Elevated IgE Levels—An Allergy or an Underlying Inborn Error of Immunity in Children with Recurrent Infections?
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joshi, A.Y.; Iyer, V.N.; Boyce, T.G.; Hagan, J.B.; Park, M.A.; Abraham, R.S. Elevated Serum Immunoglobulin E (IgE): When to Suspect Hyper-IgE Syndrome—A 10-Year Pediatric Tertiary Care Center Experience. Allergy Asthma Proc. 2009, 30, 23–27. [Google Scholar] [CrossRef]
- Castagnoli, R.; Lougaris, V.; Giardino, G.; Volpi, S.; Leonardi, L.; La Torre, F.; Federici, S.; Corrente, S.; Cinicola, B.L.; Soresina, A.; et al. Inborn Errors of Immunity with Atopic Phenotypes: A Practical Guide for Allergists. World Allergy Organ. J. 2021, 14, 100513. [Google Scholar] [CrossRef] [PubMed]
- Notarangelo, L.D.; Bacchetta, R.; Casanova, J.-L.; Su, H.C. Human Inborn Errors of Immunity: An Expanding Universe. Sci. Immunol. 2020, 5, eabb1662. [Google Scholar] [CrossRef]
- Chan, S.K.; Gelfand, E.W. Primary Immunodeficiency Masquerading as Allergic Disease. Immunol. Allergy Clin. N. Am. 2015, 35, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Milner, J.D. Primary Atopic Disorders. Annu. Rev. Immunol. 2020, 38, 785–808. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, Z.A.; El-Ghoneimy, D.H.; Ortega-Martell, J.A.; Radwan, N.; Aldave, J.C.; Al-Herz, W.; Al-Nesf, M.A.; Condino-Neto, A.; Cole, T.; Eley, B.; et al. Allergic Manifestations of Inborn Errors of Immunity and Their Impact on the Diagnosis: A Worldwide Study. World Allergy Organ. J. 2022, 15, 100657. [Google Scholar] [CrossRef]
- Abraham, R.S.; Butte, M.J. The New “Wholly Trinity” in the Diagnosis and Management of Inborn Errors of Immunity. J. Allergy Clin. Immunol. Pract. 2021, 9, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Vaseghi-Shanjani, M.; Smith, K.L.; Sara, R.J.; Modi, B.P.; Branch, A.; Sharma, M.; Lu, H.Y.; James, E.L.; Hildebrand, K.J.; Biggs, C.M.; et al. Inborn Errors of Immunity Manifesting as Atopic Disorders. J. Allergy Clin. Immunol. 2021, 148, 1130–1139. [Google Scholar] [CrossRef]
- Stadler, P.-C.; Renner, E.D.; Milner, J.; Wollenberg, A. Inborn Error of Immunity or Atopic Dermatitis: When to Be Concerned and How to Investigate. J. Allergy Clin. Immunol. Pract. 2021, 9, 1501–1507. [Google Scholar] [CrossRef]
