Abstract
Human cytomegalovirus (HCMV) is an important pathogen that infects the majority of the population worldwide, yet, currently, there is no licensed vaccine. Despite HCMV encoding at least seven Natural Killer (NK) cell evasion genes, NK cells remain critical for the control of infection in vivo. Classically Antibody-Dependent Cellular Cytotoxicity (ADCC) is mediated by CD16, which is found on the surface of the NK cell in a complex with FcεRI-γ chains and/or CD3ζ chains. Ninety percent of NK cells express the Fc receptor CD16; thus, they have the potential to initiate ADCC. HCMV has a profound effect on the NK cell repertoire, such that up to 10-fold expansions of NKG2C+ cells can be seen in HCMV seropositive individuals. These NKG2C+ cells are reported to be FcεRI-γ deficient and possess variable levels of CD16+, yet have striking ADCC functions. A subset of HCMV cell surface proteins will induce robust antibody responses that could render cells susceptible to ADCC. We will consider how the strong anti-HCMV function of NKG2C+ FcεRI-γ-deficient NK cells could potentially be harnessed in the clinic to treat patients suffering from HCMV disease and in the development of an efficacious HCMV vaccine.
Keywords:
HCMV; Cytomegalovirus; ADCC; antibody-dependent cellular cytotoxicity; NK cells; NKG2C; UL40; HLA-E; vaccine 1. Introduction
Human cytomegalovirus (HCMV), the prototype member of the β-herpesvirus family, establishes lifelong infections in immunocompetent individuals, which only rarely results in overt disease. However, in certain populations, HCMV is a major cause of clinical problems. Allograft and AIDS patients with an active HCMV infection often suffer from end organ disease, such as pneumonitis, hepatitis, gastrointestinal ulceration and retinitis. Targeted use of antivirals can significantly reduce disease burden; however, drugs can be toxic and resistance can occur, reducing their effectiveness [1,2,3]. The transmission of HCMV from mother to foetus during gestation occurs in 0.5%–2% of births and is the leading infectious cause of congenital birth defects worldwide, including sensorineural hearing loss (SHNL) and neurodevelopmental delay [4]. As a result of the lifelong burden of congenital HCMV infection, the U.S. institute of medicine has designated a HCMV vaccine as highest priority [5].
CD8+ T-cells are implicated in the control of HCMV in vivo. Healthy HCMV seropositive individuals have large clonal expansions of CD8+ T-cells, and bone marrow transplantation studies demonstrate a strong correlation between the recovery of the CD8+ T-cell population and protection from HCMV disease [6,7,8,9,10]. Furthermore, adoptive transfer studies of HCMV-specific T-cell clones or polyclonal T-cell lines into T-cell-suppressed transplant patients is associated with decreased HCMV viraemia and disease [11,12,13,14]. Natural Killer (NK) cells are also important for the control of HCMV in vivo, and individuals with impaired NK cell-mediated immunity are particularly susceptible to HCMV disease [15,16,17,18,19,20,21]. NK cells express a plethora of activating and inhibitory receptors, and it is the balance of signals received by these receptors that determines the NK cell response to a pathogen or a transformed cell [22]. Members of the Killer Immunoglobulin Receptor (KIR) family may be stimulatory or inhibitory and, along with the inhibitory receptor LIR-1, recognise the classical major histocompatibility complex (MHC) class Ia molecules, Human Leukocyte Antigen (HLA)-A, -B and -C on the surface of cells. Downregulation of MHC class I by HCMV should render infected cells more susceptible to lysis by NK cells. However, HCMV encodes a number of NK immunomodulatory functions that protect infected cells from NK cell attack (reviewed in Table 1). It has also been known for some time that HCMV has profound effects on the NK cell repertoire: expansions of NK cells expressing the activating lectin-like receptor, CD94-NKG2C, are readily detectable in a large proportion of HCMV seropositive individuals [23]. However, the role of these cells in controlling infection remains poorly understood. This review will focus on new and exciting findings suggesting that NKG2C+ NK cells are highly functional and recognise HCMV infected target cells through Antibody-Dependent Cellular Cytotoxicity (ADCC).
2. NK Cell Control of HCMV
Patients suffering from reduced NK cell numbers as a result of Absolute NK Cell Deficiency (ANKD) or because of unknown aetiology are reported to suffer from recurrent HCMV infections and potentially life-threatening HCMV disease [15,19,20,21]. Similarly, individuals who have normal numbers of NK cells, but whose NK cells are deficient in their cytotoxic capacity, such as patients suffering from hypohidrotic ectodermal dysplasia with immunodeficiency (HED-ID), are also more susceptible to HCMV disease [17,24]. Alterations in the NK cell phenotype may also impact on NK cell function in response to HCMV: a patient who had normal numbers of NK cells, but every NK cell expressed the inhibitory Killer Immunoglobulin-like Receptor (KIR), 2DL1, suffered from recurrent HCMV infections, suggesting that the prevalent expression of this inhibitory receptor resulted in a reduced functional response to HCMV [25]. The importance of NK cells is further demonstrated in a severe combined immunodeficiency (SCID) patient, who was T-cell-deficient, but whose NK cells were able to effectively control HCMV infection [26].

Table 1.
Natural Killer (NK) immunomodulatory functions encoded by Human cytomegalovirus (HCMV). MHC, major histocompatibility complex; MICB, major histocompatibility complex (MHC) class I chain related protein B; ULBP, UL16 binding protein; PVR, poliiovirus receptor; TRAILR, tumour necrosis factor-related apoptosis inducing ligand receptor.
| HCMV Gene/locus | Comment |
|---|---|
| UL18 | MHC class I homologue. Binds to inhibitory receptor LIR-1, as well as an unknown NK activating receptor [27,28] |
| SP-UL40 | Upregulates HLA-E, the ligand for the inhibitory receptor, CD94-NKG2A [29,30,31] |
| UL83 | Directly binds to the activating receptor, NKp30 [32] |
| UL16 | Retains the NKG2D ligands, MICB, ULBP1, 2, 4, 5 (immature form) and 6, intracellularly [33,34,35,36,37,38,39] |
| miR-UL112 | Suppresses gene expression of the NKG2D ligand, MICB [40,41] |
| UL142 | Retains the NKG2D ligands, MICA and ULBP3, intracellularly [42,43,44,45] |
| UL141 | Downregulates surface expression of PVR (CD155) and Nectin-2 (CD112), ligands for DNAM-1 (CD226), TACTILE (CD96) and TIGIT [46,47] |
| Retains TRAILR2, the ligand for TRAIL, intracellularly [48,49] |
3. Mechanisms of Action of NK Cells
In humans, NK cells are classically divided into two subsets, immature CD56bright and mature CD56dim NK cells. Ninety percent of circulating NK cells are CD56 dim; they secrete high levels of cytokines and cytotoxic granules when stimulated through their activating receptors and are capable of ADCC. In contrast, the CD56 bright population, which make up the remaining 10% of circulating NK cells, are not as cytotoxic, but produce high levels of interferon-γ (IFNγ) and interleukin-12 (IL-12) in response to stimulation [50]. NK cells express activating and inhibitory receptors on their surface, and it is the balance of these signals, brought about by interaction with ligands, that determines the NK cell’s response; if the NK cell receives an overall inhibitory signal, it will leave a potential target cell intact; however, if the overall signal is activatory, it will set in motion a cascade of events that lead to the death of the target cell [50]. NK cells have a number of ways in which they can kill targets: (1) the perforin/granzyme pathway brought about by ligation of activating NK receptors such as NKG2D; (2) ligation of death receptors which leads to activation of the death inducing signal complex (DISC) complex and caspases in the target cell; and (3) perforin/granzyme release brought about by CD16 ligation by antibody bound to cell surface antigens, causing ADCC.
