Optimization of an Antibody Light Chain Framework Enhances Expression, Biophysical Properties and Pharmacokinetics
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibody and Antigen Construction, Expression, and Purification
2.2. Biochemical Analytics
2.3. Pharmacokinetics
2.4. Statistics
3. Results
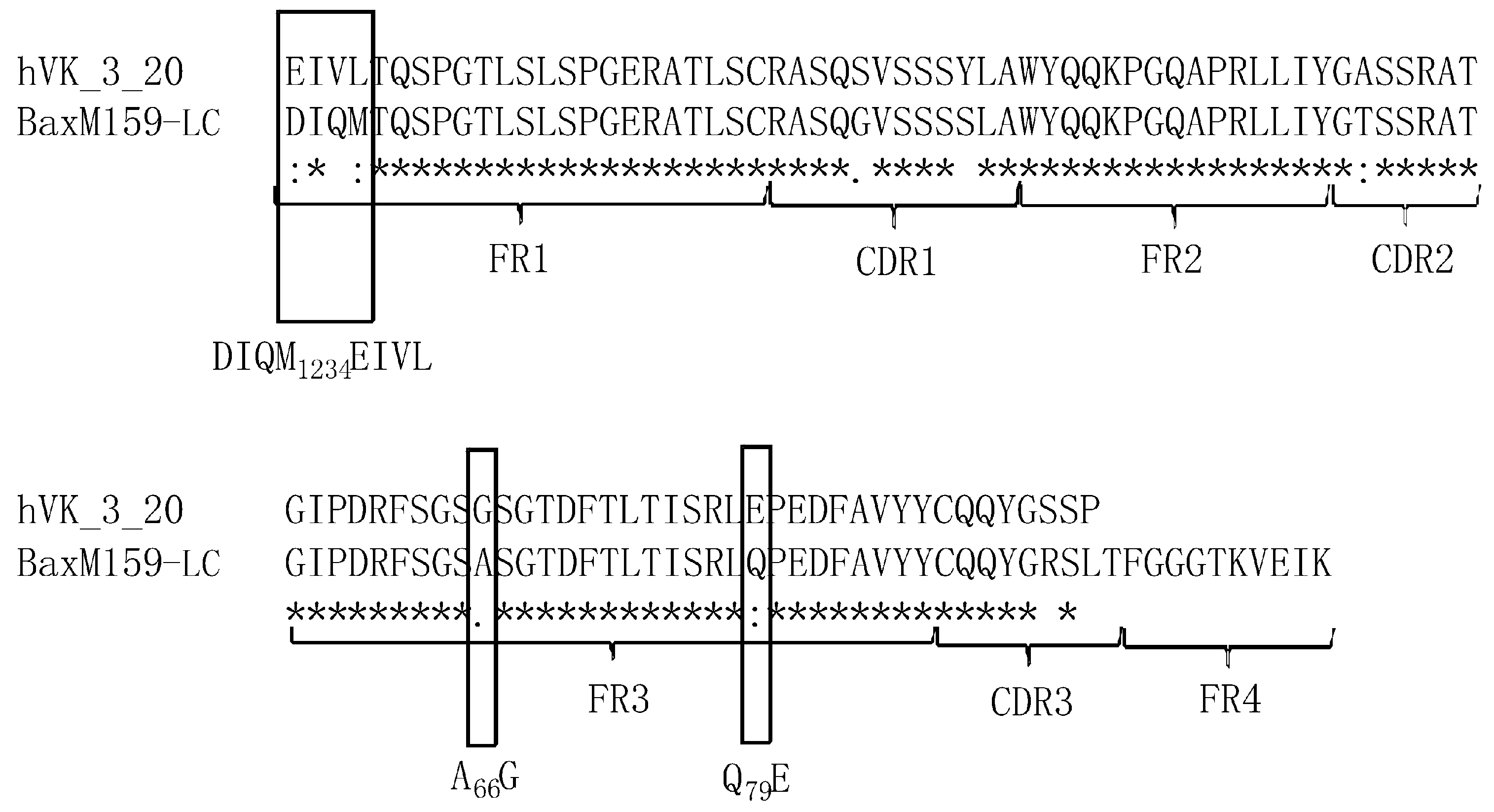
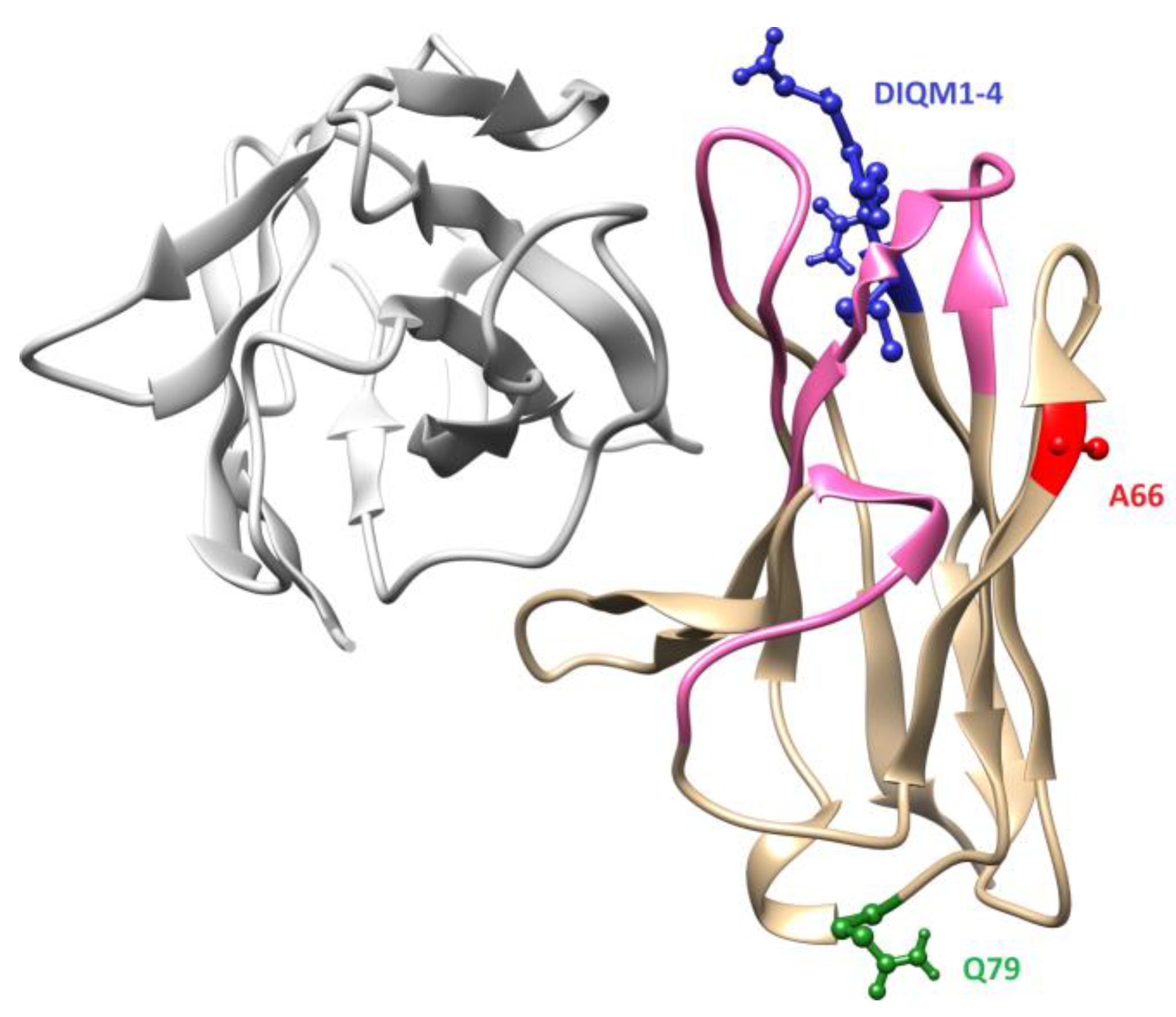
3.1. Design and Generation of Anti-oxMIF Antibody BaxM159 Variants
3.2. Framework Optimized BaxM159 Variants Have Unaltered Epitope Binding Properties
3.3. Framework Optimization of Anti-oxMIF Antibody BaxM159 Increases Expression
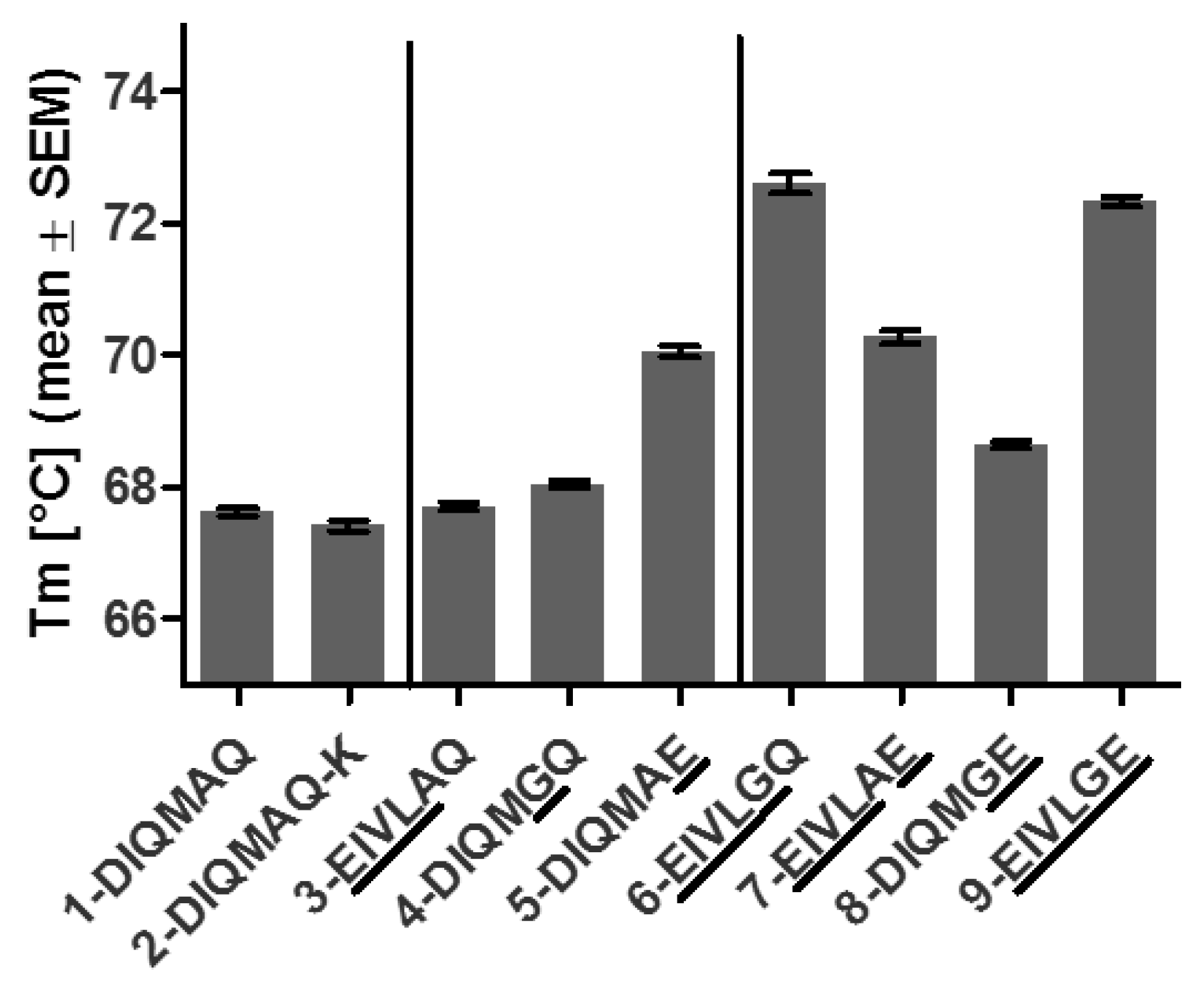
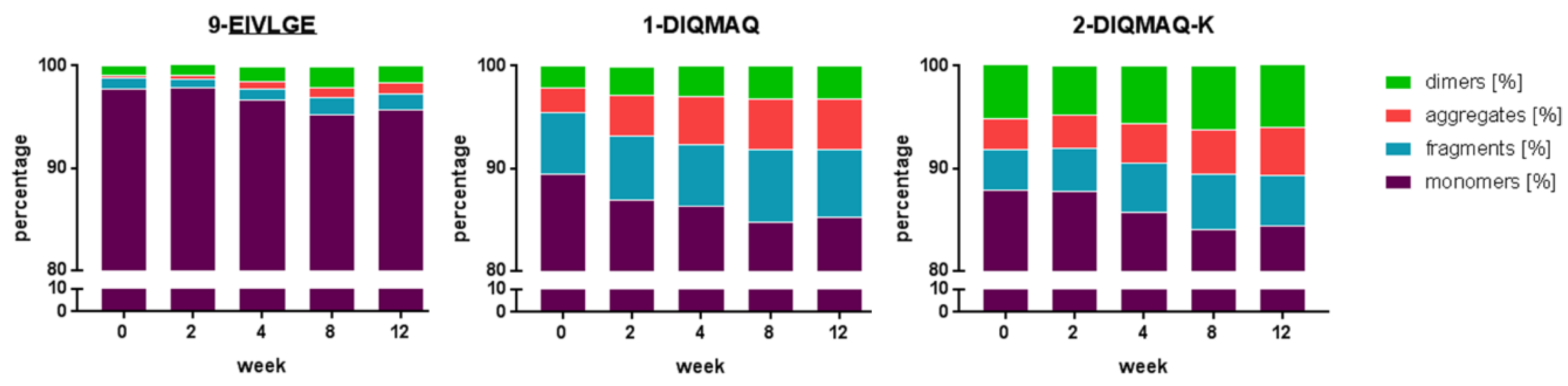
3.4. Framework Optimization of Anti-oxMIF Antibody BaxM159 Increases Stability and Decreases Aggregation Propensity
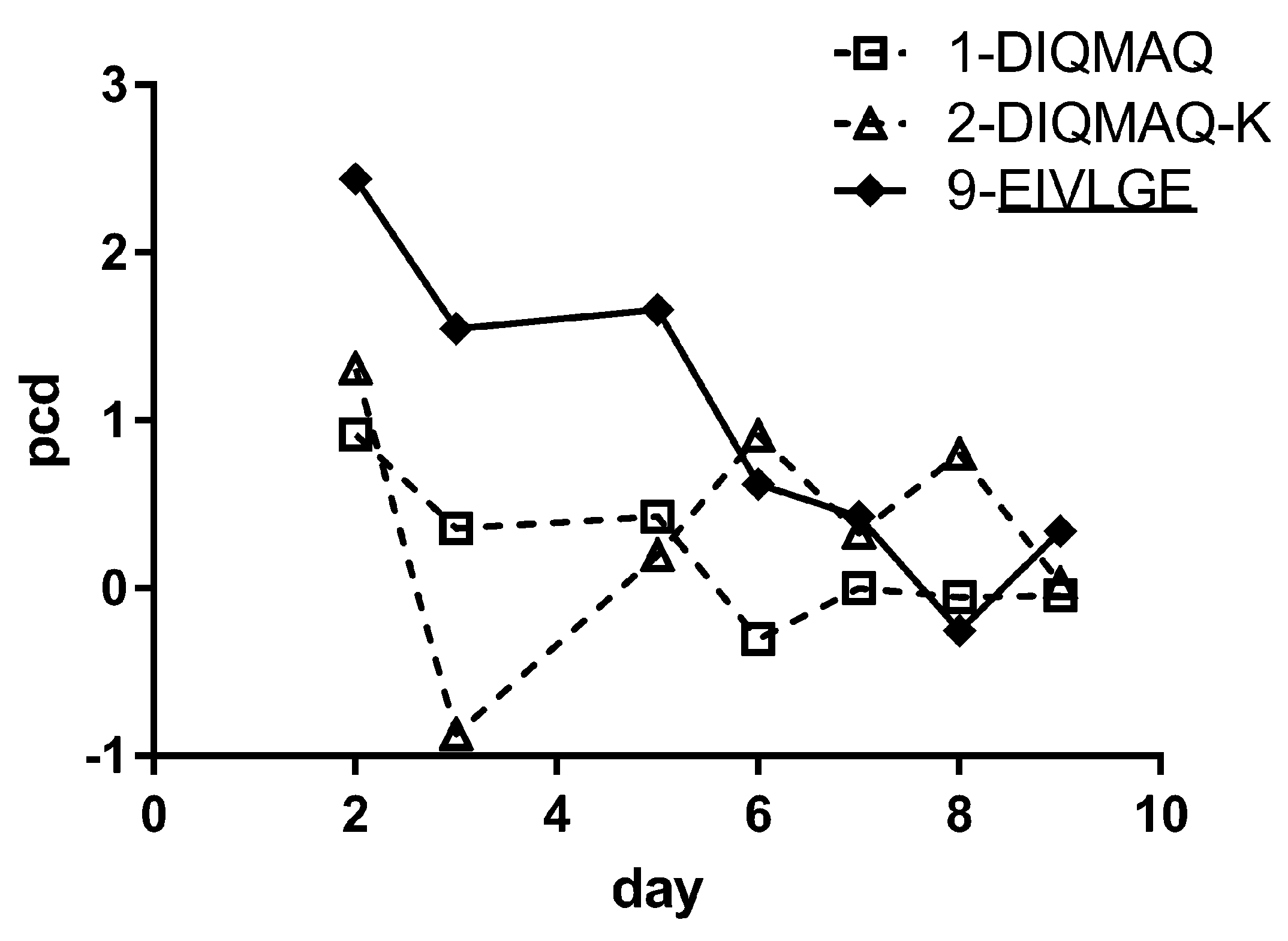
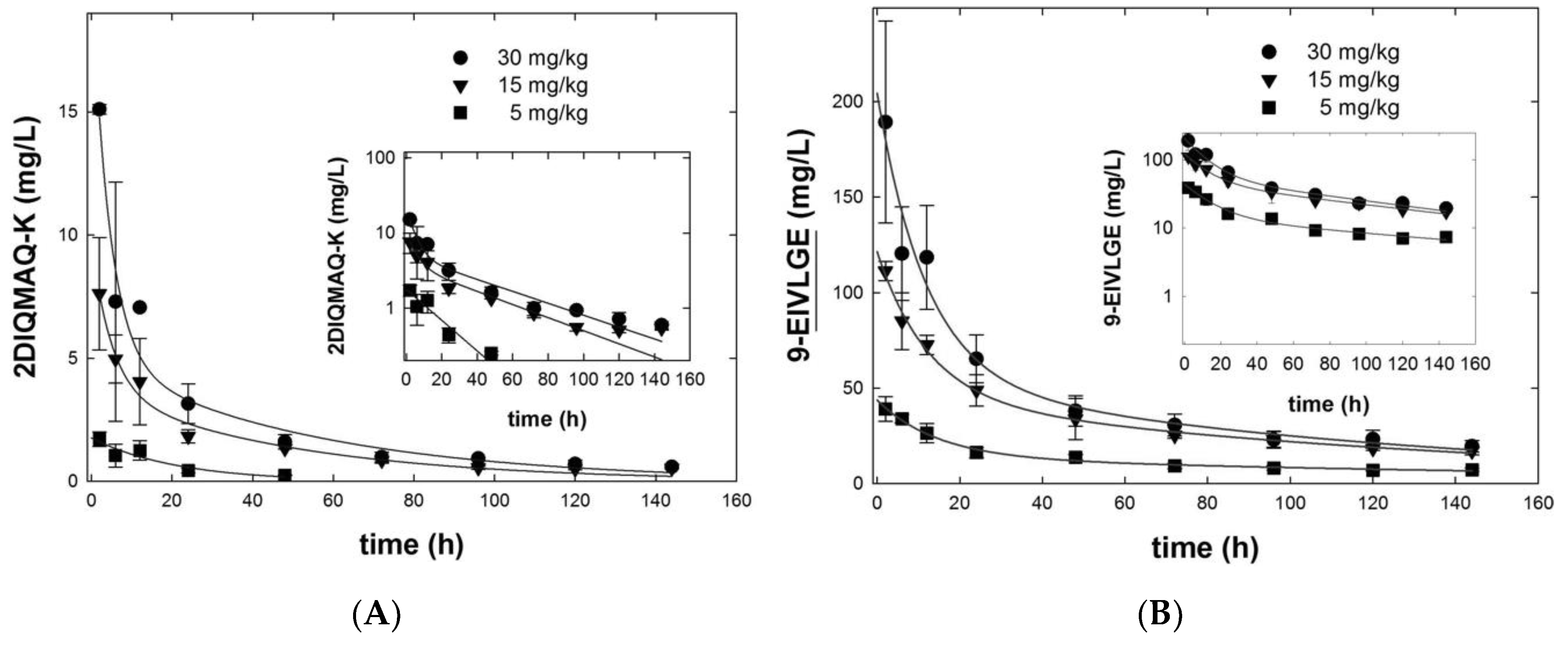
3.5. Framework Optimization of BaxM159 Does not Alter the In Vitro Functionality but Improves Its Pharmacokinetics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ecker, D.M.; Jones, S.D.; Levine, H.L. The therapeutic monoclonal antibody market. mAbs 2015, 7, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Rita Costa, A.; Elisa Rodrigues, M.; Henriques, M.; Azeredo, J.; Oliveira, R. Guidelines to cell engineering for monoclonal antibody production. Eur. J. Pharm. Biopharm. 2010, 74, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.J.; Deng, S.J.; Huang, D.G.; He, Y.; Lei, M.; Zhou, L.; Jin, P. Frontier of therapeutic antibody discovery: The challenges and how to face them. World J. Biol. Chem. 2012, 3, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Buttel, I.C.; Chamberlain, P.; Chowers, Y.; Ehmann, F.; Greinacher, A.; Jefferis, R.; Kramer, D.; Kropshofer, H.; Lloyd, P.; Lubiniecki, A.; et al. Taking immunogenicity assessment of therapeutic proteins to the next level. Biologicals 