- Khan, Y.W.; Williams, K.W. Inborn Errors of Immunity Associated with Elevated Immunoglobulin E. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2022, 129, 552–561. [Google Scholar] [CrossRef]
- Tsilifis, C.; Freeman, A.F.; Gennery, A.R. STAT3 Hyper-IgE Syndrome—An Update and Unanswered Questions. J. Clin. Immunol. 2021, 41, 864–880. [Google Scholar] [CrossRef] [PubMed]
- Kandilarova, S.M.; Lesichkova, S.S.; Gesheva, N.T.; Yankova, P.S.; Ivanov, N.H.; Stoyanova, G.P.; Perenovska, P.I.; Baleva, M.P.; Naumova, E.J. On Two Cases with Autosomal Dominant Hyper IgE Syndrome: Importance of Immunological Parameters for Clinical Course and Follow-Up. Case Rep. Immunol. 2020, 2020, 6694957. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.Y.; Bauman, B.M.; Arjunaraja, S.; Dorjbal, B.; Milner, J.D.; Snow, A.L.; Turvey, S.E. The CBM-Opathies—A Rapidly Expanding Spectrum of Human Inborn Errors of Immunity Caused by Mutations in the CARD11-BCL10-MALT1 Complex. Front. Immunol. 2018, 9, 2078. [Google Scholar] [CrossRef] [PubMed]
- Vaseghi-Shanjani, M.; Yousefi, P.; Sharma, M.; Samra, S.; Sifuentes, E.; Turvey, S.E.; Biggs, C.M. Transcription Factor Defects in Inborn Errors of Immunity with Atopy. Front. Allergy 2023, 4, 1237852. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, Y. The Signal Transducer and Activator of Transcription 3 at the Center of the Causative Gene Network of the Hyper-IgE Syndrome. Curr. Opin. Immunol. 2023, 80, 102264. [Google Scholar] [CrossRef]
- Wang, N.; Shen, N.; Vyse, T.J.; Anand, V.; Gunnarson, I.; Sturfelt, G.; Rantapää-Dahlqvist, S.; Elvin, K.; Truedsson, L.; Andersson, B.A.; et al. Selective IgA Deficiency in Autoimmune Diseases. Mol. Med. Camb. Mass. 2011, 17, 1383–1396. [Google Scholar] [CrossRef]
- Jorgensen, G.H.; Gardulf, A.; Sigurdsson, M.I.; Sigurdardottir, S.T.; Thorsteinsdottir, I.; Gudmundsson, S.; Hammarström, L.; Ludviksson, B.R. Clinical Symptoms in Adults with Selective IgA Deficiency: A Case-Control Study. J. Clin. Immunol. 2013, 33, 742–747. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Palacios-Kibler, T.V.; Workman, L.J.; Schuyler, A.J.; Steinke, J.W.; Payne, S.C.; McGowan, E.C.; Patrie, J.; Fuleihan, R.L.; Sullivan, K.E.; et al. Low Serum IgE Is a Sensitive and Specific Marker for Common Variable Immunodeficiency (CVID). J. Clin. Immunol. 2018, 38, 225–233. [Google Scholar] [CrossRef]
