A Coarse-to-Fine Fully Convolutional Neural Network for Fundus Vessel Segmentation
Abstract
1. Introduction
1.1. Related Works
1.2. Our Motivations and Contributions
- (1)
- Different from previous methods, we utilize RGB images instead of using the gray green channel image only to retain as much as possible raw information inside the Field of View (FOV). Morphological transformations are also applied to enhance the contrast of RGB input images for accurate vessel segmentation.
- (2)
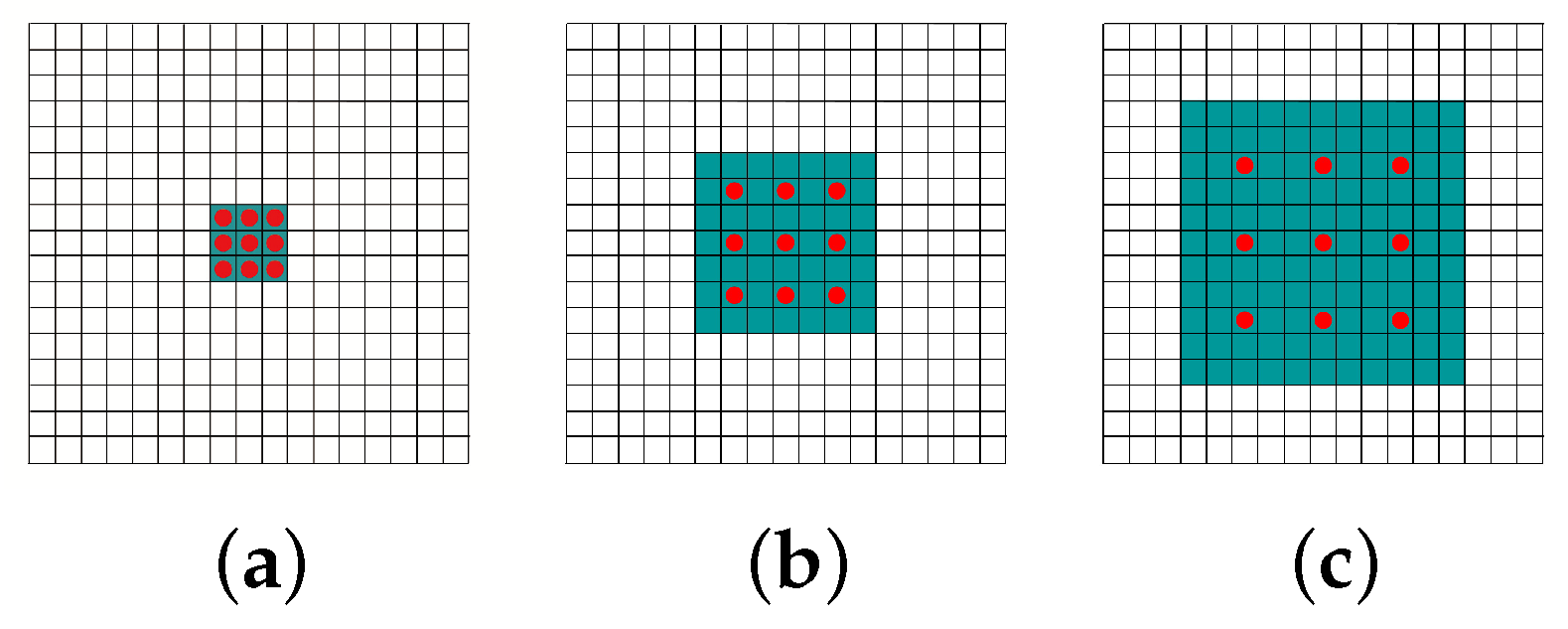
- We propose a specially designed network structure for full gauge fundus vessel segmentation named FG-FCN which replaces pooling layers in original FCN with dilated convolution layers to keep the spatial and semantic information with large receptive fields.
- (3)
- We integrate CRF as recurrent neural networks (RNN) into our structure to adapt the weights of FG-FCN during the training stage for refining its coarse output which do not consider non-local correlations.