4. HCMV UL40 and HLA-E
HCMV encodes several proteins that reduce surface expression levels of MHC class Ia proteins and interfere with transporter associated with antigen processing (TAP) function, limiting the CD8+ T-cell response [51,52,53,54,55]. The non-classical MHC Ib molecule, HLA-E, is stabilised on the surface of the cell by binding to conserved nonameric signal peptides normally derived from the MHC class Ia in a TAP-dependent manner. HLA-E, therefore, provides an additional mechanism for surveying endogenous MHC class Ia and TAP function [56]. NK cells are thought to be highly sensitive to MHC class Ia downregulation because of the loss of inhibitory signals via KIR. However, during HCMV infection, HLA-E is preserved on the surface of the infected cell by the HCMV UL40 protein leader sequence (SP-UL40), which shows consensus with the canonical HLA-E binding peptide (VMAPRTLIL) and is delivered to HLA-E in a TAP-independent manner [29,31,57].
CD94-NKG2A is a member of the CD94-NKG2 family that consists of an invariant CD94 subunit covalently associated with either inhibitory (NKG2-A or -B) or activating (NKG2-C, -E or -F) molecules [58,59,60]. The inhibitory CD94-NKG2A (herein referred to as NKG2A) and the activating CD94-NKG2C (herein referred to as NKG2C) bind to peptide stabilised HLA-E. The leader sequence of SP-UL40 is polymorphic between strains of HCMV, and this has been shown to impact on the binding affinities of NKG2A and NKG2C to HLA-E [61,62,63,64]. In the context of an HCMV infection, HCMV strain AD169 SP-UL40 has been shown to preferentially elicit protection against NKG2A+ NK cells, which are able to recognise peptide stabilised HLA-E [30]. The functional effect of SP-UL40 stabilised HLA-E on activating NKG2C+ cells is complicated; however, in expression systems, the peptide sequence and surface levels of NKG2C impact the functional responses of these cells [65,66].
5. NKG2C+ NK Cell Expansions in HCMV
Expansions of NKG2C+ NK cells in healthy HCMV seropositive donors were first described in 2004 [23]. Subsequently, a number of papers have described expansions of NKG2C+ cells occurring in both healthy HCMV seropositive individuals and in patients infected with Hantavirus, chikungunya virus (CHIKV), HIV, hepatitis B and hepatitis C and in individuals suffering from B-cell chronic lymphocytic leukaemia (B-CLL), but only in those who had prior exposure to HCMV [67,68,69,70,71,72,73]. Additionally, children with symptomatic congenital HCMV infections have higher proportions of NKG2C+ NK cells than asymptomatic or non-infected children [74]. In the clinical setting, NKG2C+ cells have been shown to expand during the acute phase of HCMV reactivation after haematopoietic stem cell transplantation (HSCT) and umbilical cord blood transplantation (UCBT), and increased percentages of NKG2C+ CD57+ cells are detected in solid organ transplant (SOT) recipients shortly after the detection of HCMV viraemia [75,76,77]. More recently, a number of studies have described associations with HCMV seropositivity and NK cells expressing KIR [70,78,79,80].
All these data indicate that the NK cell repertoire can be profoundly altered by HCMV infection; however, the mechanism responsible for the expansion of NKG2C+ cells in HCMV seropositive donors is currently not known. Indeed, the role and importance of NKG2C+ NK cells in control of HCMV is an area of current debate; whilst complete deletion of the NKG2C gene occurs with approximately 4%–8% homozygosity in Dutch, Japanese and Spanish cohorts, there have been no reports correlating the NKG2Cnull genotype with overt HCMV disease [81,82,83,84,85]. Similarly, no differences in the NKG2C genotype distribution between HCMV+ and HCMV− donors have been observed [82]. However, in allogeneic stem cell transplant recipients, peak numbers of NKG2C+ NK cells correlated with the resolution of HCMV DNAemia, suggesting that NKG2C+ NK cells may be involved in the clearance of HCMV [86].
6. Functional Responses of NKG2C+ NK Cells
NKG2C+ NK cells show enhanced degranulation and IFNγ production in response to HCMV strain TB40/E infected autologous macrophages in the presence of human serum containing HCMV antibodies [87]. However, whilst NKG2C+ NK cells have been shown to expand in vitro in response to HCMV infected fibroblast cells, it is not clear whether this is in response to UL40 stabilised HLA-E, or whether they can lyse HCMV infected cells through ligation of NKG2C with HLA-E [74,88]. In response to HLA-E expressing, but otherwise MHC class I-deficient, targets, a higher percentage of NKG2C+ NK cells degranulate compared to their NKG2C− counterparts, whilst in response to the MHC class I-deficient targets, K562, NKG2C+ NK cells degranulate less well, but produce more IFNγ and tumour necrosis factor-α (TNFα) than NKG2C− NK cells [67,70,75,87]. NKG2C+ cells also show more robust cytokine and degranulation responses to plate-bound anti-CD16 antibody and anti-NKG2C antibody and to B-cells coated with an anti-CD20 antibody, than NKG2C− cells [70,77]. In response to antibody coated B-cells not only did a greater percentage of NKG2C+ cells degranulate, but a higher proportion released IFNγ and TNFα, and overall, a higher proportion of these cells were “tri-functional” compared to their NKG2C− counterparts [70]. In redirected killing assays using P815 cells coated with an anti-CD16 antibody, NKG2Chi NK cells were more responsive than NKG2C− cells despite having similar levels of surface CD16, and crosslinking NKG2C enhanced the responsiveness of these cells [87]. Taken together, these data suggest that NKG2C+ NK cells are cytotoxic, but are triggered into action by a mechanism that is not measured in standard NK assay protocols that measure responses to MHC class I-deficient target cells in the absence of antibody.
Receptor expression levels and ligation of receptors by cellular ligands can alter the threshold required for the activation of NK cells. In this respect, ligation of NKG2A on NKG2C+NKG2A+KIR−cells has been shown to significantly decrease the ADCC responsiveness and proliferative potential of these cells in response to NKG2C crosslinking, suggesting that in the absence of inhibitory KIR, NKG2A could provide a regulatory feedback mechanism for controlling the activating NKG2C receptor, thereby preventing potential autoreactivity against self-HLA-E+ cells [89]. It is therefore interesting that a number of groups have observed that in HCMV seropositive donors with expansions of NKG2C+ NK cells, a higher percentage of these cells are NKG2A−, but express a KIR that specifically recognises self-MHC class-I molecules [87,89,90]. Engagement of these inhibitory KIR in a redirected killing assay using anti-NKG2C antibody-coated P815 cells significantly decreased degranulation, suggesting that a bias towards self-KIR expression may dampen the responsiveness of NKG2C+ self KIR+ cells to normal tissues with intact MHC class I expression [70].
7. Role of CD16 in NK Cell Responses
CD16, also known as FcεRI-γIII, is a multichain receptor consisting of an α-chain that is associated in the membrane with either an FcεRI-γ or CD3ζ homodimer or a heterodimer comprised of FcεRI-γ and CD3ζ and confers ADCC to CD56l° NK cells on which it is predominantly found (Figure 1) [91]. The extracellular domain of the α-chain has two Ig-like subunits that bind to the Fc portion of Immunoglobulin-G (IgG) molecules with medium to low affinity. A bi-allelic functional polymorphism at position 158 in the membrane proximal Ig-like domain of the α-chain results in either a valine (V) or phenylalanine (F) and determines the level of receptor interaction with different IgG subclasses, as well as the efficiency of the IgG-induced effector functions [92]. Crosslinking of CD16 by IgG causes activation of src family protein tyrosine kinases (PTKs) and phosphorylation of tyrosine residues on the FcεRI-γ and CD3ζ chain immunoreceptor tyrosine-based activation motifs (ITAMs), setting in motion a biochemical cascade that results in NK cell activation and the release of cytotoxic granules [93]. Whilst there is little work studying the effect of FcεRI-γ or CD3ζ deletions in humans, FcRγ-deficient mice have impaired ADCC activity [94,95].
