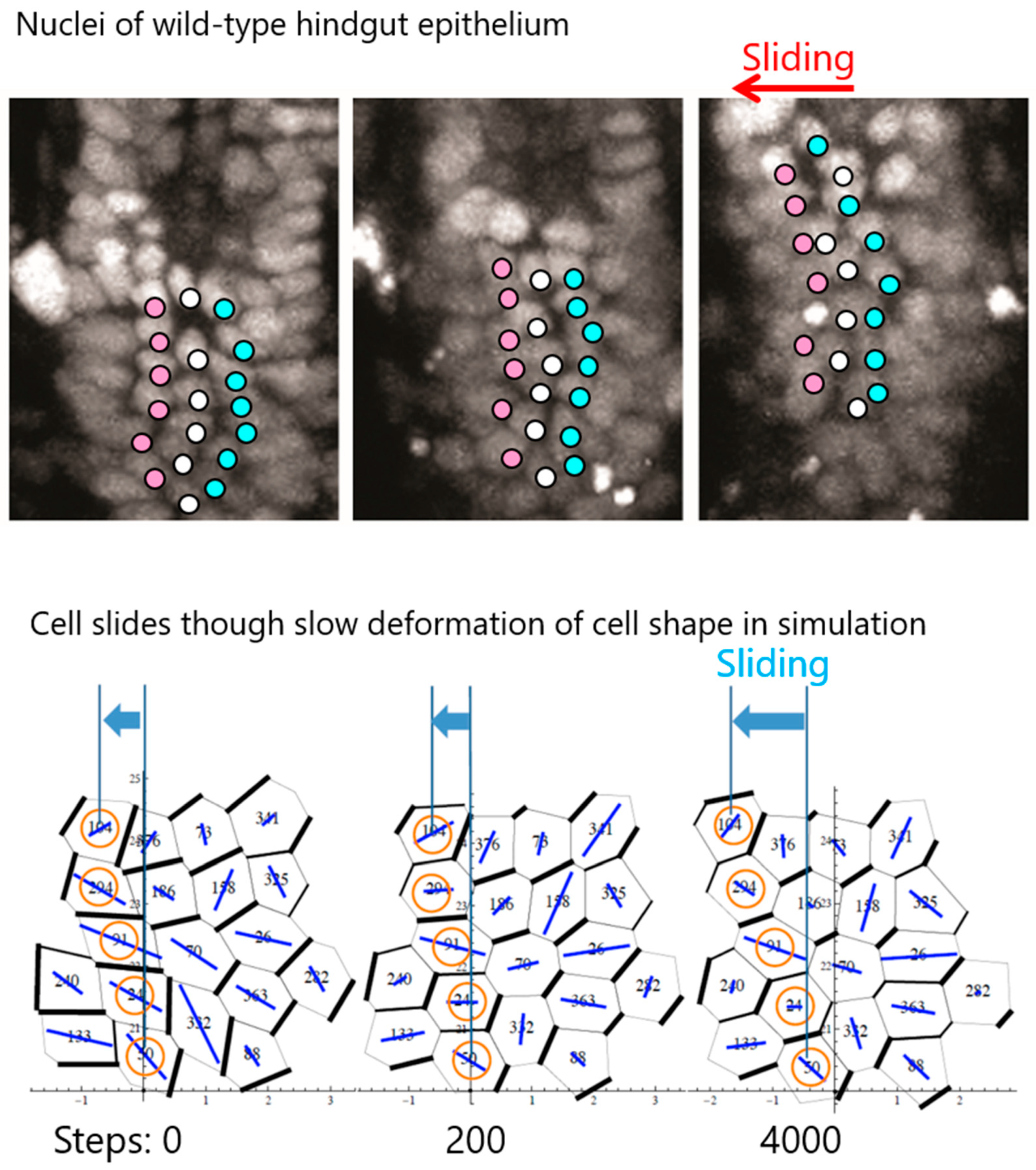
5. Cell Chirality Induces LR-Directional Cell Sliding in the Hindgut Epithelium
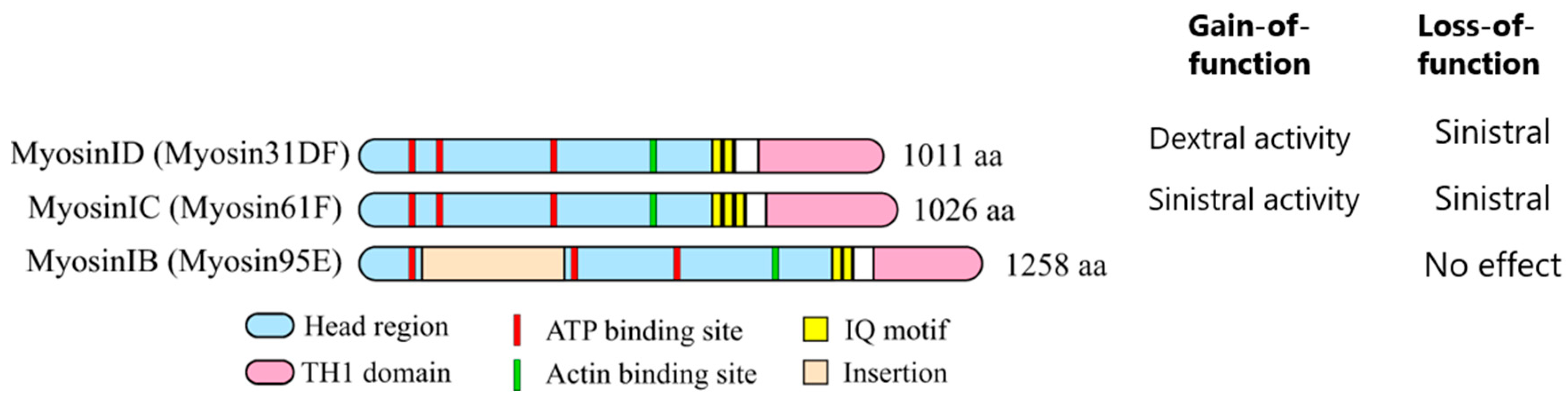
6. Mechanisms of Cell Chirality Formation
7. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Genchi, G. An overview on D-amino acids. Amino Acids 2017, 49, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Blackmond, D.G. The origin of biological homochirality. Cold Spring Harb Perspect. Biol. 2010, 2, a002147. [Google Scholar] [CrossRef] [PubMed]
- Watson, T. An account of some cases of transposition observed in the human body. Lond. Med. Gaz. 1836, 18, 393–403. [Google Scholar]
- Spemann, H.; Falkenberg, H. Uber Asymmetrische Entwicklung und Situs inversus viscerum bei Zwillingen und Doppelbildungen. Wilhelm Roux’ Arch. Entwicklungsmech. Org. 1919, 45, 371–422. [Google Scholar] [CrossRef]
- Okumura, T.; Utsuno, H.; Kuroda, J.; Gittenberger, E.; Asami, T.; Matsuno, K. The development and evolution of left-right asymmetry in invertebrates: Lessons from Drosophila and snails. Dev. Dyn. 2008, 237, 3497–3515. [Google Scholar] [CrossRef]
- Sturtevant, A.H. Inheritance of direction of coilling in limnaea. Science 1923, 58, 269–270. [Google Scholar] [CrossRef]
- Nonaka, S.; Yoshiba, S.; Watanabe, D.; Ikeuchi, S.; Goto, T.; Marshall, W.F.; Hamada, H. De novo formation of left-right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 2005, 3, e268. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.; McDowell, G.S.; Holden, J.M.; Johnson, H.F.; Koutsovoulos, G.D.; Liu, M.M.; Hulpiau, P.; Van Roy, F.; Wade, C.M.; Banerjee, R.; et al. Formin Is Associated with Left-Right Asymmetry in the Pond Snail and the Frog. Curr. Biol. 2016, 26, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, R.; Fujikura, K.; Abe, M.; Hosoiri, Y.; Asakawa, S.; Shimizu, M.; Umeda, S.; Ichikawa, F.; Takahashi, H. Diaphanous gene mutation affects spiral cleavage and chirality in snails. Sci. Rep. 2016, 6, 34809. [Google Scholar] [CrossRef]
- Lebreton, G.; Géminard, C.; Lapraz, F.; Pyrpassopoulos, S.; Cerezo, D.; Spéder, P.; Ostap, E.M.; Noselli, S. Molecular to organismal chirality is induced by the conserved myosin 1D. Science 2018, 362, 949–952. [Google Scholar] [CrossRef]
- Taniguchi, K.; Maeda, R.; Ando, T.; Okumura, T.; Nakazawa, N.; Hatori, R.; Nakamura, M.; Hozumi, S.; Fujiwara, H.; Matsuno, K. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science 2011, 333, 339–341. [Google Scholar] [CrossRef]
- González-Morales, N.; Géminard, C.; Lebreton, G.; Cerezo, D.; Coutelis, J.-B.; Noselli, S. The Atypical Cadherin Dachsous Controls Left-Right Asymmetry in Drosophila. Dev. Cell 2015, 33, 675–689. [Google Scholar] [CrossRef]