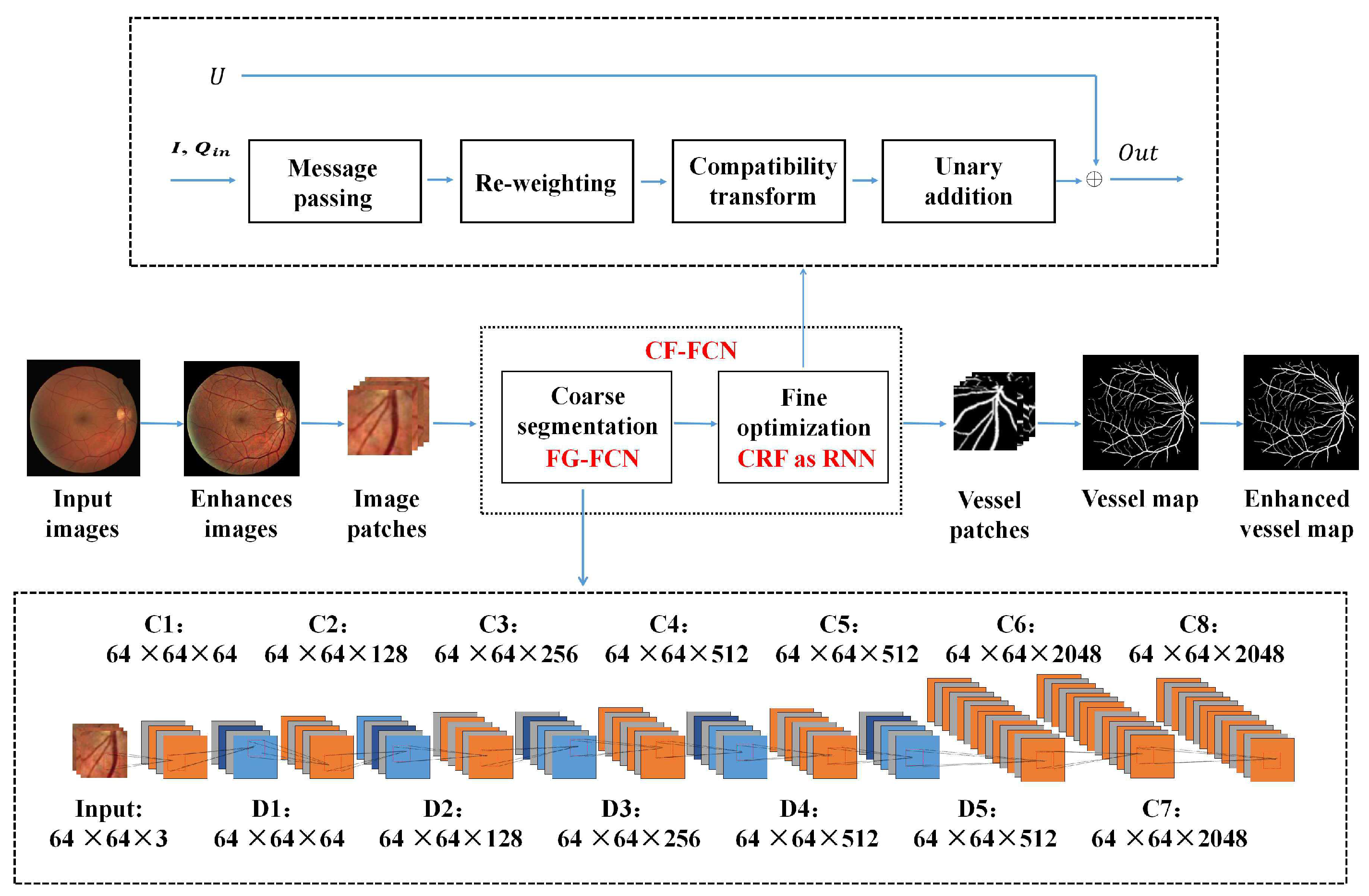
2. CF-FCN: A Coarse-to-Fine FCN for Fundus Vessel Segmentation
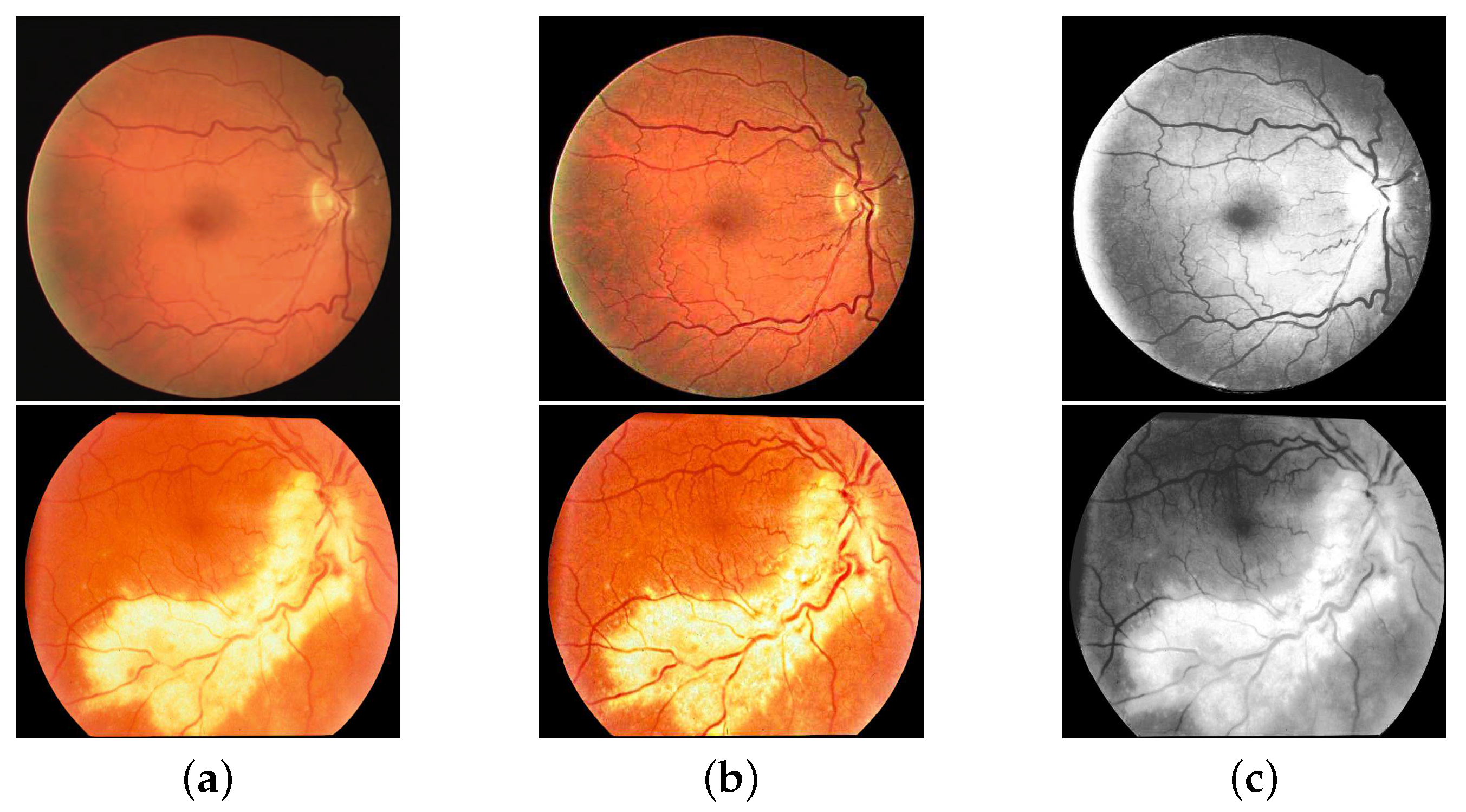
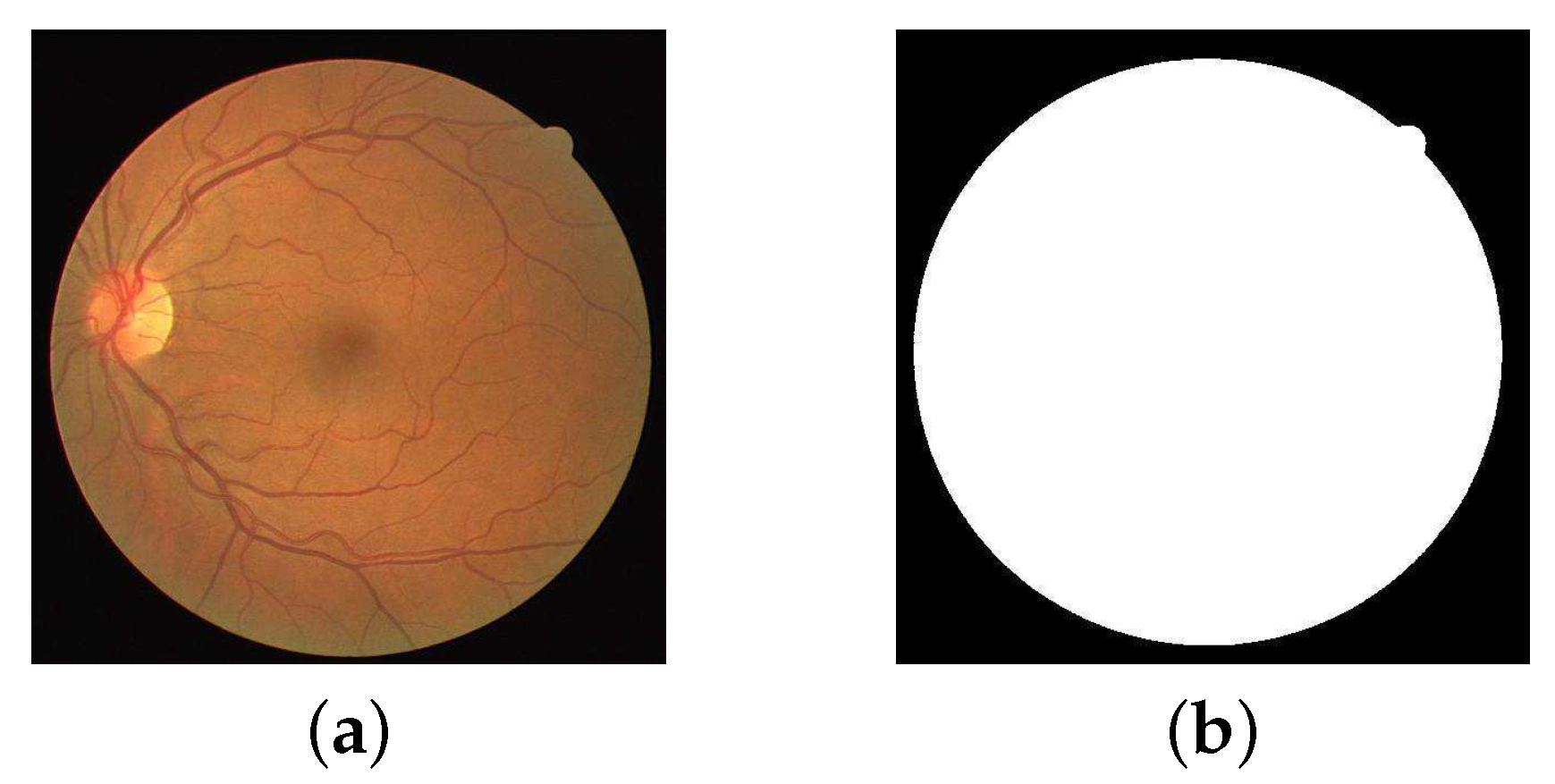
2.1. Data Pre-Processing
| Algorithm 1 Pre-processing algorithm. |
| Input: origin fundus images X Output: pre-processed fundus images Y
|
2.2. Coarse Vessel Segmentation
2.3. Fine Optimization
| Algorithm 2 CRF as RNN. |
|
2.4. Morphological Post-Processing
3. Experiments
3.1. Dataset
3.2. Evaluation Metrics
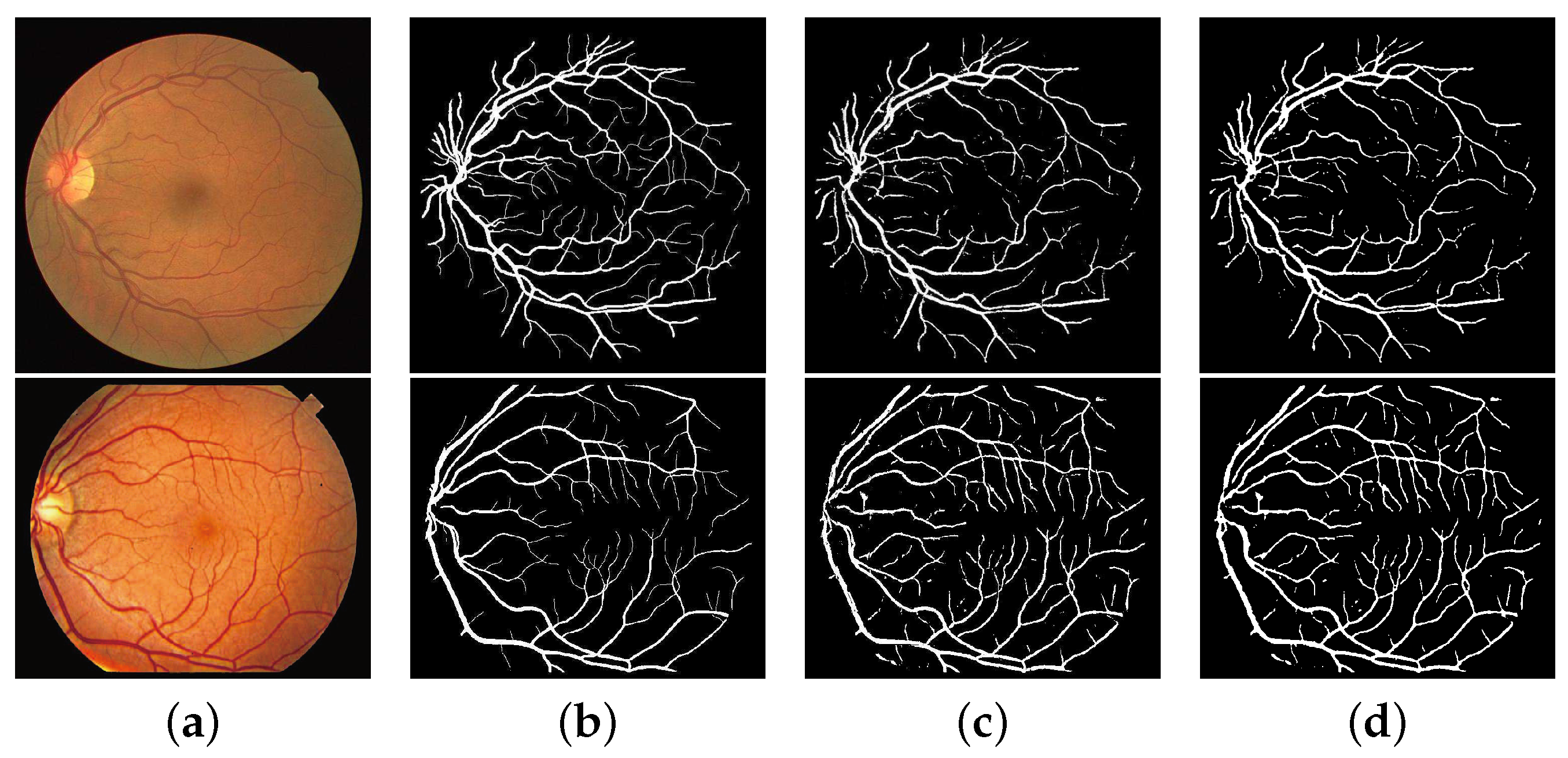
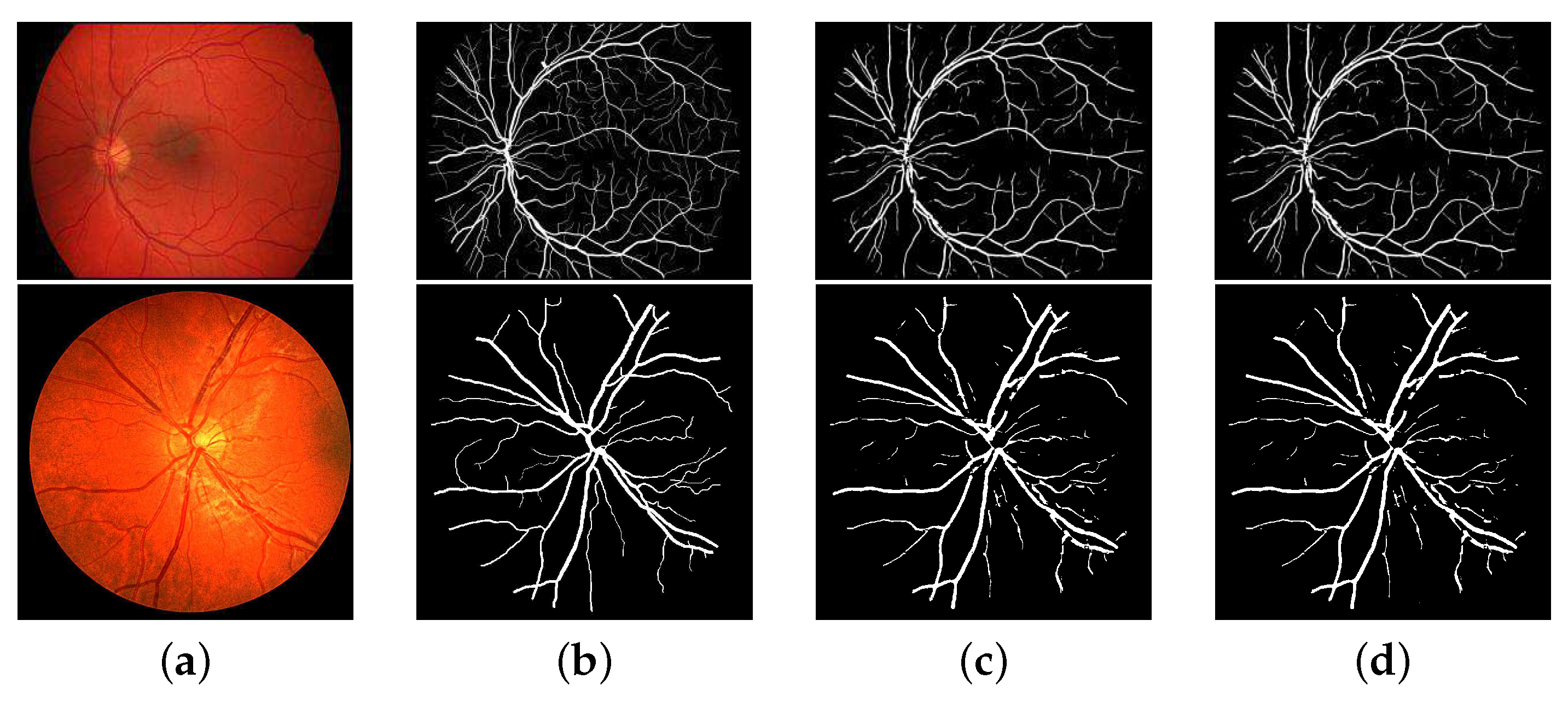
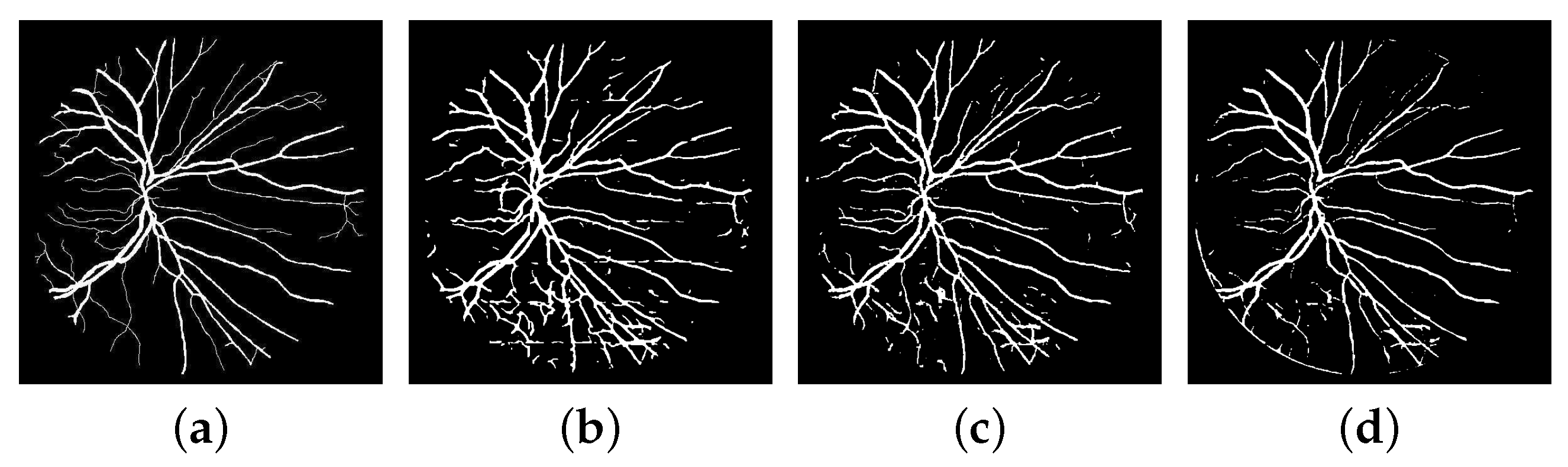
3.3. Experimental Results
3.3.1. The Improvement of the Data Quality
3.3.2. Comparison with the State-of-the-Art Works
3.3.3. Improvement of the Network’s Structure
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, J.; Hu, Q.; Imran, A.; Zhang, L.; Yang, J.; Wang, Q. Vessel Recognition of Retinal Fundus Images Based on Fully Convolutional Network. In Proceedings of the 2018 IEEE 42nd Annual Computer Software and Applications Conference (COMPSAC), Tokyo, Japan, 23–27 July 2018; pp. 413–418. [Google Scholar] [CrossRef]
- Poplin, R.; Varadarajan, A.V.; Blumer, K.; Liu, Y.; Mcconnell, M.V.; Corrado, G.S.; Peng, L.; Webster, D.R. Predicting Cardiovascular Risk Factors from Retinal Fundus Photographs using Deep Learning. arXiv, 2017; arXiv:1708.09843. [Google Scholar]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. Blood vessel segmentation methodologies in retinal images—A survey. Comput. Methods Prog. Biomed. 