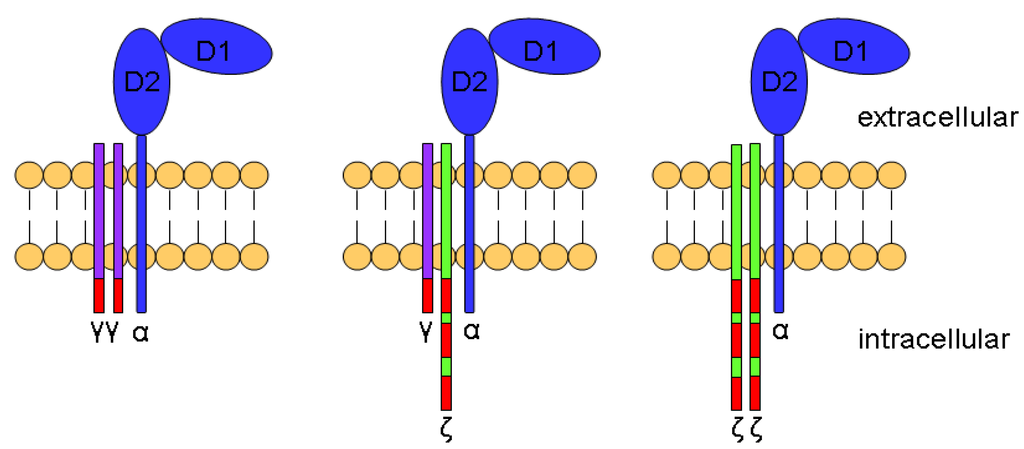
Figure 1.
Structure of CD16. A schematic drawing of CD16 showing the two Ig-like extracellular subunits, D1 and D2, and the α-chain (blue) in complex with FcεRI-γ (purple) or CD3ζ (green) chains. Immunoreceptor tyrosine-based activation motifs (ITAMs) motifs are shown in red.
8. FcεRI-γ Chain-Deficient NK Cells
A novel population of NK cells that express surface CD16, but lack the FcεRI-γ chain, are present in approximately one third of healthy individuals and, despite lacking the FcεRI-γ chain, are capable of ADCC [96]. Phenotypic analysis showed that these cells share similar receptor expression patterns to the NKG2C+ NK cells previously described in HCMV seropositive donors; low levels of NKp30 and NKp46, normal levels of NKp44, CD69, CD25, DNAM1, NKG2D, 2B4 and perforins and granzymes. Analysis of KIR expression on the FcεRI-γ-deficient NK cells suggested that approximately 50% of the individuals analysed had skewed KIR expression patterns. In contrast to NKG2C+ NK cells, which have normal levels of CD16, levels of CD16 on the FcεRI-γ deficient cells was approximately 60% lower than on FcεRI-γ+ NK cells [87,96]. However, despite the reported reduced expression of CD16, the FcεRI-γ-deficient cells were able to produce more IFNγ and TNFα in response to plate-bound anti-CD16 antibody and degranulated better and produced more of these cytokines in response to antibody-coated P815 cells than FcεRI-γ+ cells [96]. Like the NKG2C+ NK cells, FcεRI-γ-deficient cells were not as responsive towards the classic MHC class I-deficient target K562 or 721.221 cells. These data suggested that the functional capacity of FcεRI-γ-deficient NK cells differed markedly from FcεRI-γ+ NK cells and shared many characteristics with the NKG2C+ NK cells previously described in HCMV seropositive individuals. Significantly, it was shown that the presence of FcεRI-γ-deficient NK cells correlated with previous exposure to HCMV [96]. Like NKG2C+ cells, the FcεRI-γ-deficient cells produced more IFNγ and TNFα and were able to degranulate in response to HCMV infected targets, but only in the presence of HCMV-specific antibodies [87,97]. In light of the expansion of FcεRI-γ-deficient cells in HCMV seropositive donors, it is of particular interest that that these cells were also able to respond to target cells infected with herpes simplex virus-1 (HSV-1), but only in the presence of HSV-1-specific antibodies [97]. This suggests that whilst HCMV infection can drive the expansion of this cell subset, they are still able to act with innate characteristics and clear viral infections that they may not necessarily have expanded in response to.
Both FcεRI-γ-deficient and NKG2C+ NK cells express low levels of NKp46, an NK phenotype known to be associated with HCMV seropositivity [23,87,96]. NKp46 is an activating receptor found on a high proportion of NK cells and recognises viral haemagglutinin (HA) and highly charged heparin sulphate/heparin epitopes [98,99]. In standard degranulation assays, blocking antibodies have shown that NKp46 is important for the recognition of HCMV strain TB40/E-infected macrophages [100,101]. Perhaps surprisingly, given the lower surface expression levels of NKp46 on NKG2C+ cells, crosslinking of this receptor with a plate-bound monoclonal antibody elicited a better degranulation response than crosslinking NKp46 on NKG2C− cells, which have much higher levels of NKp46 on their surface [87]. However, it is interesting to note that NKp46 and CD16 both use FcεRI-γ and CD3ζ as part of their intracellular signalling machinery, and it is therefore possible that in FcεRI-γ-deficient cells, CD16 and NKp46 have to rely solely on the CD3ζ chain for their intracellular signalling cascade [102,103]. In this scenario, it may be that signalling solely through the CD3ζ chain, which has three ITAMs compared to the single ITAM in the FcεRI-γ chain, may be enhanced.
9. Clinical Importance of NK Cell-Mediated ADCC in Vaccine Design and Therapeutics
Since NKG2C+ cells are capable of ADCC, administration of antibody preparations or vaccines that induce robust antibody responses may offer advantages in the treatment of HCMV disease. The use of HCMV-specific immunoglobulins (Iv-Ig) to treat transplant patients and mothers who have active HCMV infections indicate that Iv-Ig has a clinical benefit [104,105,106,107,108]. However, the results of these studies have been met with a certain amount of controversy [109,110]. Because of the large economic burden incurred by HCMV, there is now a real need for the development of an effective HCMV vaccine, and this will probably involve inducing both the humoral and cellular arms of the immune response to work in concert.
All sera from HCMV seropositive individuals contain antibodies to gB, and up to 70% of the neutralising antibody responses have been reported to be gB-specific [111]. Virion glycoproteins and complexes involved in entry, such as gH/gL/gO or gH/gL/pUL131A/pUL30/pUL128, are also major targets for neutralising antibodies in vivo [112,113,114,115,116,117,118,119,120,121]. However, as naturally-induced immunity to HCMV from pre-existing infections is not enough to provide complete protection from re-infection, it is extremely unlikely that vaccines using attenuated strains of HCMV can provide a sufficient immunogenic stimulus to provide protection from HCMV disease [122,123,124]. Research is therefore focussing on subunit vaccines that provide specific immunogenic viral proteins designed to elicit excellent cellular and humoral immune responses in the absence of HCMV’s immunomodulatory functions.
The recombinant subunit vaccine, gB + M59 adjuvant, has been shown to induce neutralising gB antibody levels similar to that observed in naturally acquired infection and confer protection against congenital HCMV infection in mothers who acquired primary HCMV infection during gestation and in those who had pre-existing immunity to HCMV [125,126,127]. Furthermore, in transplant patients, the vaccine was able to induce significant levels of neutralising gB antibody, and this had an inverse correlation with the duration of viraemia [128]. However, whilst neutralising antibodies do not always lead to reduced viral load, Iv-Ig, which contains both neutralising and non-neutralising antibodies, can reduce disease burden, suggesting that CMV-specific IgG can elicit effective immune responses besides virus neutralization [104,108].
MSL-109 is a naturally occurring human IgG antibody that recognises the surface gH antigen complex and is able to block infection of fibroblast, epithelial and endothelial cell lines by both laboratory and clinical strains of HCMV in vitro [129,130]. However, phase 2/3 clinical trials in AIDS patients demonstrated that the in vivo efficacy of MSL-109 was not sufficient to prevent disease [131,132]. Development of resistance to neutralising antibody therapies is well documented and often occurs after genetic mutations in one or more proteins, rendering the virus insensitive to the antibody neutralisation. The rapid nature and reversibility of the resistance to MSL-109 suggested that this was not the underlying mechanism, and it was shown that a gH-MSL-109 complex was formed that was incorporated into gH/gL complexes, leaving the Fc portion of MSL-109 decorating the virion, thereby allowing the MSL-109 Fc domain to play a role in the infection of target cells [130]. This meant that not only was MSL-109 ineffective as a neutralising antibody, but that it enhanced the virus’s ability to infect fibroblasts. These data, and the often lethal Antibody-Dependent Enhancement (ADE) after re-infection with dengue virus, suggest that there is much to learn regarding antibody therapies for viral infections. Indeed, recent work on HIV and influenza has begun to focus on the interplay between humoral and cellular immunity [133,134]. Taken together, processes, such as the MSL-109 effect and ADE, need to be considered along with cell-mediated responses, such as ADCC, when developing antibody therapies/immunisation protocols for HCMV.