- Sato, K.; Hiraiwa, T.; Maekawa, E.; Isomura, A.; Shibata, T.; Kuranaga, E. Left-right asymmetric cell intercalation drives directional collective cell movement in epithelial morphogenesis. Nat. Commun. 2015, 6, 10074. [Google Scholar] [CrossRef]
- Inaki, M.; Hatori, R.; Nakazawa, N.; Okumura, T.; Ishibashi, T.; Kikuta, J.; Ishii, M.; Matsuno, K.; Honda, H. Chiral cell sliding drives left-right asymmetric organ twisting. eLife 2018, 7. [Google Scholar] [CrossRef]
- Inaki, M.; Liu, J.; Matsuno, K. Cell chirality: Its origin and roles in left-right asymmetric development. Philos. Trans. R. Soc. Lond B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Inaki, M.; Sasamura, T.; Matsuno, K. Cell Chirality Drives Left-Right Asymmetric Morphogenesis. Front Cell Dev. Biol. 2018, 6, 34. [Google Scholar] [CrossRef]
- Ray, P.; Chin, A.S.; Worley, K.E.; Fan, J.; Kaur, G.; Wu, M.; Wan, L.Q. Intrinsic cellular chirality regulates left-right symmetry breaking during cardiac looping. Proc. Natl. Acad. Sci. USA 2018. [Google Scholar] [CrossRef] [PubMed]
- Gail, M.H.; Boone, C.W. The locomotion of mouse fibroblasts in tissue culture. Biophys. J. 1970, 10, 980–993. [Google Scholar] [CrossRef]
- Hagmann, J. Pattern formation and handedness in the cytoskeleton of human platelets. Proc. Natl. Acad. Sci. USA 1993, 90, 3280–3283. [Google Scholar] [CrossRef]
- Xu, J.; Van Keymeulen, A.; Wakida, N.M.; Carlton, P.; Berns, M.W.; Bourne, H.R. Polarity reveals intrinsic cell chirality. Proc. Natl. Acad. Sci. USA 2007, 104, 9296–9300. [Google Scholar] [CrossRef] [PubMed]
- Tamada, A.; Kawase, S.; Murakami, F.; Kamiguchi, H. Autonomous right-screw rotation of growth cone filopodia drives neurite turning. J. Cell Biol. 2010, 188, 429–441. [Google Scholar] [CrossRef]
- Wan, L.Q.; Ronaldson, K.; Park, M.; Taylor, G.; Zhang, Y.; Gimble, J.M.; Vunjak-Novakovic, G. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc. Natl. Acad. Sci. USA 2011, 108, 12295–12300. [Google Scholar] [CrossRef]
- Chen, T.-H.; Hsu, J.J.; Zhao, X.; Guo, C.; Wong, M.N.; Huang, Y.; Li, Z.; Garfinkel, A.; Ho, C.-M.; Tintut, Y.; et al. Left-Right Symmetry Breaking in Tissue Morphogenesis via Cytoskeletal MechanicsNovelty and Significance. Circ. Res. 2012, 110, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, H.; Kondo, S. Rotating pigment cells exhibit an intrinsic chirality. Genes Cells 2015, 20, 29–35. [Google Scholar] [CrossRef]
- Tee, Y.H.; Shemesh, T.; Thiagarajan, V.; Hariadi, R.F.; Anderson, K.L.; Page, C.; Volkmann, N.; Hanein, D.; Sivaramakrishnan, S.; Kozlov, M.M.; et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 2015, 17, 445–457. [Google Scholar] [CrossRef]