2012, 108, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Azzopardi, G.; Strisciuglio, N.; Vento, M.; Petkov, N. Trainable COSFIRE filters for vessel delineation with application to retinal images. Med. Image Anal. 2015, 19, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Mojon, D. Adaptive local thresholding by verification-based multithreshold probing with application to vessel detection in retinal images. IEEE Trans. Patt. Anal. Mach. Intell. 2015, 25, 131–137. [Google Scholar] [CrossRef]
- Ramalho, G.L.B.; Ramalho, G.L.B.; Veras, R.M.S.; Veras, R.M.S.; Medeiros, F.N.S. An unsupervised coarse-to-fine algorithm for blood vessel segmentation in fundus images. Expert Syst. Appl. 2017, 78, 182–192. [Google Scholar] [CrossRef]
- Zhu, C.; Zou, B.; Xiang, Y.; Cui, J.; Hui, W.U. An ensemble retinal vessel segmentation based on supervised learning in fundus images. Chin. J. Electron. 2016, 25, 503–511. [Google Scholar] [CrossRef]
- Xiao, Z.; Wang, M.; Zhang, F.; Geng, L.; Wu, J.; Su, L.; Tong, J. Retinal vessel segmentation based on adaptive difference of Gauss filter. In Proceedings of the IEEE International Conference on Digital Signal Processing, Beijing, China, 16–18 October 2016; pp. 15–19. [Google Scholar]
- Staal, J.; Abramoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Hoover, A.D.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Budai, A.; Bock, R.; Maier, A.; Hornegger, J.; Michelson, G. Robust vessel segmentation in fundus images. Int. J. Biomed. Imaging 2013, 2013, 154860. [Google Scholar] [CrossRef] [PubMed]
- Fraz, M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, R.A.; Owen, G.C.; Barman, S.A. An Ensemble Classification-Based Approach Applied to Retinal Blood Vessel Segmentation. IEEE Trans. Biomed. Eng. 2012, 59, 2538–2548. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Dharmveer, S.; Singh, B. A new morphology based approach for blood vessel segmentation in retinal images. In Proceedings of the 2014 Annual IEEE India Conference (INDICON), Pune, India, 11–13 December 2014; pp. 1–6. [Google Scholar]