10. Conclusions
Recent advances in the understanding of the function of NKG2C+ NK cells in ADCC opens the path to a new area of HCMV vaccine research. Whilst neutralising antibodies have been studied and their formation has been considered in vaccine design strategies, the protection generated is only partial. The fact that individuals can be superinfected with a number of HCMV strains indicates that neutralising antibodies do not provide complete protection [135]. One potential explanation could be that the spread of HCMV in vivo is primarily by cell-to-cell contact, rather than the release of free virions. By generating a vaccine that is able to harness the ability of the immune system to clear HCMV-infected cells by ADCC, clinicians would be able to target infected cells, as well as to neutralise free virions. A vaccine capable of expanding and/or maintaining a pool of NKG2C+ NK cells, whilst also generating a strong, appropriately targeted, antibody response, could provide a novel and efficacious system of protection against this virus. Excitingly, the fact that these NKG2C+ NK cells are also able to respond to other viral infections suggests that these cell populations may have the potential to treat other diseases, such as cancer and HIV, provided antibodies of the right specificity are present.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Patel, R.; Snydman, D.R.; Rubin, R.H.; Ho, M.; Pescovitz, M.; Martin, M.; Paya, C.V. Cytomegalovirus prophylaxis in solid organ transplant recipients. Transplantation 1996, 61, 1279–1289. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Thomas, S.T.; Griffiths, P.; Pass, R.F. Cytomegaloviruses in Fields Virology; Knipe, D.M., Howley, P., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Baldanti, F.; Lurain, N.; Gerna, G. Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs. Hum. Immunol. 2004, 65, 403–409. [Google Scholar] [CrossRef]
- Dollard, S.C.; Grosse, S.D.; Ross, D.S. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev. Med. Virol. 2007, 17, 355–363. [Google Scholar] [CrossRef]
- Stratton, K.R.; Durch, J.S.; Lawrence, R.S. Vaccines for the 21st Century; National Academies Press: Washington, DC, USA, 2000. [Google Scholar]
- Avetisyan, G.; Aschan, J.; Hagglund, H.; Ringden, O.; Ljungman, P. Evaluation of intervention strategy based on CMV-specific immune responses after allogeneic SCT. Bone Marrow Transplant. 2007, 40, 865–869. [Google Scholar] [CrossRef]
- Barron, M.A.; Gao, D.; Springer, K.L.; Patterson, J.A.; Brunvand, M.W.; McSweeney, P.A.; Zeng, C.; Baron, A.E.; Weinberg, A. Relationship of reconstituted adaptive and innate cytomegalovirus (CMV)-specific immune responses with CMV viremia in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 2009, 49, 1777–1783. [Google Scholar] [CrossRef]
- Cwynarski, K.; Ainsworth, J.; Cobbold, M.; Wagner, S.; Mahendra, P.; Apperley, J.; Goldman, J.; Craddock, C.; Moss, P.A. Direct visualization of cytomegalovirus-specific T-cell reconstitution after allogeneic stem cell transplantation. Blood 2001, 97, 1232–1240. [Google Scholar] [CrossRef]
- Tormo, N.; Solano, C.; Benet, I.; Clari, M.A.; Nieto, J.; de la Camara, R.; Lopez, J.; Lopez-Aldeguer, N.; Hernandez-Boluda, J.C.; Remigia, M.J.; et al. Lack of prompt expansion of cytomegalovirus pp65 and IE-1-specific IFNgamma CD8+ and CD4+ T cells is associated with rising levels of pp65 antigenemia and DNAemia during pre-emptive therapy in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2010, 45, 543–549. [Google Scholar] [CrossRef]
- Sylwester, A.W.; Mitchell, B.L.; Edgar, J.B.; Taormina, C.; Pelte, C.; Ruchti, F.; Sleath, P.R.; Grabstein, K.H.; Hosken, N.A.; Kern, F.; et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005, 202, 673–685. [Google Scholar] [CrossRef]
- Einsele, H.; Roosnek, E.; Rufer, N.; Sinzger, C.; Riegler, S.; Loffler, J.; Grigoleit, U.; Moris, A.; Rammensee, H.G.; Kanz, L.; et al. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood 2002, 99, 3916–3922. [Google Scholar] [CrossRef]
- Peggs, K.S.; Verfuerth, S.; Pizzey, A.; Khan, N.; Guiver, M.; Moss, P.A.; Mackinnon, S. Adoptive cellular therapy for early cytomegalovirus infection after allogeneic stem-cell transplantation with virus-specific T-cell lines. Lancet 2003, 362, 1375–1377. [Google Scholar] [CrossRef]
- Riddell, S.R.; Watanabe, K.S.; Goodrich, J.M.; Li, C.R.; Agha, M.E.; Greenberg, P.D. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 1992, 257, 238–241. [Google Scholar]
- Walter, E.A.; Greenberg, P.D.; Gilbert, M.J.; Finch, R.J.; Watanabe, K.S.; Thomas, E.D.; Riddell, S.R. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 1995, 333, 1038–1044. [Google Scholar] [CrossRef]
- Biron, C.A.; Byron, K.S.; Sullivan, J.L. Severe Herpesvirus Infections in an Adolescent without Natural-Killer Cells. N. Engl. J. Med. 1989, 320, 1731–1735. [Google Scholar] [CrossRef]
- Orange, J.S. Natural killer cell deficiency. J. Allergy Clin. Immunol. 2013, 132, 515–525. [Google Scholar] [CrossRef]
- Orange, J.S.; Brodeur, S.R.; Jain, A.; Bonilla, F.A.; Schneider, L.C.; Kretschmer, R.; Nurko, S.; Koehler, J.R.; Rasmussen, W.L.; Fergusson, B.M.; et al. Deficiency of natural cytotoxicity in patients with IKK gamma/NEMO mutations. Faseb J. 2002, 16, A1242–A1242. [Google Scholar] [CrossRef]
- Cunningham-Rundles, C. Human B cell defects in perspective. Immunol. Res. 2012, 54, 227–232. [Google Scholar] [CrossRef]
- Aspalter, R.M.; Sewell, W.A.C.; Dolman, K.; Farrant, J.; Webster, A.D.B. Deficiency in circulating natural killer (NK) cell subsets in common variable immunodeficiency and X-linked agammaglobulinaemia. Clin. Exp. Immunol. 2000, 121, 506–514. [Google Scholar]
- Witte, T.; Werwitzke, S.; Schmidt, R.E. CMV complications in common variable immunodeficiency. Immunobiology 2000, 202, 194–198. [Google Scholar] [CrossRef]
- Rai, N.; Thakur, N. Congenital CMV With LAD Type 1 and NK Cell Deficiency. J. Pediatr. Hematol. Oncol. 2013, 35, 468–469. [Google Scholar] [CrossRef]
- Long, E.O.; Kim, H.S.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef]
- Guma, M.; Angulo, A.; Vilches, C.; Gomez-Lozano, N.; Malats, N.; Lopez-Botet, M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef]
- Orange, J.S.; Brodeur, S.R.; Jain, A.; Bonilla, F.A.; Schneider, L.C.; Kretschmer, R.; Nurko, S.; Rasmussen, W.L.; Kohler, J.R.; Gellis, S.E.; et al. Deficient natural killer cell cytotoxicity in patients with IKK-gamma/NEMO mutations. J. Clin. Invest. 2002, 109, 1501–1509. [Google Scholar]