2011, 39, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Moussa, E.M.; Panchal, J.P.; Moorthy, B.S.; Blum, J.S.; Joubert, M.K.; Narhi, L.O.; Topp, E.M. Immunogenicity of Therapeutic Protein Aggregates. J. Pharm. Sci. 2016, 105, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Perchiacca, J.M.; Tessier, P.M. Engineering aggregation-resistant antibodies. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 263–286. [Google Scholar] [CrossRef] [PubMed]
- Ratanji, K.D.; Derrick, J.P.; Dearman, R.J.; Kimber, I. Immunogenicity of therapeutic proteins: Influence of aggregation. J. Immunotoxicol. 2014, 11, 99–109. [Google Scholar] [CrossRef]
- Igawa, T.; Tsunoda, H.; Kuramochi, T.; Sampei, Z.; Ishii, S.; Hattori, K. Engineering the variable region of therapeutic IgG antibodies. mAbs 2011, 3, 243–252. [Google Scholar] [CrossRef]
- Van der Kant, R.; Karow-Zwick, A.R.; Van Durme, J.; Blech, M.; Gallardo, R.; Seeliger, D.; Assfalg, K.; Baatsen, P.; Compernolle, G.; Gils, A.; et al. Prediction and Reduction of the Aggregation of Monoclonal Antibodies. J. Mol. Biol. 2017, 429, 1244–1261. [Google Scholar] [CrossRef]
- Fukuda, J.; Iwura, T.; Yanagihara, S.; Kano, K. Factors to Govern Soluble and Insoluble Aggregate-formation in Monoclonal Antibodies. Anal. Sci. 2015, 31, 1233–1240. [Google Scholar] [CrossRef]
- Buchanan, A.; Clementel, V.; Woods, R.; Harn, N.; Bowen, M.A.; Mo, W.; Popovic, B.; Bishop, S.M.; Dall’Acqua, W.; Minter, R.; et al. Engineering a therapeutic IgG molecule to address cysteinylation, aggregation and enhance thermal stability and expression. mAbs 2013, 5, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Ewert, S.; Honegger, A.; Pluckthun, A. Structure-based improvement of the biophysical properties of immunoglobulin VH domains with a generalizable approach. Biochemistry 2003, 42, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