- Dash, J.; Bhoi, N. A thresholding based technique to extract retinal blood vessels from fundus images. Future Comput. Inf. J. 2017, 2, 103–109. [Google Scholar] [CrossRef]
- Odstrcilik, J.; Kolar, R.; Budai, A.; Hornegger, J. Retinal vessel segmentation by improved matched filtering: Evaluation on a new high-resolution fundus image database. IET Image Process. 2013, 7, 373–383. [Google Scholar] [CrossRef]
- Osareh, A.; Shadgar, B. Automatic blood vessel segmentation in color images of retina. Iran. J. Sci. Technol. Trans. B 2009, 33, 191–206. [Google Scholar]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.; Larochelle, H. Brain Tumor Segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Dan, C.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Deep neural networks segment neuronal membranes in electron microscopy images. Adv. Neural Inf. Process. Syst. 2012, 25, 2852–2860. [Google Scholar]
- Li, Q.; Feng, B.; Xie, L.P.; Liang, P.; Zhang, H.; Wang, T. A cross-modality learning approach for vessel segmentation in retinal images. IEEE Trans. Med. Imaging 2016, 35, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Boreom, L. Development of automatic retinal vessel segmentation method in fundus images via convolutional neural networks. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Seogwipo, Korea, 11–15 July 2017; pp. 681–684. [Google Scholar] [CrossRef]