- Gazit, R.; Garty, B.Z.; Monselise, Y.; Hoffer, V.; Finkelstein, Y.; Markel, G.; Katz, G.; Hanna, J.; Achdout, H.; Gruda, R.; et al. Expression of KIR2DL1 on the entire NK cell population: a possible novel immunodeficiency syndrome. Blood 2004, 103, 1965–1966. [Google Scholar] [CrossRef]
- Kuijpers, T.W.; Baars, P.A.; Dantin, C.; van den Burg, M.; van Lier, R.A.; Roosnek, E. Human NK cells can control CMV infection in the absence of T cells. Blood 2008, 112, 914–915. [Google Scholar] [CrossRef]
- Prod’homme, V.; Griffin, C.; Aicheler, R.J.; Wang, E.C.; McSharry, B.P.; Rickards, C.R.; Stanton, R.J.; Borysiewicz, L.K.; Lopez-Botet, M.; Wilkinson, G.W.; et al. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1- NK cells. J. Immunol. 2007, 178, 4473–4481. [Google Scholar]
- Cosman, D.; Fanger, N.; Borges, L.; Kubin, M.; Chin, W.; Peterson, L.; Hsu, M.L. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunobiology 1997, 7, 273–282. [Google Scholar]
- Tomasec, P.; Braud, V.M.; Rickards, C.; Powell, M.B.; McSharry, B.P.; Gadola, S.; Cerundolo, V.; Borysiewicz, L.K.; McMichael, A.J.; Wilkinson, G.W. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000, 287, 1031. [Google Scholar] [CrossRef]
- Wang, E.C.; McSharry, B.; Retiere, C.; Tomasec, P.; Williams, S.; Borysiewicz, L.K.; Braud, V.M.; Wilkinson, G.W. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2002, 99, 7570–7575. [Google Scholar] [CrossRef]
- Ulbrecht, M.; Martinozzi, S.; Grzeschik, M.; Hengel, H.; Ellwart, J.W.; Pla, M.; Weiss, E.H. Cutting edge: The human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 2000, 164, 5019–5022. [Google Scholar]
- Arnon, T.I.; Achdout, H.; Levi, O.; Markel, G.; Saleh, N.; Katz, G.; Gazit, R.; Gonen-Gross, T.; Hanna, J.; Nahari, E.; et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 2005, 6, 515–523. [Google Scholar]
- Welte, S.A.; Sinzger, C.; Lutz, S.Z.; Singh-Jasuja, H.; Sampaio, K.L.; Eknigk, U.; Rammensee, H.G.; Steinle, A. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 2003, 33, 194–203. [Google Scholar] [CrossRef]
- Wu, J.; Chalupny, N.J.; Manley, T.J.; Riddell, S.R.; Cosman, D.; Spies, T. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J. Immunol. 2003, 170, 4196–4200. [Google Scholar]
- Dunn, C.; Chalupny, N.J.; Sutherland, C.L.; Dosch, S.; Sivakumar, P.V.; Johnson, D.C.; Cosman, D. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 2003, 197, 1427–1439. [Google Scholar] [CrossRef]
- Rolle, A.; Mousavi-Jazi, M.; Eriksson, M.; Odeberg, J.; Soderberg-Naucler, C.; Cosman, D.; Karre, K.; Cerboni, C. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J. Immunol. 2003, 171, 902–908. [Google Scholar]
- Vales-Gomez, M.; Browne, H.; Reyburn, H.T. Expression of the UL16 glycoprotein of Human Cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol. 2003, 4, 4. [Google Scholar] [CrossRef]
- Kubin, M.; Cassiano, L.; Chalupny, J.; Chin, W.; Cosman, D.; Fanslow, W.; Mullberg, J.; Rousseau, A.M.; Ulrich, D.; Armitage, R. ULBP1, 2, 3: Novel MHC class I-related molecules that bind to human cytomegalovirus glycoprotein UL16, activate NK cells. Eur. J. Immunol. 2001, 31, 1428–1437. [Google Scholar] [CrossRef]
- Spreu, J.; Stehle, T.; Steinle, A. Human cytomegalovirus-encoded UL16 discriminates MIC molecules by their alpha2 domains. J. Immunol. 2006, 177, 3143–3149. [Google Scholar]
- Stern-Ginossar, N.; Elefant, N.; Zimmermann, A.; Wolf, D.G.; Saleh, N.; Biton, M.; Horwitz, E.; Prokocimer, Z.; Prichard, M.; Hahn, G.; et al. Host immune system gene targeting by a viral miRNA. Science 2007, 317, 376–381. [Google Scholar] [CrossRef]
- Nachmani, D.; Lankry, D.; Wolf, D.G.; Mandelboim, O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 2010, 11, 806–813. [Google Scholar]
- Wills, M.R.; Ashiru, O.; Reeves, M.B.; Okecha, G.; Trowsdale, J.; Tomasec, P.; Wilkinson, J.; Sinclair, G.W.; Sissons, J.G. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 2005, 175, 7457–7465. [Google Scholar]
- Chalupny, N.J.; Rein-Weston, A.; Dosch, S.; Cosman, D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biophys. Res. Commun. 2006, 346, 175–181. [Google Scholar] [CrossRef]
- Ashiru, O.; Bennett, N.J.; Boyle, L.H.; Thomas, M.; Trowsdale, J.; Wills, M.R. NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J. Virol. 2009, 83, 12345–12354. [Google Scholar] [CrossRef]
- Bennett, N.J.; Ashiru, O.; Morgan, F.J.; Pang, Y.; Okecha, G.; Eagle, R.A.; Trowsdale, J.; Sissons, J.G.; Wills, M.R. Intracellular sequestration of the NKG2D ligand ULBP3 by human cytomegalovirus. J. Immunol. 2010, 185, 1093–1102. [Google Scholar]
- Tomasec, P.; Wang, E.C.; Davison, A.J.; Vojtesek, B.; Armstrong, M.; Griffin, C.; McSharry, B.P.; Morris, R.J.; Llewellyn-Lacey, S.; Rickards, C.; et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 2005, 6, 181–188. [Google Scholar]
- Prod’homme, V.; Sugrue, D.M.; Stanton, R.J.; Nomoto, A.; Davies, J.; Rickards, C.R.; Cochrane, D.; Moore, M.; Wilkinson, G.W.; Tomasec, P. Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J. Gen. Virol. 2010, 91, 2034–2039. [Google Scholar] [CrossRef]
- Smith, W.; Tomasec, P.; Aicheler, R.; Loewendorf, A.; Nemcovicova, I.; Wang, E.C.; Stanton, R.J.; Macauley, M.; Norris, P.; Willen, L.; et al. Human cytomegalovirus glycoprotein UL141 targets the TRAIL death receptors to thwart host innate antiviral defenses. Cell Host Microbe 2013, 13, 324–335. [Google Scholar] [CrossRef]
- Nemcovicova, I.; Benedict, C.A.; Zajonc, D.M. Structure of human cytomegalovirus UL141 binding to TRAIL-R2 reveals novel, non-canonical death receptor interactions. PLoS Pathog. 2013, 9, e1003224. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar]
- Jones, T.R.; Wiertz, E.J.; Sun, L.; Fish, K.N.; Nelson, J.A.; Ploegh, H.L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 1996, 93, 11327–11333. [Google Scholar] [CrossRef]
- Wiertz, E.J.; Jones, T.R.; Sun, L.; Bogyo, M.; Geuze, H.J.; Ploegh, H.L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 1996, 84, 769–779. [Google Scholar] [CrossRef]
- Jones, T.R.; Hanson, L.K.; Sun, L.; Slater, J.S.; Stenberg, R.M.; Campbell, A.E. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 1995, 69, 4830–4841. [Google Scholar]
- Machold, R.P.; Wiertz, E.J.; Jones, T.R.; Ploegh, H.L. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J. Exp. Med. 1997, 185, 363–366. [Google Scholar] [CrossRef]
- Ahn, K.; Gruhler, A.; Galocha, B.; Jones, T.R.; Wiertz, E.J.; Ploegh, H.L.; Peterson, P.A.; Yang, Y.; Fruh, K. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 1997, 6, 613–621. [Google Scholar] [CrossRef]