- Tamada, A.; Igarashi, M. Revealing chiral cell motility by 3D Riesz transform-differential interference contrast microscopy and computational kinematic analysis. Nat. Commun. 2017, 8, 2194. [Google Scholar] [CrossRef]
- Wan, L.Q.; Chin, A.S.; Worley, K.E.; Ray, P. Cell chirality: Emergence of asymmetry from cell culture. Philos. Trans. R. Soc. Lond B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Satir, P. Chirality of the cytoskeleton in the origins of cellular asymmetry. Philos. Trans. R. Soc. Lond B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Kuroda, R.; Endo, B.; Abe, M.; Shimizu, M. Chiral blastomere arrangement dictates zygotic left-right asymmetry pathway in snails. Nature 2009, 462, 790–794. [Google Scholar] [CrossRef]
- Naganathan, S.R.; Fürthauer, S.; Nishikawa, M.; Jülicher, F.; Grill, S.W. Active torque generation by the actomyosin cell cortex drives left–right symmetry breaking. eLife 2014, 3, e04165. [Google Scholar] [CrossRef]
- Hozumi, S.; Maeda, R.; Taniguchi, K.; Kanai, M.; Shirakabe, S.; Sasamura, T.; Spéder, P.; Noselli, S.; Aigaki, T.; Murakami, R.; et al. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature 2006, 440, 798–802. [Google Scholar] [CrossRef]
- Speder, P.; Adam, G.; Noselli, S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature 2006, 440, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Kozopas, K.M.; Samos, C.H.; Nusse, R. DWnt-2, a Drosophila Wnt gene required for the development of the male reproductive tract, specifies a sexually dimorphic cell fate. Genes Dev. 1998, 12, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Adám, G.; Perrimon, N.; Noselli, S. The retinoic-like juvenile hormone controls the looping of left-right asymmetric organs in Drosophila. Development 2003, 130, 2397–2406. [Google Scholar] [CrossRef]
- Pascual, A.; Huang, K.L.; Neveu, J.; Préat, T. Neuroanatomy: Brain asymmetry and long-term memory. Nature 2004, 427, 605–606. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, J.A.; Iwaki, D.D. It takes guts: The Drosophila hindgut as a model system for organogenesis. Dev. Biol. 2002, 243, 1–19. [Google Scholar] [CrossRef]
- Wells, R.E.; Barry, J.D.; Warrington, S.J.; Cuhlmann, S.; Evans, P.; Huber, W.; Strutt, D.; Zeidler, M.P. Control of tissue morphology by Fasciclin III-mediated intercellular adhesion. Development 2013, 140, 3858–3868. [Google Scholar] [CrossRef] [PubMed]
- Bertet, C.; Sulak, L.; Lecuit, T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 2004, 429, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, J.T.; Backovic, S.T.; Sanny, J.S.; Weitz, O.; Zallen, J.A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 2006, 11, 459–470. [Google Scholar] [CrossRef]