- Tiller, K.E.; Tessier, P.M. Advances in Antibody Design. Annu. Rev. Biomed. Eng. 2015, 17, 191–216. [Google Scholar] [CrossRef] [PubMed]
- Monsellier, E.; Bedouelle, H. Improving the stability of an antibody variable fragment by a combination of knowledge-based approaches: Validation and mechanisms. J. Mol. Biol. 2006, 362, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Potapov, V.; Cohen, M.; Schreiber, G. Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng. Des. Sel. 2009, 22, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.; Sweeney, B.; Cain, K.; Stephens, P.; Sharfstein, S.T. Identifying bottlenecks in transient and stable production of recombinant monoclonal-antibody sequence variants in Chinese hamster ovary cells. Biotechnol. Prog. 2012, 28, 846–855. [Google Scholar] [CrossRef]
- Miller, B.R.; Demarest, S.J.; Lugovskoy, A.; Huang, F.; Wu, X.; Snyder, W.B.; Croner, L.J.; Wang, N.; Amatucci, A.; Michaelson, J.S.; et al. Stability engineering of scFvs for the development of bispecific and multivalent antibodies. Protein Eng. Des. Sel. 2010, 23, 549–557. [Google Scholar] [CrossRef]
- Steipe, B.; Schiller, B.; Pluckthun, A.; Steinbacher, S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 1994, 240, 188–192. [Google Scholar] [CrossRef]
- Thiele, M.; Kerschbaumer, R.J.; Tam, F.W.; Volkel, D.; Douillard, P.; Schinagl, A.; Kuhnel, H.; Smith, J.; McDaid, J.P.; Bhangal, G.; et al. Selective Targeting of a Disease-Related Conformational Isoform of Macrophage Migration Inhibitory Factor Ameliorates Inflammatory Conditions. J. Immunol. 2015, 195, 2343–2352. [Google Scholar] [CrossRef]
- Schinagl, A.; Kerschbaumer, R.J.; Sabarth, N.; Douillard, P.; Scholz, P.; Voelkel, D.; Hollerweger, J.C.; Goettig, P.; Brandstetter, H.; Scheiflinger, F.; et al. Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch. Biochemistry 2018, 57, 1523–1532. [Google Scholar] [CrossRef]