- Liskowski, P.; Krawiec, K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans. Med. Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Xu, Y.; Lin, S.; Wong, D.W.K.; Liu, J. DeepVessel: Retinal Vessel Segmentation via Deep Learning and Conditional Random Field. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; pp. 132–139. [Google Scholar]
- Dasgupta, A.; Singh, S. A fully convolutional neural network based structured prediction approach towards the retinal vessel segmentation. In Proceedings of the ISBI, Melbourne, VIC, Australia, 18–21 April 2017; pp. 248–251. [Google Scholar] [CrossRef]
- Li, Q.; Xie, L.; Zhang, Q.; Qi, S.; Liang, P.; Zhang, H.; Wang, T. A supervised method using convolutional neural networks for retinal vessel delineation. In Proceedings of the 2015 8th International Congress on Image and Signal Processing (CISP), Shenyang, China, 14–16 October 2015; pp. 418–422. [Google Scholar]
- Fu, H.; Xu, Y.; Wong, D.W.K.; Liu, J. Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech, 13–16 April 2016; pp. 698–701. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv, 2015; arXiv:1511.07122. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv, 2014; arXiv:1409.1556. [Google Scholar]
- Wang, P.; Chen, P.; Yuan, Y.; Liu, D.; Huang, Z.; Hou, X.; Cottrell, G. Understanding convolution for semantic segmentation. WACV 2017, 2018, 1451–1460. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv, 2014; arXiv:1412.6980. [Google Scholar]
- Zheng, S.; Jayasumana, S.; Romeraparedes, B.; Vineet, V.; Su, Z.; Du, D.; Huang, C.; Torr, P.H.S. Conditional random fields as recurrent neural networks. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1529–1537. [Google Scholar]
- Lafferty, J.D.; Mccallum, A.; Pereira, F.C.N. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In Proceedings of the Eighteenth International Conference on Machine Learning, Williamstown, MA, USA, 28 June–1 July 2001; pp. 282–289. [Google Scholar]