- Ogershok, P.R.; Hogan, M.B.; Welch, J.E.; Corder, W.T.; Wilson, N.W. Spectrum of Illness in Pediatric Common Variable Immunodeficiency. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2006, 97, 653–656. [Google Scholar] [CrossRef]
- Yong, P.L.; Orange, J.S.; Sullivan, K.E. Pediatric Common Variable Immunodeficiency: Immunologic and Phenotypic Associations with Switched Memory B Cells. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2010, 21, 852–858. [Google Scholar] [CrossRef]
- Ghaini, M.; Jamee, M.; Mahdaviani, S.A.; Mesdaghi, M.; Eskandarzadeh, S.; Rae, W.; Eslami, N.; Eslamian, G.; Mansouri, M.; Babaei, D.; et al. The Prevalence of Atopic Manifestations in 313 Iranian Patients with Inborn Errors of Immunity. Int. Arch. Allergy Immunol. 2021, 182, 1122–1126. [Google Scholar] [CrossRef]
- Zarezadeh Mehrabadi, A.; Aghamohamadi, N.; Abolhassani, H.; Aghamohammadi, A.; Rezaei, N.; Yazdani, R. Comprehensive Assessment of Skin Disorders in Patients with Common Variable Immunodeficiency (CVID). J. Clin. Immunol. 2022, 42, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Cinicola, B.L.; Corrente, S.; Castagnoli, R.; Lougaris, V.; Giardino, G.; Leonardi, L.; Volpi, S.; La Torre, F.; Federici, S.; Soresina, A.; et al. Primary Atopic Disorders and Chronic Skin Disease. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2022, 33, 65–68. [Google Scholar] [CrossRef]
- Matricardi, P.M. The Very Low Ige Producer: Allergology, Genetics, Immunodeficiencies, and Oncology. Biomedicines 2023, 11, 1378. [Google Scholar] [CrossRef]
- Elkuch, M.; Greiff, V.; Berger, C.T.; Bouchenaki, M.; Daikeler, T.; Bircher, A.; Navarini, A.A.; Heijnen, I.; Recher, M. Low Immunoglobulin E Flags Two Distinct Types of Immune Dysregulation. Clin. Exp. Immunol. 2017, 187, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Leung, D.; Momenilandi, M.; Jones, L.C.W.; Pacillo, L.; James, A.E.; Murrell, J.R.; Delafontaine, S.; Maimaris, J.; Vaseghi-Shanjani, M.; et al. Human Germline Heterozygous Gain-of-Function STAT6 Variants Cause Severe Allergic Disease. J. Exp. Med. 2023, 220, e20221755. [Google Scholar] [CrossRef]
- Schwitzguébel, A.J.-P.; Jandus, P.; Lacroix, J.-S.; Seebach, J.D.; Harr, T. Immunoglobulin Deficiency in Patients with Chronic Rhinosinusitis: Systematic Review of the Literature and Meta-Analysis. J. Allergy Clin. Immunol. 2015, 136, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Plebani, A.; Monafo, V.; Avanzini, M.A.; Ugazio, A.G.; Burgio, G.R. Relationship between IgA and IgG Subclass Deficiencies: A Reappraisal. Monogr. Allergy 1986, 20, 171–178. [Google Scholar] [PubMed]