- Schinagl, A.; Thiele, M.; Douillard, P.; Volkel, D.; Kenner, L.; Kazemi, Z.; Freissmuth, M.; Scheiflinger, F.; Kerschbaumer, R.J. Oxidized macrophage migration inhibitory factor is a potential new tissue marker and drug target in cancer. Oncotarget 2016, 7, 73486–73496. [Google Scholar] [CrossRef] [PubMed]
- Kerschbaumer, R.J.; Rieger, M.; Volkel, D.; Le Roy, D.; Roger, T.; Garbaraviciene, J.; Boehncke, W.H.; Mullberg, J.; Hoet, R.M.; Wood, C.R.; et al. Neutralization of macrophage migration inhibitory factor (MIF) by fully human antibodies correlates with their specificity for the beta-sheet structure of MIF. J. Biol. Chem. 2012, 287, 7446–7455. [Google Scholar] [CrossRef] [PubMed]
- Hussain, F.; Freissmuth, M.; Volkel, D.; Thiele, M.; Douillard, P.; Antoine, G.; Thurner, P.; Ehrlich, H.; Schwarz, H.P.; Scheiflinger, F.; et al. Human anti-macrophage migration inhibitory factor antibodies inhibit growth of human prostate cancer cells in vitro and in vivo. Mol. Cancer Ther. 2013, 12, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Kabat, E.A.; Wu, T.T. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J. Immunol. 1991, 147, 1709–1719. [Google Scholar] [PubMed]
- Hollriegl, W.; Bauer, A.; Baumgartner, B.; Dietrich, B.; Douillard, P.; Kerschbaumer, R.J.; Hobarth, G.; McKee, J.S.; Schinagl, A.; Tam, F.W.K.; et al. Pharmacokinetics, disease-modifying activity, and safety of an experimental therapeutic targeting an immunological isoform of macrophage migration inhibitory factor, in rat glomerulonephritis. Eur. J. Pharmacol. 2018, 820, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Ikeda, K.; Amada, K.; Liang, S.; Tsuchiya, Y.; Nakamura, H.; Shirai, H.; Standley, D.M. Kotai Antibody Builder: Automated high-resolution structural modeling of antibodies. Bioinformatics 2014, 30, 3279–3280. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Honegger, A.; Pluckthun, A. Yet another numbering scheme for immunoglobulin variable domains: An automatic modeling and analysis tool. J. Mol. Biol. 2001, 309, 657–670. [Google Scholar] [CrossRef]