- Soares, J.V.B.; Leandro, J.J.G.; Cesar, R.M.; Jelinek, H.F.; Cree, M.J. Retinal vessel segmentation using the 2-D gabor wavelet and supervised classification. IEEE Trans. Med. Imaging 2006, 25, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Ngo, L.; Han, J.H. Multi-level deep neural network for efficient segmentation of blood vessels in fundus images. Electron. Lett. 2017, 53, 1096–1098. [Google Scholar] [CrossRef]
- Niemeijer, M.; Ginneken, B.V.; Loog, M. Comparative study of retinal vessel segmentation methods on a new publicly available database. In Proceedings of the Medical Imaging 2004: Image Processing, San Diego, CA, USA, 12 May 2004; pp. 648–656. [Google Scholar] [CrossRef]
- Fraz, M.M.; Barman, S.A.; Remagnino, P.; Hoppe, A.; Basit, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G. An approach to localize the retinal blood vessels using bit planes and centerline detection. Comput. Meth. Prog. Biomed. 2017, 108, 600–616. [Google Scholar] [CrossRef] [PubMed]
- Budai, A.; Hornegger, J.; Michelson, G. Multiscale approach for blood vessel segmentation on retinal fundus images. Arvo Meet. Abst. 2009, 50, 325. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv, 2017; arXiv:1706.05587. [Google Scholar]



| Layer | Type | Maps | Kernel | Para. | Layer | Type | Maps | Kernel | Para. |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Input | 3 | - | - | 11 | Conv | 512 | 3 × 3 | 1 |
| 1 | Conv | 64 | 3 × 3 | 1 | 12 | Conv | 512 | 3 × 3 | 1 |
| 2 | Conv | 64 | 3 × 3 | 1 | 13 | Conv | 512 | 3 × 3 | 1 |
| 3 | DConv | 64 | 3 × 3 | 1 | 14 | DConv | 512 | 3 × 3 | 5 |