- Braud, V.; Jones, E.Y.; McMichael, A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 1997, 27, 1164–1169. [Google Scholar] [CrossRef]
- Millo, E.; Pietra, G.; Armirotti, A.; Vacca, P.; Mingari, M.C.; Moretta, L.; Damonte, G. Purification and HPLC-MS analysis of a naturally processed HCMV-derived peptide isolated from the HEK-293T/HLA-E+/Ul40+ cell transfectants and presented at the cell surface in the context of HLA-E. J. Immunol. Meth. 2007, 322, 128–136. [Google Scholar] [CrossRef]
- Borrego, F.; Ulbrecht, M.; Weiss, E.H.; Coligan, J.E.; Brooks, A.G. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 1998, 187, 813–818. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Lee, N.; Llano, M.; Carretero, M.; Ishitani, A.; Navarro, F.; Lopez-Botet, M.; Geraghty, D.E. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 1998, 95, 5199–5204. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Barahmand-Pour, F.; Paulsene, W.; Medley, S.; Geraghty, D.E.; Strong, R.K. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 2005, 174, 2878–2884. [Google Scholar]
- Miller, J.D.; Weber, D.A.; Ibegbu, C.; Pohl, J.; Altman, J.D.; Jensen, P.E. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J. Immunol. 2003, 171, 1369–1375. [Google Scholar]
- Sullivan, L.C.; Clements, C.S.; Beddoe, T.; Johnson, D.; Hoare, H.L.; Lin, J.; Huyton, T.; Hopkins, E.J.; Reid, H.H.; Wilce, M.C.; et al. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity 2007, 27, 900–911. [Google Scholar] [CrossRef]
- Vales-Gomez, M.; Reyburn, H.T.; Erskine, R.A.; Lopez-Botet, M.; Strominger, J.L. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999, 18, 4250–4260. [Google Scholar] [CrossRef]
- Heatley, S.L.; Pietra, G.; Lin, J.; Widjaja, J.M.; Harpur, C.M.; Lester, S.; Rossjohn, J.; Szer, J.; Schwarer, A.; Bradstock, K.; et al. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. J. Biol. Chem. 2013, 288, 8679–8690. [Google Scholar] [CrossRef]
- Llano, M.; Lee, N.; Navarro, F.; Garcia, P.; Albar, J.P.; Geraghty, D.E.; Lopez-Botet, M. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur. J. Immunol. 1998, 28, 2854–2863. [Google Scholar] [CrossRef]
- Bjorkstrom, N.K.; Lindgren, T.; Stoltz, M.; Fauriat, C.; Braun, M.; Evander, M.; Michaelsson, J.; Malmberg, K.J.; Klingstrom, J.; Ahlm, C.; et al. Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 2011, 208, 13–21. [Google Scholar] [CrossRef]
- Petitdemange, C.; Becquart, P.; Wauquier, N.; Beziat, V.; Debre, P.; Leroy, E.M.; Vieillard, V. Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog. 2011, 7, e1002268. [Google Scholar] [CrossRef]
- Brunetta, E.; Fogli, M.; Varchetta, S.; Bozzo, L.; Hudspeth, K.L.; Marcenaro, E.; Moretta, A.; Mavilio, D. Chronic HIV-1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co-infection. AIDS 2010, 24, 27–34. [Google Scholar] [CrossRef]
- Beziat, V.; Dalgard, O.; Asselah, T.; Halfon, P.; Bedossa, P.; Boudifa, A.; Hervier, B.; Theodorou, I.; Martinot, M.; Debre, P.; et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 2012, 42, 447–457. [Google Scholar] [CrossRef]
- Monsivais-Urenda, A.; Noyola-Cherpitel, D.; Hernandez-Salinas, A.; Garcia-Sepulveda, C.; Romo, N.; Baranda, L.; Lopez-Botet, M.; Gonzalez-Amaro, R. Influence of human cytomegalovirus infection on the NK cell receptor repertoire in children. Eur. J. Immunol. 2010, 40, 1418–1427. [Google Scholar] [CrossRef]
- Petersen, L.; Roug, A.S.; Skovbo, A.; Thysen, A.H.; Eskelund, C.W.; Hokland, M.E. The CD94/NKG2C-expressing NK cell subset is augmented in chronic lymphocytic leukemia patients with positive human cytomegalovirus serostatus. Viral. Immunol. 2009, 22, 333–337. [Google Scholar] [CrossRef]
- Guma, M.; Cabrera, C.; Erkizia, I.; Bofill, M.; Clotet, B.; Ruiz, L.; Lopez-Botet, M. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J. Infect. Dis. 2006, 194, 38–41. [Google Scholar] [CrossRef]
- Noyola, D.E.; Fortuny, C.; Muntasell, A.; Noguera-Julian, A.; Munoz-Almagro, C.; Alarcon, A.; Juncosa, T.; Moraru, M.; Vilches, C.; Lopez-Botet, M. Influence of congenital human cytomegalovirus infection and the NKG2C genotype on NK-cell subset distribution in children. Eur. J. Immunol. 2012, 42, 3256–3266. [Google Scholar] [CrossRef]
- Foley, B.; Cooley, S.; Verneris, M.R.; Pitt, M.; Curtsinger, J.; Luo, X.; Lopez-Verges, S.; Lanier, L.L.; Weisdorf, D.; Miller, J.S. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 2012, 119, 2665–2674. [Google Scholar] [CrossRef]
- Chiesa, D.M.; Falco, M.; Podesta, M.; Locatelli, F.; Moretta, L.; Frassoni, F.; Moretta, A. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: A role for human cytomegalovirus? Blood 2012, 119, 399–410. [Google Scholar]
- Lopez-Verges, S.; Milush, J.M.; Schwartz, B.S.; Pando, M.J.; Jarjoura, J.; York, V.A.; Houchins, J.P.; Miller, S.; Kang, S.M.; Norris, P.J.; et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 14725–14732. [Google Scholar]
- Charoudeh, H.N.; Terszowski, G.; Czaja, K.; Gonzalez, A.; Schmitter, K.; Stern, M. Modulation of the natural killer cell KIR repertoire by cytomegalovirus infection. Eur. J. Immunol. 2013, 43, 480–487. [Google Scholar] [CrossRef]
- Beziat, V.; Liu, L.L.; Malmberg, J.A.; Ivarsson, M.A.; Sohlberg, E.; Bjorklund, A.T.; Retiere, C.; Sverremark-Ekstrom, E.; Traherne, J.; Ljungman, P.; et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013, 121, 2678–2688. [Google Scholar] [CrossRef]
- Djaoud, Z.; David, G.; Bressollette, C.; Willem, C.; Rettman, P.; Gagne, K.; Legrand, N.; Mehlal, S.; Cesbron, A.; Imbert-Marcille, B.M.; et al. Amplified NKG2C+ NK cells in cytomegalovirus (CMV) infection preferentially express killer cell Ig-like receptor 2DL: functional impact in controlling CMV-infected dendritic cells. J. Immunol. 2013, 191, 2708–2716. [Google Scholar] [CrossRef]
- Moraru, M.; Cisneros, E.; Gomez-Lozano, N.; de Pablo, R.; Portero, F.; Canizares, M.; Vaquero, M.; Roustan, G.; Millan, I.; Lopez-Botet, M.; et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: contribution of polymorphic genes at the interface of innate and adaptive immunity. J. Immunol. 2012, 188, 4412–4420. [Google Scholar] [CrossRef]
- Muntasell, A.; Vilches, C.; Angulo, A.; Lopez-Botet, M. Adaptive reconfiguration of the human NK-cell compartment in response to cytomegalovirus: a different perspective of the host-pathogen interaction. Eur. J. Immunol. 2013, 43, 1133–1141. [Google Scholar] [CrossRef]
- Hikami, K.; Tsuchiya, N.; Yabe, T.; Tokunaga, K. Variations of human killer cell lectin-like receptors: Common occurrence of NKG2-C deletion in the general population. Genes Immun. 2003, 4, 160–167. [Google Scholar] [CrossRef]