- Morgan, N.S.; Skovronsky, D.M.; Artavanis-Tsakonas, S.; Mooseker, M.S. The molecular cloning and characterization of Drosophila melanogaster myosin-IA and myosin-IB. J. Mol. Biol. 1994, 239, 347–356. [Google Scholar] [CrossRef]
- Amcheslavsky, A.; Wang, S.; Fogarty, C.E.; Lindblad, J.L.; Fan, Y.; Bergmann, A. Plasma Membrane Localization of Apoptotic Caspases for Non-apoptotic Functions. Dev. Cell 2018, 45, 450–464.e3. [Google Scholar] [CrossRef]
- Packard, M.; Jokhi, V.; Ding, B.; Ruiz-Canada, C.; Ashley, J.; Budnik, V. Nucleus to Synapse Nesprin1 Railroad Tracks Direct Synapse Maturation through RNA Localization. Neuron 2015, 86, 1015–1028. [Google Scholar] [CrossRef]
- Hatori, R.; Ando, T.; Sasamura, T.; Nakazawa, N.; Nakamura, M.; Taniguchi, K.; Hozumi, S.; Kikuta, J.; Ishii, M.; Matsuno, K. Left-right asymmetry is formed in individual cells by intrinsic cell chirality. Mech. Dev. 2014, 133, 146–162. [Google Scholar] [CrossRef]
- Iwaki, D.D.; Lengyel, J.A. A Delta-Notch signaling border regulated by Engrailed/Invected repression specifies boundary cells in the Drosophila hindgut. Mech. Dev. 2002, 114, 71–84. [Google Scholar] [CrossRef]
- Okumura, T.; Sasamura, T.; Inatomi, M.; Hozumi, S.; Nakamura, M.; Hatori, R.; Taniguchi, K.; Nakazawa, N.; Suzuki, E.; Maeda, R.; et al. Class I Myosins Have Overlapping and Specialized Functions in Left-Right Asymmetric Development in Drosophila. Genetics 2015, 199, 1183–1199. [Google Scholar] [CrossRef]
- Tzolovsky, G.; Millo, H.; Pathirana, S.; Wood, T.; Bownes, M. Identification and phylogenetic analysis of Drosophila melanogaster myosins. Mol. Biol. Evol. 2002, 19, 1041–1052. [Google Scholar] [CrossRef][Green Version]
- Petzoldt, A.G.; Coutelis, J.B.; Géminard, C.; Spéder, P.; Suzanne, M.; Cerezo, D.; Noselli, S. DE-Cadherin regulates unconventional Myosin ID and Myosin IC in Drosophila left-right asymmetry establishment. Development 2012, 139, 1874–1884. [Google Scholar] [CrossRef]
- Hozumi, S.; Maeda, R.; Taniguchi-Kanai, M.; Okumura, T.; Taniguchi, K.; Kawakatsu, Y.; Nakazawa, N.; Hatori, R.; Matsuno, K. Head region of unconventional myosin I family members is responsible for the organ-specificity of their roles in left-right polarity in Drosophila. Dev. Dyn. 2008, 237, 3528–3537. [Google Scholar] [CrossRef]
- Pyrpassopoulos, S.; Feeser, E.A.; Mazerik, J.N.; Tyska, M.J.; Ostap, E.M. Membrane-bound myo1c powers asymmetric motility of actin filaments. Curr. Biol. 2012, 22, 1688–1692. [Google Scholar] [CrossRef]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers—Assembly, dynamics and biological roles. J. Cell Sci. 2012, 125 Pt 8, 1855–1864. [Google Scholar] [CrossRef]