| 4 | Conv | 128 | 3 × 3 | 1 | 15 | Conv | 512 | 3 × 3 | 1 |
| 5 | Conv | 128 | 3 × 3 | 1 | 16 | Conv | 512 | 3 × 3 | 1 |
| 6 | DConv | 128 | 3 × 3 | 2 | 17 | Conv | 512 | 3 × 3 | 1 |
| 7 | Conv | 256 | 3 × 3 | 1 | 18 | DConv | 512 | 3 × 3 | 7 |
| 8 | Conv | 256 | 3 × 3 | 1 | 19 | Conv | 2048 | 7 × 7 | 1 |
| 9 | Conv | 256 | 3 × 3 | 1 | 20 | Conv | 2048 | 1 × 1 | 1 |
| 10 | DConv | 256 | 3 × 3 | 3 | 21 | Conv | 2048 | 1 × 1 | 1 |
| Type | Se | Sp | Acc |
|---|---|---|---|
| Green channel images | 0.6309 | 0.9891 | 0.9556 |
| RGB images | 0.7941 | 0.9870 | 0.9634 |
| Type | Method | Se | Sp | Acc | AUC |
|---|---|---|---|---|---|
| Unsupervised | Singh et al. [13] | 0.7138 | 0.9801 | 0.9460 | - |
| methods | Odstrcilik et al. [15] | 0.7060 | 0.9693 | 0.9340 | 0.9519 |
| Xiao et al. [8] | 0.8127 | 0.9786 | 0.9580 | - | |
| Neto et al. [6] | 0.7806 | 0.9629 | 0.8718 | - | |
| Supervised | Li et al. [19] | 0.7569 | 0.9816 | 0.9527 | 0.9738 |
| methods | Song et al. [20] | 0.7501 | 0.9795 | 0.9499 | - |
| Ngo et al. [33] | 0.7464 | 0.9836 | 0.9533 | 0.9752 | |
| Zhu et al. [7] | 0.7462 | 0.9838 | 0.9618 | - | |
| Li et al. [24] | 0.7659 | 0.9797 | 0.9522 | - | |
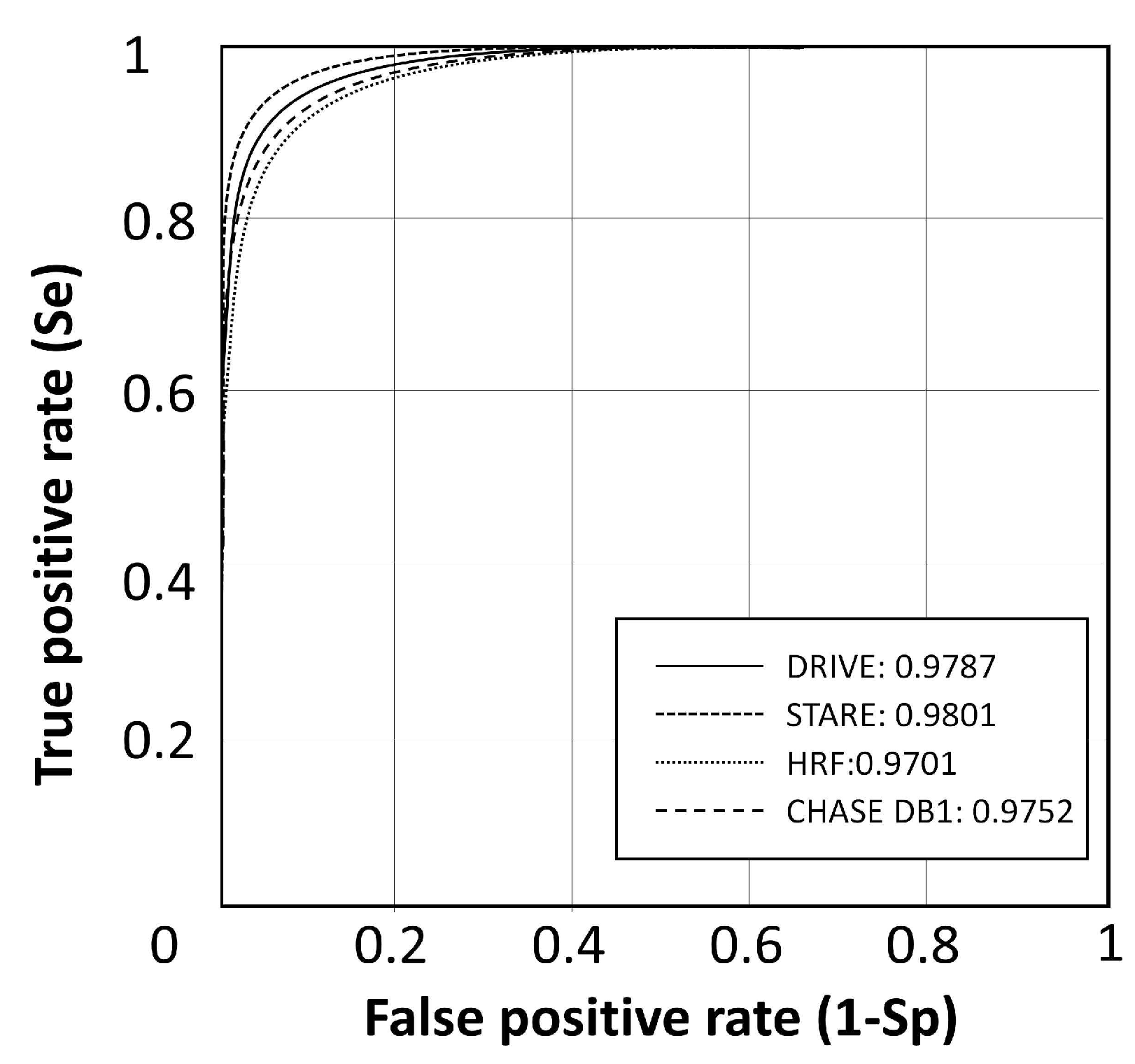
| CF-FCN-post | 0.7941 | 0.9870 | 0.9634 | 0.9787 |
| Type | Method | Se | Sp | Acc | AUC |
|---|---|---|---|---|---|
| Unsupervised | Niemeijer et al. [34] | 0.6898 | 0.9696 | 0.9417 | - |
| methods | Odstrcilik et al. [15] | 0.7847 | 0.9512 | 0.9341 | 0.9569 |
| Xiao et al. [8] | 0.7641 | 0.9757 | 0.9569 | - | |
| Neto et al. [6] | 0.8344 | 0.9443 | 0.8894 | - | |
| Supervised | Fraz et al. [12] | 0.7548 | 0.9763 | 0.9534 | 0.9768 |
| methods | Fu et al. [25] | 0.7140 | - | 0.9545 | - |