- Miyashita, R.; Tsuchiya, N.; Hikami, K.; Kuroki, K.; Fukazawa, T.; Bijl, M.; Kallenberg, C.G.; Hashimoto, H.; Yabe, T.; Tokunaga, K. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int. Immunol. 2004, 16, 163–168. [Google Scholar] [CrossRef]
- Moraru, M.; Canizares, M.; Muntasell, A.; de Pablo, R.; Lopez-Botet, M.; Vilches, C. Assessment of copy-number variation in the NKG2C receptor gene in a single-tube and characterization of a reference cell panel, using standard polymerase chain reaction. Tissue Antigens 2012, 80, 184–187. [Google Scholar] [CrossRef]
- Munoz-Cobo, B.; Solano, C.; Benet, I.; Costa, E.; Remigia, M.J.; de la Camara, R.; Nieto, J.; Lopez, J.; Amat, P.; Garcia-Noblejas, A.; et al. Functional profile of cytomegalovirus (CMV)-specific CD8+ T cells and kinetics of NKG2C+ NK cells associated with the resolution of CMV DNAemia in allogeneic stem cell transplant recipients. J. Med. Virol. 2012, 84, 259–267. [Google Scholar] [CrossRef]
- Wu, Z.; Sinzger, C.; Frascaroli, G.; Reichel, J.; Bayer, C.; Wang, L.; Schirmbeck, R.; Mertens, T. Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J. Virol. 2013, 87, 7717–7725. [Google Scholar] [CrossRef]
- Guma, M.; Budt, M.; Saez, A.; Brckalo, T.; Hengel, H.; Angulo, A.; Lopez-Botet, M. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 2006, 107, 3624–3631. [Google Scholar] [CrossRef]
- Beziat, V.; Hervier, B.; Achour, A.; Boutolleau, D.; Marfain-Koka, A.; Vieillard, V. Human NKG2A overrides NKG2C effector functions to prevent autoreactivity of NK cells. Blood 2011, 117, 4394–4396. [Google Scholar] [CrossRef]
- Foley, B.; Cooley, S.; Verneris, M.R.; Curtsinger, J.; Luo, X.; Waller, E.K.; Anasetti, C.; Weisdorf, D.; Miller, J.S. Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J. Immunol. 2012, 189, 5082–5088. [Google Scholar] [CrossRef]
- Daeron, M. Fc receptor biology. Annu. Rev. Immunol. 1997, 15, 203–234. [Google Scholar] [CrossRef]
- Ravetch, J.V.; Perussia, B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J. Exp. Med. 1989, 170, 481–497. [Google Scholar] [CrossRef]
- Wirthmueller, U.; Kurosaki, T.; Murakami, M.S.; Ravetch, J.V. Signal transduction by Fc gamma RIII (CD16) is mediated through the gamma chain. J. Exp. Med. 1992, 175, 1381–1390. [Google Scholar] [CrossRef]
- Takai, T.; Li, M.; Sylvestre, D.; Clynes, R.; Ravetch, J.V. FcR gamma chain deletion results in pleiotrophic effector cell defects. Cell 1994, 76, 519–529. [Google Scholar] [CrossRef]
- Hazenbos, W.L.; Gessner, J.E.; Hofhuis, F.M.; Kuipers, H.; Meyer, D.; Heijnen, I.A.; Schmidt, R.E.; Sandor, M.; Capel, P.J.; Daeron, M.; et al. Impaired IgG-dependent anaphylaxis and Arthus reaction in Fc gamma RIII (CD16) deficient mice. Immunity 1996, 5, 181–188. [Google Scholar] [CrossRef]
- Hwang, I.; Zhang, T.; Scott, J.M.; Kim, A.R.; Lee, T.; Kakarla, T.; Kim, A.; Sunwoo, J.B.; Kim, S. Identification of human NK cells that are deficient for signaling adaptor FcRgamma and specialized for antibody-dependent immune functions. Int. Immunol. 2012, 24, 793–802. [Google Scholar] [CrossRef]
- Zhang, T.; Scott, J.M.; Hwang, I.; Kim, S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 2013, 190, 1402–1406. [Google Scholar] [CrossRef]
- Mandelboim, O.; Lieberman, N.; Lev, M.; Paul, L.; Arnon, T.I.; Bushkin, Y.; Davis, D.M.; Strominger, J.L.; Yewdell, J.W.; Porgador, A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 2001, 409, 1055–1060. [Google Scholar] [CrossRef]
- Hecht, M.L.; Rosental, B.; Horlacher, T.; Hershkovitz, O.; de Paz, J.L.; Noti, C.; Schauer, S.; Porgador, A.; Seeberger, P.H. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J. Proteome Res. 2009, 8, 712–720. [Google Scholar] [CrossRef]
- Romo, N.; Magri, G.; Muntasell, A.; Heredia, G.; Baia, D.; Angulo, A.; Guma, M.; Lopez-Botet, M. Natural killer cell-mediated response to human cytomegalovirus-infected macrophages is modulated by their functional polarization. J. Leukoc. Biol. 2011, 90, 717–726. [Google Scholar] [CrossRef]
- Magri, G.; Muntasell, A.; Romo, N.; Saez-Borderias, A.; Pende, D.; Geraghty, D.E.; Hengel, H.; Angulo, A.; Moretta, A.; Lopez-Botet, M. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood 2011, 117, 848–856. [Google Scholar] [CrossRef]
- Pessino, A.; Sivori, S.; Bottino, C.; Malaspina, A.; Morelli, L.; Moretta, L.; Biassoni, R.; Moretta, A. Molecular cloning of NKp46: A novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998, 188, 953–960. [Google Scholar]
- Moretta, A.; Biassoni, R.; Bottino, C.; Mingari, M.C.; Moretta, L. Natural cytotoxicity receptors that trigger human NK-cell-mediated cytolysis. Immunol. Today 2000, 21, 228–234. [Google Scholar] [CrossRef]
- Nigro, G.; Adler, S.P.; Torre, L.R.; Best, A.M. Passive immunization during pregnancy for congenital cytomegalovirus infection. N. Engl. J. Med. 2005, 353, 1350–1362. [Google Scholar] [CrossRef]
- Snydman, D.R.; Werner, B.G.; Dougherty, N.N.; Griffith, J.; Rubin, R.H.; Dienstag, J.L.; Rohrer, R.H.; Freeman, R.; Jenkins, R.; Lewis, W.D.; et al. Cytomegalovirus immune globulin prophylaxis in liver transplantation. A randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 1993, 119, 984–991. [Google Scholar] [CrossRef]
- Snydman, D.R.; Werner, B.G.; Heinze-Lacey, B.; Berardi, V.P.; Tilney, N.L.; Kirkman, R.L.; Milford, E.L.; Cho, S.I.; Bush, H.L.J.; Levey, A.S.; et al. Use of cytomegalovirus immune globulin to prevent cytomegalovirus disease in renal-transplant recipients. N. Engl. J. Med. 1987, 317, 1049–1054. [Google Scholar] [CrossRef]
- Metselaar, H.J.; Rothbarth, P.H.; Brouwer, R.M.; Wenting, G.J.; Jeekel, J.; Weimar, W. Prevention of cytomegalovirus-related death by passive immunization. A double-blind placebo-controlled study in kidney transplant recipients treated for rejection. Transplantation 1989, 48, 264–266. [Google Scholar] [CrossRef]
- Alexander, B.T.; Hladnik, L.M.; Augustin, K.M.; Casabar, E.; McKinnon, P.S.; Reichley, R.M.; Ritchie, D.J.; Westervelt, P.; Dubberke, E.R. Use of cytomegalovirus intravenous immune globulin for the adjunctive treatment of cytomegalovirus in hematopoietic stem cell transplant recipients. Pharmacotherapy 2010, 30, 554–561. [Google Scholar] [CrossRef]
- Tsevat, J.; Snydman, D.R.; Pauker, S.G.; Durand-Zaleski, I.; Werner, B.G.; Levey, A.S. Which renal transplant patients should receive cytomegalovirus immune globulin? A cost-effectiveness analysis. Transplantation 1991, 52, 259–265. [Google Scholar]