- de Moraes Lui, C.; Oliveira, L.C.; Diogo, C.L.; Kirschfink, M.; Grumach, A.S. Immunoglobulin G Subclass Concentrations and Infections in Children and Adolescents with Severe Asthma. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2002, 13, 195–202. [Google Scholar] [CrossRef]
- Jensen-Jarolim, E.; Bax, H.J.; Bianchini, R.; Crescioli, S.; Daniels-Wells, T.R.; Dombrowicz, D.; Fiebiger, E.; Gould, H.J.; Irshad, S.; Janda, J.; et al. AllergoOncology: Opposite Outcomes of Immune Tolerance in Allergy and Cancer. Allergy 2018, 73, 328–340. [Google Scholar] [CrossRef]
- Karagiannis, P.; Villanova, F.; Josephs, D.H.; Correa, I.; Van Hemelrijck, M.; Hobbs, C.; Saul, L.; Egbuniwe, I.U.; Tosi, I.; Ilieva, K.M.; et al. Elevated IgG4 in Patient Circulation Is Associated with the Risk of Disease Progression in Melanoma. Oncoimmunology 2015, 4, e1032492. [Google Scholar] [CrossRef]
- Komulainen, J.; Siiskonen, H.; Haimakainen, S.; Kanasuo, E.; Harvima, R.J.; Harvima, I.T. Patients with a History of Atopy Have Fewer Cutaneous Melanomas than Those without Atopy: A Cross-Sectional Study in 496 Patients at Risk of Skin Cancers. Melanoma Res. 2023, 33, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Singer, J.; Achatz-Straussberger, G.; Bentley-Lukschal, A.; Fazekas-Singer, J.; Achatz, G.; Karagiannis, S.N.; Jensen-Jarolim, E. AllergoOncology: High Innate IgE Levels Are Decisive for the Survival of Cancer-Bearing Mice. World Allergy Organ. J. 2019, 12, 100044. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Jarolim, E.; Bax, H.J.; Bianchini, R.; Capron, M.; Corrigan, C.; Castells, M.; Dombrowicz, D.; Daniels-Wells, T.R.; Fazekas, J.; Fiebiger, E.; et al. AllergoOncology—The Impact of Allergy in Oncology: EAACI Position Paper. Allergy 2017, 72, 866–887. [Google Scholar] [CrossRef] [PubMed]
- Ferastraoaru, D.; Gross, R.; Rosenstreich, D. Increased Malignancy Incidence in IgE Deficient Patients Not Due to Concomitant Common Variable Immunodeficiency. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2017, 119, 267–273. [Google Scholar] [CrossRef]