- Swindells, M.B.; Porter, C.T.; Couch, M.; Hurst, J.; Abhinandan, K.R.; Nielsen, J.H.; Macindoe, G.; Hetherington, J.; Martin, A.C. abYsis: Integrated Antibody Sequence and Structure-Management, Analysis, and Prediction. J. Mol. Biol. 2017, 429, 356–364. [Google Scholar] [CrossRef]
- Harris, R.J. Processing of C-terminal lysine and arginine residues of proteins isolated from mammalian cell culture. J. Chromatogr. A 1995, 705, 129–134. [Google Scholar] [CrossRef]
- Foote, J.; Winter, G. Antibody framework residues affecting the conformation of the hypervariable loops. J. Mol. Biol. 1992, 224, 487–499. [Google Scholar] [CrossRef]
- Ewert, S.; Huber, T.; Honegger, A.; Pluckthun, A. Biophysical properties of human antibody variable domains. J. Mol. Biol. 2003, 325, 531–553. [Google Scholar] [CrossRef]
- Honegger, A. Engineering antibodies for stability and efficient folding. Handb. Exp. Pharmacol. 2008, 181, 47–68. [Google Scholar] [CrossRef]
- Jain, T.; Sun, T.; Durand, S.; Hall, A.; Houston, N.R.; Nett, J.H.; Sharkey, B.; Bobrowicz, B.; Caffry, I.; Yu, Y.; et al. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. USA 2017, 114, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; Schulz, P.; Litzenburger, T.; Spitz, J.; Hoerer, S.; Blech, M.; Enenkel, B.; Studts, J.M.; Garidel, P.; Karow, A.R. Boosting antibody developability through rational sequence optimization. mAbs 2015, 7, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Dudgeon, K.; Rouet, R.; Kokmeijer, I.; Schofield, P.; Stolp, J.; Langley, D.; Stock, D.; Christ, D. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 10879–10884. [Google Scholar] [CrossRef] [PubMed]
- Perchiacca, J.M.; Bhattacharya, M.; Tessier, P.M. Mutational analysis of domain antibodies reveals aggregation hotspots within and near the complementarity determining regions. Proteins 2011, 79, 2637–2647. [Google Scholar] [CrossRef] [PubMed]