| Fraz et al. [35] | 0.7311 | 0.9680 | 0.9442 | - | |
| Soares et al. [32] | 0.7103 | 0.9737 | 0.9480 | - | |
| CF-FCN-post | 0.8090 | 0.9770 | 0.9628 | 0.9801 |
| Dataset | Method | Se | Sp | Acc | AUC |
|---|---|---|---|---|---|
| HRF | Odstrcilik et al. [15] | 0.7741 | 0.9669 | 0.9494 | 0.9667 |
| Budai et al. [36] | 0.7099 | 0.9745 | 0.9481 | - | |
| CF-FCN-post | 0.7762 | 0.9760 | 0.9608 | 0.9701 | |
| CHASE DB1 | Li et al. [19] | 0.7569 | 0.9816 | 0.9527 | 0.9712 |
| Fu et al. [22] | 0.7130 | - | 0.9489 | - | |
| CF-FCN-post | 0.7571 | 0.9823 | 0.9664 | 0.9752 |
| Datasets | DRIVE | STARE | HRF | CHASE DB1 |
|---|---|---|---|---|
| CI | [0.9612, 0.9641] | [0.9620, 0.9679] | [0.9584, 0.9612] | [0.9624, 0.9705] |
| Step | Se | Sp | Acc |
|---|---|---|---|
| FCN | 0.7110 | 0.9750 | 0.9508 |
| FG-FCN | 0.7481 | 0.9822 | 0.9600 |
| CF-FCN | 0.7732 | 0.9876 | 0.9623 |
| CF-FCN-post | 0.7941 | 0.9870 | 0.9634 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Xu, Y.; Chen, M.; Luo, Y. A Coarse-to-Fine Fully Convolutional Neural Network for Fundus Vessel Segmentation. Symmetry 2018, 10, 607. https://doi.org/10.3390/sym10110607
Lu J, Xu Y, Chen M, Luo Y. A Coarse-to-Fine Fully Convolutional Neural Network for Fundus Vessel Segmentation. Symmetry. 2018; 10(11):607. https://doi.org/10.3390/sym10110607
Chicago/Turabian StyleLu, Jianwei, Yixuan Xu, Mingle Chen, and Ye Luo. 2018. "A Coarse-to-Fine Fully Convolutional Neural Network for Fundus Vessel Segmentation" Symmetry 10, no. 11: 607. https://doi.org/10.3390/sym10110607
APA StyleLu, J., Xu, Y., Chen, M., & Luo, Y. (2018). A Coarse-to-Fine Fully Convolutional Neural Network for Fundus Vessel Segmentation. Symmetry, 10(11), 607. https://doi.org/10.3390/sym10110607