- Raanani, P.; Gafter-Gvili, A.; Paul, M.; Ben-Bassat, I.; Leibovici, L.; Shpilberg, O. Immunoglobulin prophylaxis in hematological malignancies and hematopoietic stem cell transplantation. Cochrane Database Syst. Rev. 2008, 4, CD006501. [Google Scholar]
- Britt, W.J.; Vugler, L.; Butfiloski, E.J.; Stephens, E.B. Cell surface expression of human cytomegalovirus (HCMV) gp55–116 (gB): Use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J. Virol. 1990, 64, 1079–1085. [Google Scholar]
- Adler, B.; Scrivano, L.; Ruzcics, Z.; Rupp, B.; Sinzger, C.; Koszinowski, U. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 2006, 87, 2451–2460. [Google Scholar] [CrossRef]
- Revello, M.G.; Gerna, G. Human cytomegalovirus tropism for endothelial/epithelial cells: Scientific background and clinical implications. Rev. Med. Virol. 2010, 20, 136–155. [Google Scholar] [CrossRef]
- Lilleri, D.; Kabanova, A.; Revello, M.G.; Percivalle, E.; Sarasini, A.; Genini, E.; Sallusto, F.; Lanzavecchia, A.; Corti, D.; Gerna, G. Fetal human cytomegalovirus transmission correlates with delayed maternal antibodies to gH/gL/pUL128–130–131 complex during primary infection. PLoS One 2013, 8, e59863. [Google Scholar]
- Macagno, A.; Bernasconi, N.L.; Vanzetta, F.; Dander, E.; Sarasini, A.; Revello, M.G.; Gerna, G.; Sallusto, F.; Lanzavecchia, A. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128–131A complex. J. Virol. 2010, 84, 1005–1013. [Google Scholar] [CrossRef]
- Hahn, G.; Revello, M.G.; Patrone, M.; Percivalle, E.; Campanini, G.; Sarasini, A.; Wagner, M.; Gallina, A.; Milanesi, G.; Koszinowski, U.; et al. Human cytomegalovirus UL131–128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 2004, 78, 10023–10033. [Google Scholar] [CrossRef]
- Ryckman, B.J.; Rainish, B.L.; Chase, M.C.; Borton, J.A.; Nelson, J.A.; Jarvis, M.A.; Johnson, D.C. Characterization of the human cytomegalovirus gH/gL/UL128–131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 2008, 82, 60–70. [Google Scholar] [CrossRef]
- Wang, D.; Shenk, T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 2005, 102, 18153–18158. [Google Scholar] [CrossRef]
- Wang, D.; Shenk, T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 2005, 79, 10330–10338. [Google Scholar] [CrossRef]
- Genini, E.; Percivalle, E.; Sarasini, A.; Revello, M.G.; Baldanti, F.; Gerna, G. Serum antibody response to the gH/gL/pUL128–131 five-protein complex of human cytomegalovirus (HCMV) in primary and reactivated HCMV infections. J. Clin. Virol. 2011, 52, 113–118. [Google Scholar] [CrossRef]
- Fouts, A.E.; Chan, P.; Stephan, J.P.; Vandlen, R.; Feierbach, B. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J. Virol. 2012, 86, 7444–7447. [Google Scholar] [CrossRef]
- Plotkin, S.A.; Starr, S.E.; Friedman, H.M.; Brayman, K.; Harris, S.; Jackson, S.; Tustin, N.B.; Grossman, R.; Dafoe, D.; Barker, C. Effect of Towne live virus vaccine on cytomegalovirus disease after renal transplant. A controlled trial. Ann. Intern. Med. 1991, 114, 525–531. [Google Scholar] [CrossRef]
- Plotkin, S.A.; Higgins, R.; Kurtz, J.B.; Morris, P.J.; Campbell, D.A.J.; Shope, T.C.; Spector, S.A.; Dankner, W.M. Multicenter trial of Towne strain attenuated virus vaccine in seronegative renal transplant recipients. Transplantation 1994, 58, 1176–1178. [Google Scholar]
- Adler, S.P.; Starr, S.E.; Plotkin, S.A.; Hempfling, S.H.; Buis, J.; Manning, M.L.; Best, A.M. Immunity induced by primary human cytomegalovirus infection protects against secondary infection among women of childbearing age. J. Infect. Dis. 1995, 171, 26–32. [Google Scholar] [CrossRef]
- Sabbaj, S.; Pass, R.F.; Goepfert, P.A.; Pichon, S. Glycoprotein B vaccine is capable of boosting both antibody and CD4 T-cell responses to cytomegalovirus in chronically infected women. J. Infect. Dis. 2011, 203, 1534–1541. [Google Scholar] [CrossRef]
- Pass, R.F.; Zhang, C.; Evans, A.; Simpson, T.; Andrews, W.; Huang, M.L.; Corey, L.; Hill, J.; Davis, E.; Flanigan, C.; et al. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 2009, 360, 1191–1199. [Google Scholar] [CrossRef]
- Pass, R.F.; Duliege, A.M.; Boppana, S.; Sekulovich, R.; Percell, S.; Britt, W.; Burke, R.L. A subunit cytomegalovirus vaccine based on recombinant envelope glycoprotein B and a new adjuvant. J. Infect. Dis. 1999, 180, 970–975. [Google Scholar] [CrossRef]
- Griffiths, P.D.; Stanton, A.; McCarrell, E.; Smith, C.; Osman, M.; Harber, M.; Davenport, A.; Jones, G.; Wheeler, D.C.; O’Beirne, J.; et al. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. Lancet 2011, 377, 1256–1263. [Google Scholar] [CrossRef]
- Nokta, M.; Tolpin, M.D.; Nadler, P.I.; Pollard, R.B. Human monoclonal anti-cytomegalovirus (CMV) antibody (MSL 109): Enhancement of in vitro foscarnet- and ganciclovir-induced inhibition of CMV replication. Antivir. Res. 1994, 24, 17–26. [Google Scholar] [CrossRef]
- Manley, K.; Anderson, J.; Yang, F.; Szustakowski, J.; Oakeley, E.J.; Compton, T.; Feire, A.L. Human cytomegalovirus escapes a naturally occurring neutralizing antibody by incorporating it into assembling virions. Cell Host Microbe 2011, 10, 197–209. [Google Scholar] [CrossRef]
- Jabs, D.A.; Gilpin, A.M.; Min, Y.I.; Erice, A.; Kempen, J.H.; Quinn, T.C. HIV and cytomegalovirus viral load and clinical outcomes in AIDS and cytomegalovirus retinitis patients: Monoclonal Antibody Cytomegalovirus Retinitis Trial. AIDS 2002, 16, 877–887. [Google Scholar] [CrossRef]
- Boeckh, M.; Bowden, R.A.; Storer, B.; Chao, N.J.; Spielberger, R.; Tierney, D.K.; Gallez-Hawkins, G.; Cunningham, T.; Blume, K.G.; Levitt, D.; et al. Randomized, placebo-controlled, double-blind study of a cytomegalovirus-specific monoclonal antibody (MSL-109) for prevention of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2001, 7, 343–351. [Google Scholar] [CrossRef]
- Jegaskanda, S.; Job, E.R.; Kramski, M.; Laurie, K.; Isitman, G.; de Rose, R.; Winnall, W.R.; Stratov, I.; Brooks, A.G.; Reading, P.C.; et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J. Immunol. 2013, 190, 1837–1848. [Google Scholar] [CrossRef]
- Jia, M.; Li, D.; He, X.; Zhao, Y.; Peng, H.; Ma, P.; Hong, K.; Liang, H.; Shao, Y. Impaired natural killer cell-induced antibody-dependent cell-mediated cytotoxicity is associated with human immunodeficiency virus-1 disease progression. Clin. Exp. Immunol. 2013, 171, 107–116. [Google Scholar]
- Hansen, S.G.; Powers, C.J.; Richards, R.; Ventura, A.B.; Ford, J.C.; Siess, D.; Axthelm, M.K.; Nelson, J.A.; Jarvis, M.A.; Picker, L.J.; et al. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 2010, 328, 102–106. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).