- Jespers, L.; Schon, O.; Famm, K.; Winter, G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat. Biotechnol. 2004, 22, 1161–1165. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Stefani, M.; Taddei, N.; Ramponi, G.; Dobson, C.M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 2003, 424, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, F.; Tsao, C.; Liu, S.; Jain, P.; Sinclair, A.; Hung, H.C.; Bai, T.; Wu, K.; Jiang, S. Zwitterionic gel encapsulation promotes protein stability, enhances pharmacokinetics, and reduces immunogenicity. Proc. Natl. Acad. Sci. USA 2015, 112, 12046–12051. [Google Scholar] [CrossRef]
- Constantinou, A.; Chen, C.; Deonarain, M.P. Modulating the pharmacokinetics of therapeutic antibodies. Biotechnol. Lett. 2010, 32, 609–622. [Google Scholar] [CrossRef]
- Dobson, C.L.; Devine, P.W.; Phillips, J.J.; Higazi, D.R.; Lloyd, C.; Popovic, B.; Arnold, J.; Buchanan, A.; Lewis, A.; Goodman, J.; et al. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci. Rep. 2016, 6, 38644. [Google Scholar] [CrossRef] [PubMed]
- Haidar, J.N.; Yuan, Q.A.; Zeng, L.; Snavely, M.; Luna, X.; Zhang, H.; Zhu, W.; Ludwig, D.L.; Zhu, Z. A universal combinatorial design of antibody framework to graft distinct CDR sequences: A bioinformatics approach. Proteins 2012, 80, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.V.; Pluckthun, A. Engineering aggregation resistance in IgG by two independent mechanisms: Lessons from comparison of Pichia pastoris and mammalian cell expression. J. Mol. Biol. 2012, 417, 309–335. [Google Scholar] [CrossRef]