- Higgs, H.N. Formin proteins: A domain-based approach. Trends Biochem. Sci. 2005, 30, 342–353. [Google Scholar] [CrossRef]
- Mizuno, H.; Higashida, C.; Yuan, Y.; Ishizaki, T.; Narumiya, S.; Watanabe, N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science 2011, 331, 80–83. [Google Scholar] [CrossRef]
- Levin, M.; Johnson, R.L.; Stern, C.D.; Kuehn, M.; Tabin, C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 1995, 82, 803–814. [Google Scholar] [CrossRef]
- Collignon, J.; Varlet, I.; Robertson, E.J. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 1996, 381, 155–158. [Google Scholar] [CrossRef]
- Lowe, L.A.; Supp, D.M.; Sampath, K.; Yokoyama, T.; Wright, C.V.; Potter, S.S.; Overbeek, P.; Kuehn, M.R. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 1996, 381, 158–161. [Google Scholar] [CrossRef]
- Nonaka, S.; Tanaka, Y.; Okada, Y.; Takeda, S.; Harada, A.; Kanai, Y.; Kido, M.; Hirokawa, N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 1998, 95, 829–837. [Google Scholar] [CrossRef]
- Levin, M.; Thorlin, T.; Robinson, K.R.; Nogi, T.; Mercola, M. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell 2002, 111, 77–89. [Google Scholar] [CrossRef]
- Fan, J.; Ray, P.; Lu, Y.; Kaur, G.; Schwarz, J.J.; Wan, L.Q. Cell chirality regulates intercellular junctions and endothelial permeability. Sci. Adv. 2018, 4, eaat2111. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Utsunomiya, S.; Sakamura, S.; Sasamura, T.; Ishibashi, T.; Maeda, C.; Inaki, M.; Matsuno, K. Cells with Broken Left–Right Symmetry: Roles of Intrinsic Cell Chirality in Left–Right Asymmetric Epithelial Morphogenesis. Symmetry 2019, 11, 505. https://doi.org/10.3390/sym11040505
Utsunomiya S, Sakamura S, Sasamura T, Ishibashi T, Maeda C, Inaki M, Matsuno K. Cells with Broken Left–Right Symmetry: Roles of Intrinsic Cell Chirality in Left–Right Asymmetric Epithelial Morphogenesis. Symmetry. 2019; 11(4):505. https://doi.org/10.3390/sym11040505
Chicago/Turabian StyleUtsunomiya, Sosuke, So Sakamura, Takeshi Sasamura, Tomoki Ishibashi, Chinami Maeda, Mikiko Inaki, and Kenji Matsuno. 2019. "Cells with Broken Left–Right Symmetry: Roles of Intrinsic Cell Chirality in Left–Right Asymmetric Epithelial Morphogenesis" Symmetry 11, no. 4: 505. https://doi.org/10.3390/sym11040505
APA StyleUtsunomiya, S., Sakamura, S., Sasamura, T., Ishibashi, T., Maeda, C., Inaki, M., & Matsuno, K. (2019). Cells with Broken Left–Right Symmetry: Roles of Intrinsic Cell Chirality in Left–Right Asymmetric Epithelial Morphogenesis. Symmetry, 11(4), 505. https://doi.org/10.3390/sym11040505