- Jordan, J.L.; Arndt, J.W.; Hanf, K.; Li, G.; Hall, J.; Demarest, S.; Huang, F.; Wu, X.; Miller, B.; Glaser, S.; et al. Structural understanding of stabilization patterns in engineered bispecific Ig-like antibody molecules. Proteins 2009, 77, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Honegger, A.; Malebranche, A.D.; Rothlisberger, D.; Pluckthun, A. The influence of the framework core residues on the biophysical properties of immunoglobulin heavy chain variable domains. Protein Eng. Des. Sel. 2009, 22, 121–134. [Google Scholar] [CrossRef]
- Chen, W.; Li, W.; Ying, T.; Wang, Y.; Feng, Y.; Dimitrov, D.S. Germlining of the HIV-1 broadly neutralizing antibody domain m36. Antiviral Res. 2015, 116, 62–66. [Google Scholar] [CrossRef] [PubMed]
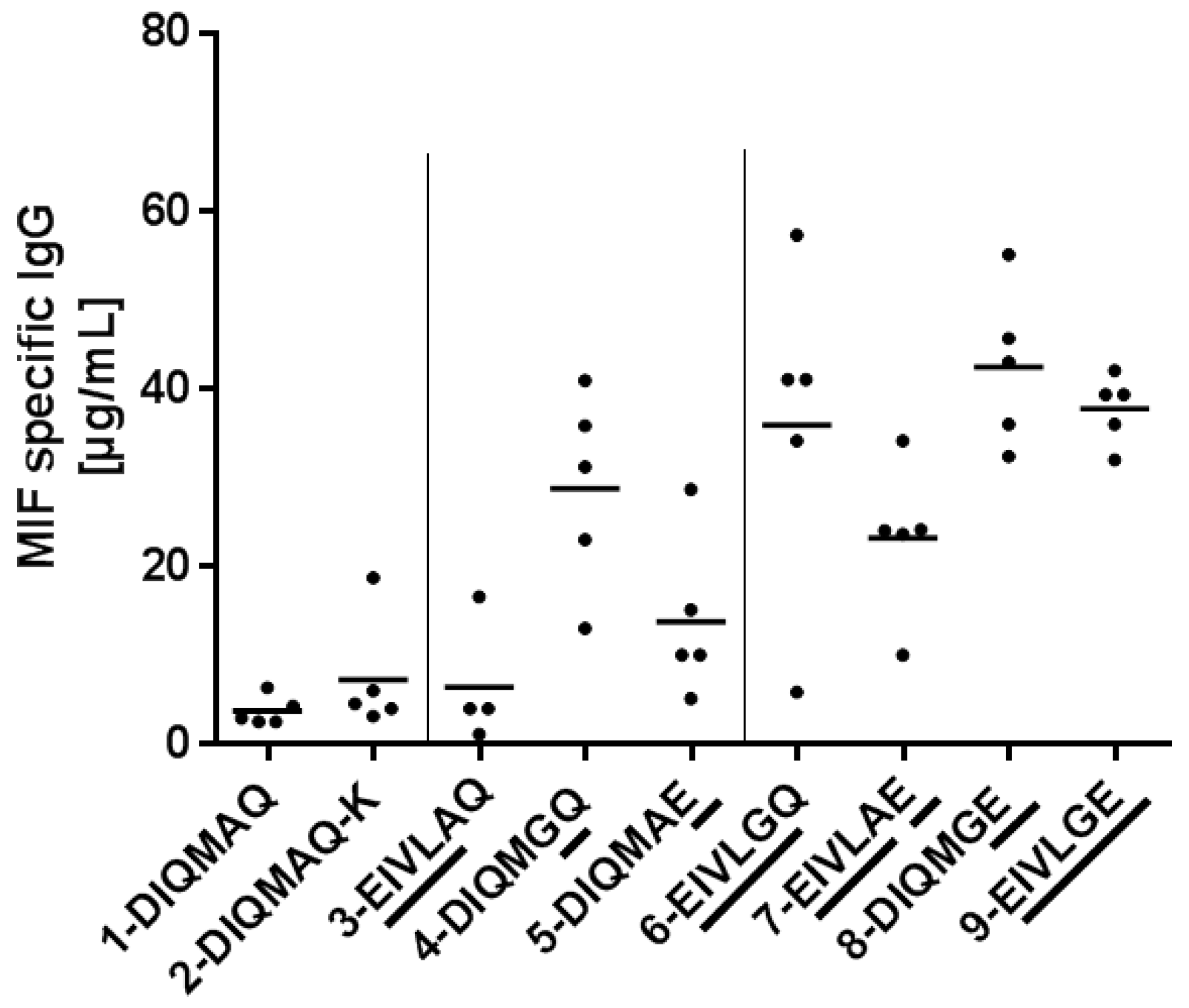

| ID | ID and Name of BaxM159 Variant | Amino Acid at Positions 1–4 | Amino Acid at Position 66 | Amino Acid at Position 79 |
|---|---|---|---|---|
| 1 | 1-DIQMAQ | DIQM | A | Q |
| 2 | 2-DIQMAQ-K | DIQM | A | Q |
| 3 | 3-EIVLAQ | EIVL | A | Q |
| 4 | 4-DIQMGQ | DIQM | G | Q |
| 5 | 5-DIQMAE | DIQM | A | E |
| 6 | 6-EIVLGQ | EIVL | G | Q |
| 7 | 7-EIVLAE | EIVL | A | E |
| 8 | 8-DIQMGE | DIQM | G | E |
| 9 | 9-EIVLGE | EIVL | G | E |
| ID | ID and Name of BaxM159 Variant | ka (M−1s−1) | kd (s−1) | KD (M) |
|---|---|---|---|---|
| 1 | 1-DIQMAQ | 1.1 × 105 | 2.9 × 10−4 | 2.6 × 10−9 |
| 2 | 2-DIQMAQ-K | 1.3 × 105 | 3.8 × 10−4 | 3.0 × 10−9 |
| 3 | 3-EIVLAQ | 2.0 × 105 | 3.1 × 10−4 | 1.5 × 10−9 |
| 4 | 4-DIQMGQ | 1.6 × 105 ± 8.7 × 104 | 5.6 × 10−4 ± 5.0 × 10−4 | 4.3 × 10−9 ± 2.6 × 10−9 |
| 5 | 5-DIQMAE | 1.6 × 105 | 4.0 × 10−4 | 2.5 × 10−9 |
| 6 | 6-EIVLGQ | 2.6 × 105 ± 1.2 × 105 | 6.4 × 10−4 ± 3.5 × 10−5 | 2.7 × 10−9 ± 1.1 × 10−9 |
| 7 | 7-EIVLAE | 1.8 × 105 ± 6.3 × 103 | 4.4 × 10−4 ± 1.5 × 10−4 | 2.4 × 10−9 ± 7.6 × 10−10 |
| 8 | 8-DIQMGE | 1.3 × 105 | 5.4 × 10−4 | 4.2 × 10−9 |
| 9 | 9-EIVLGE | 3.1 × 105 ± 6.5 × 104 | 4.4 × 10−4 ± 2.2 × 10−4 | 1.4 × 10−9 ± 4.1 × 10−10 |
| Antibody | 2-DIQMAQ-K | 9-EIVLGE | ||||
|---|---|---|---|---|---|---|
| Dose (mg/kg) | 5 | 15 | 30 | 5 | 15 | 30 |
| C0,α (mg/L) | n.d. | 6.0 ± 1.1 | 15.6 ± 3.1 | 29.7 ± 3.42 | 76.4 ± 7.3 | 149.1 ± 33.4 |
| C0,β (mg/L) | n.d. | 3.7 ± 1.1 | 5.0 ± 2.5 | 14.0 ± 3.68 | 45.1 ± 7.9 | 55.2 ± 35.9 |
| t½, β (h) | n.d. | 34.5 | 37.9 | 135.9 | 96.3 | 87.7 |
| AUC(0–144h) iv (mg/L·h) | n.d. | 209.6 | 326.8 | 1873.6 | 5036.7 | 6611.3 |
| Clearance CL (mL/h/kg) | n.d. | 72 ± 13 | 98 ± 16 | 3.5 ± 1.4 | 3.2 ± 1.4 | 3.6 ± 0.8 |
| VD,ss (mL/kg) | n.d. | 3067 ± 710 | 3757 ± 957 | 170 ± 40 | 160 ± 23 | 190 ± 33 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Douillard, P.; Freissmuth, M.; Antoine, G.; Thiele, M.; Fleischanderl, D.; Matthiessen, P.; Voelkel, D.; Kerschbaumer, R.J.; Scheiflinger, F.; Sabarth, N. Optimization of an Antibody Light Chain Framework Enhances Expression, Biophysical Properties and Pharmacokinetics. Antibodies 2019, 8, 46. https://doi.org/10.3390/antib8030046
Douillard P, Freissmuth M, Antoine G, Thiele M, Fleischanderl D, Matthiessen P, Voelkel D, Kerschbaumer RJ, Scheiflinger F, Sabarth N. Optimization of an Antibody Light Chain Framework Enhances Expression, Biophysical Properties and Pharmacokinetics. Antibodies. 2019; 8(3):46. https://doi.org/10.3390/antib8030046
Chicago/Turabian StyleDouillard, Patrice, Michael Freissmuth, Gerhard Antoine, Michael Thiele, Daniel Fleischanderl, Peter Matthiessen, Dirk Voelkel, Randolf J. Kerschbaumer, Friedrich Scheiflinger, and Nicolas Sabarth. 2019. "Optimization of an Antibody Light Chain Framework Enhances Expression, Biophysical Properties and Pharmacokinetics" Antibodies 8, no. 3: 46. https://doi.org/10.3390/antib8030046
APA StyleDouillard, P., Freissmuth, M., Antoine, G., Thiele, M., Fleischanderl, D., Matthiessen, P., Voelkel, D., Kerschbaumer, R. J., Scheiflinger, F., & Sabarth, N. (2019). Optimization of an Antibody Light Chain Framework Enhances Expression, Biophysical Properties and Pharmacokinetics. Antibodies, 8(3), 46. https://doi.org/10.3390/antib8030046