- Pate, M.B.; Smith, J.K.; Chi, D.S.; Krishnaswamy, G. Regulation and Dysregulation of Immunoglobulin E: A Molecular and Clinical Perspective. Clin. Mol. Allergy CMA 2010, 8, 3. [Google Scholar] [CrossRef]
- Ponsford, M.J.; Klocperk, A.; Pulvirenti, F.; Dalm, V.A.S.H.; Milota, T.; Cinetto, F.; Chovancova, Z.; Rial, M.J.; Sediva, A.; Litzman, J.; et al. Hyper-IgE in the Allergy Clinic—When Is It Primary Immunodeficiency? Allergy 2018, 73, 2122–2136. [Google Scholar] [CrossRef]
- Mayor, P.C.; Eng, K.H.; Singel, K.L.; Abrams, S.I.; Odunsi, K.; Moysich, K.B.; Fuleihan, R.; Garabedian, E.; Lugar, P.; Ochs, H.D.; et al. Cancer in Primary Immunodeficiency Diseases: Cancer Incidence in the United States Immune Deficiency Network Registry. J. Allergy Clin. Immunol. 2018, 141, 1028–1035. [Google Scholar] [CrossRef]
- Tak Manesh, A.; Azizi, G.; Heydari, A.; Kiaee, F.; Shaghaghi, M.; Hossein-Khannazer, N.; Yazdani, R.; Abolhassani, H.; Aghamohammadi, A. Epidemiology and Pathophysiology of Malignancy in Common Variable Immunodeficiency? Allergol. Immunopathol. 2017, 45, 602–615. [Google Scholar] [CrossRef]
- Vajdic, C.M.; Mao, L.; van Leeuwen, M.T.; Kirkpatrick, P.; Grulich, A.E.; Riminton, S. Are Antibody Deficiency Disorders Associated with a Narrower Range of Cancers than Other Forms of Immunodeficiency? Blood 2010, 116, 1228–1234. [Google Scholar] [CrossRef]
- Gangemi, S.; Allegra, A.; Musolino, C. Lymphoproliferative Disease and Cancer among Patients with Common Variable Immunodeficiency. Leuk. Res. 2015, 39, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Aghamohammadi, A.; Moin, M.; Kouhi, A.; Mohagheghi, M.-A.; Shirazi, A.; Rezaei, N.; Tavassoli, S.; Esfahani, M.; Cheraghi, T.; Dastan, J.; et al. Chromosomal Radiosensitivity in Patients with Common Variable Immunodeficiency. Immunobiology 2008, 213, 447–454. [Google Scholar] [CrossRef] [PubMed]
- de Wit, J.; van Wijck, R.T.A.; Dalm, V.A.S.H.; Snyder, K.L.; Totté, J.E.E.; Pasmans, S.G.M.A.; van der Spek, P.J.; Academic Center of Excellence (ACE) workgroups Allergic Diseases and Rare Immunological Disease Centre (RIDC). Molecular Clustering of Genes Related to the Atopic Syndrome: Towards a More Tailored Approach and Personalized Medicine? Clin. Transl. Allergy 2019, 9, 34. [Google Scholar] [CrossRef] [PubMed]

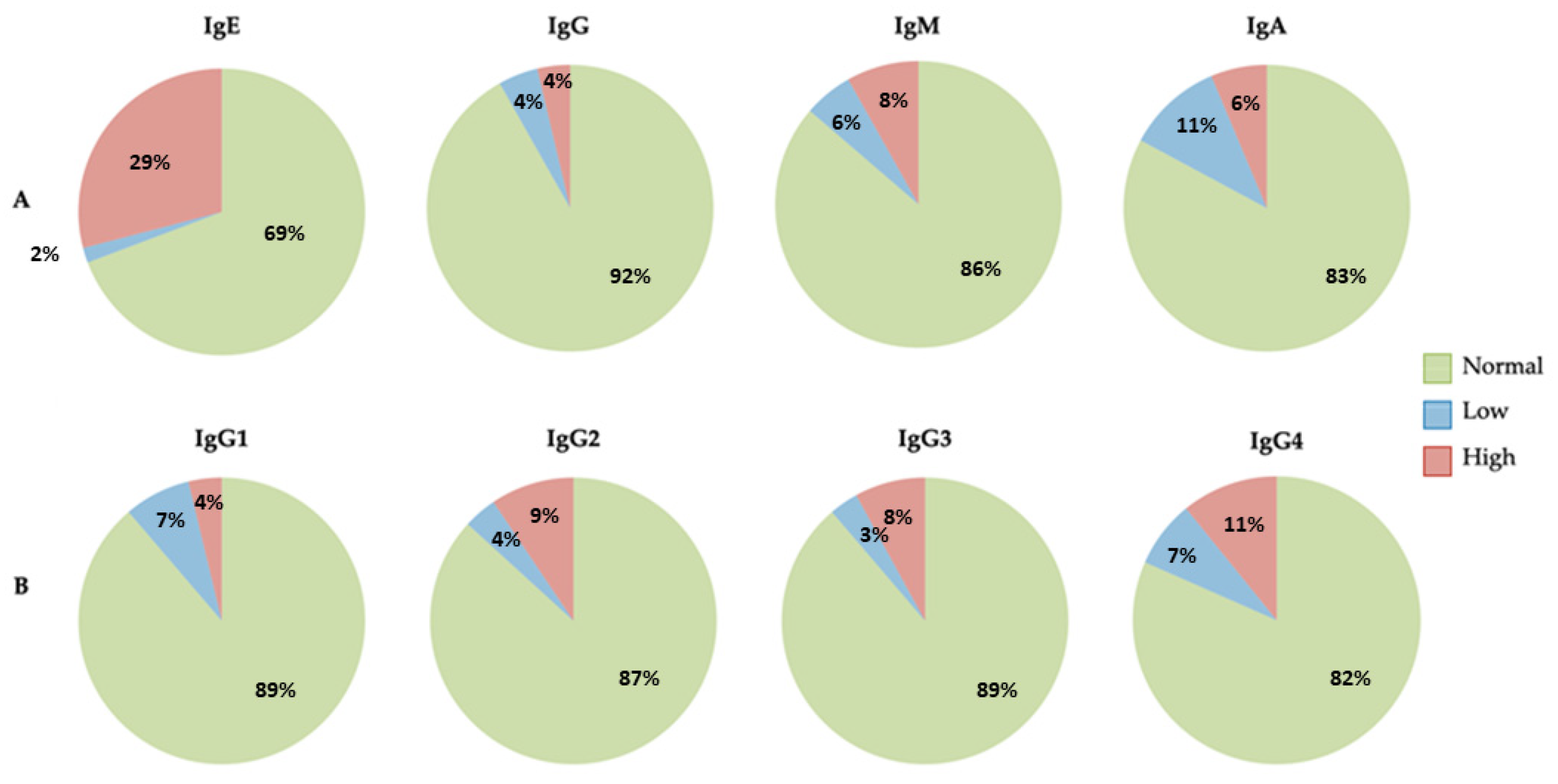
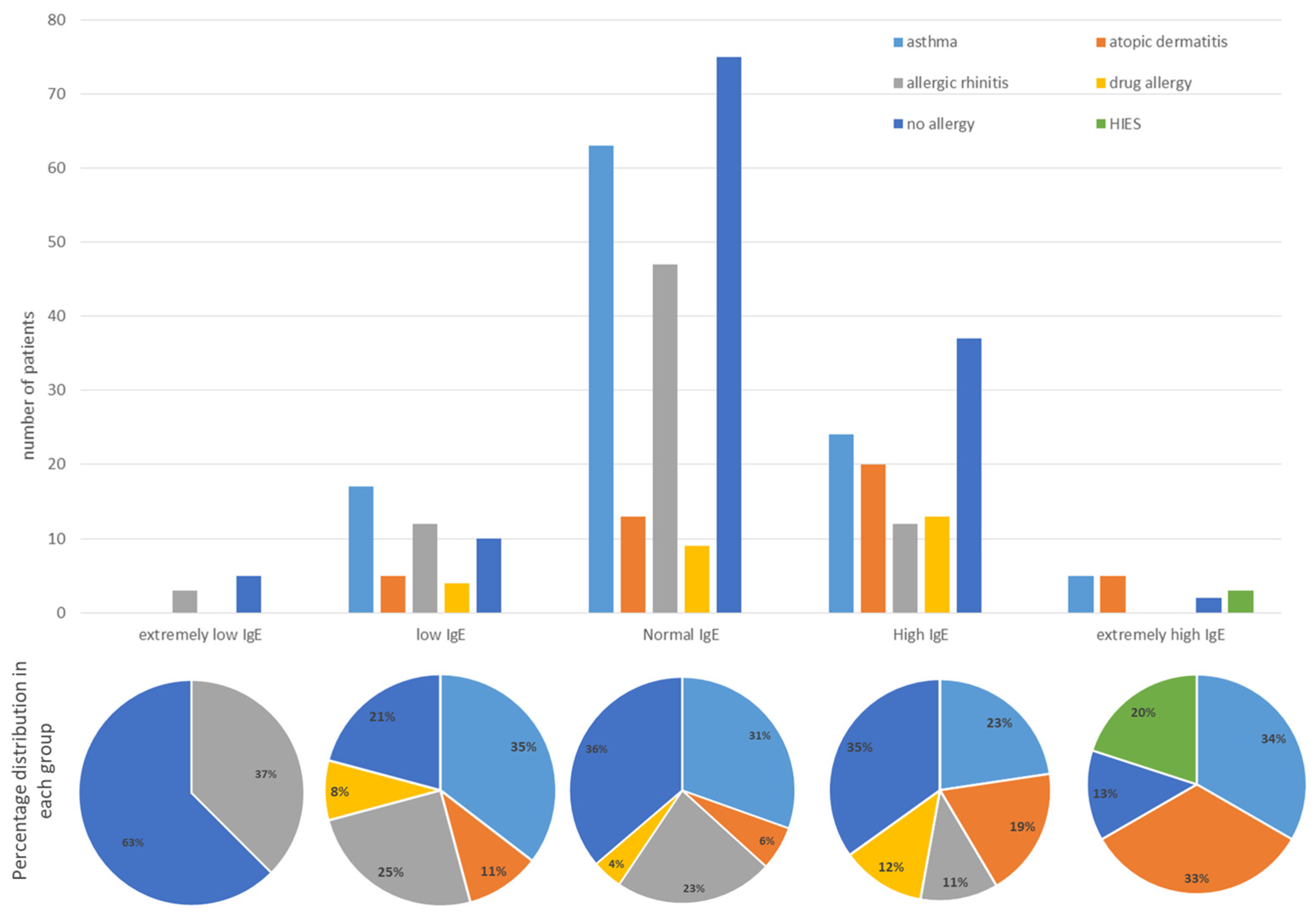
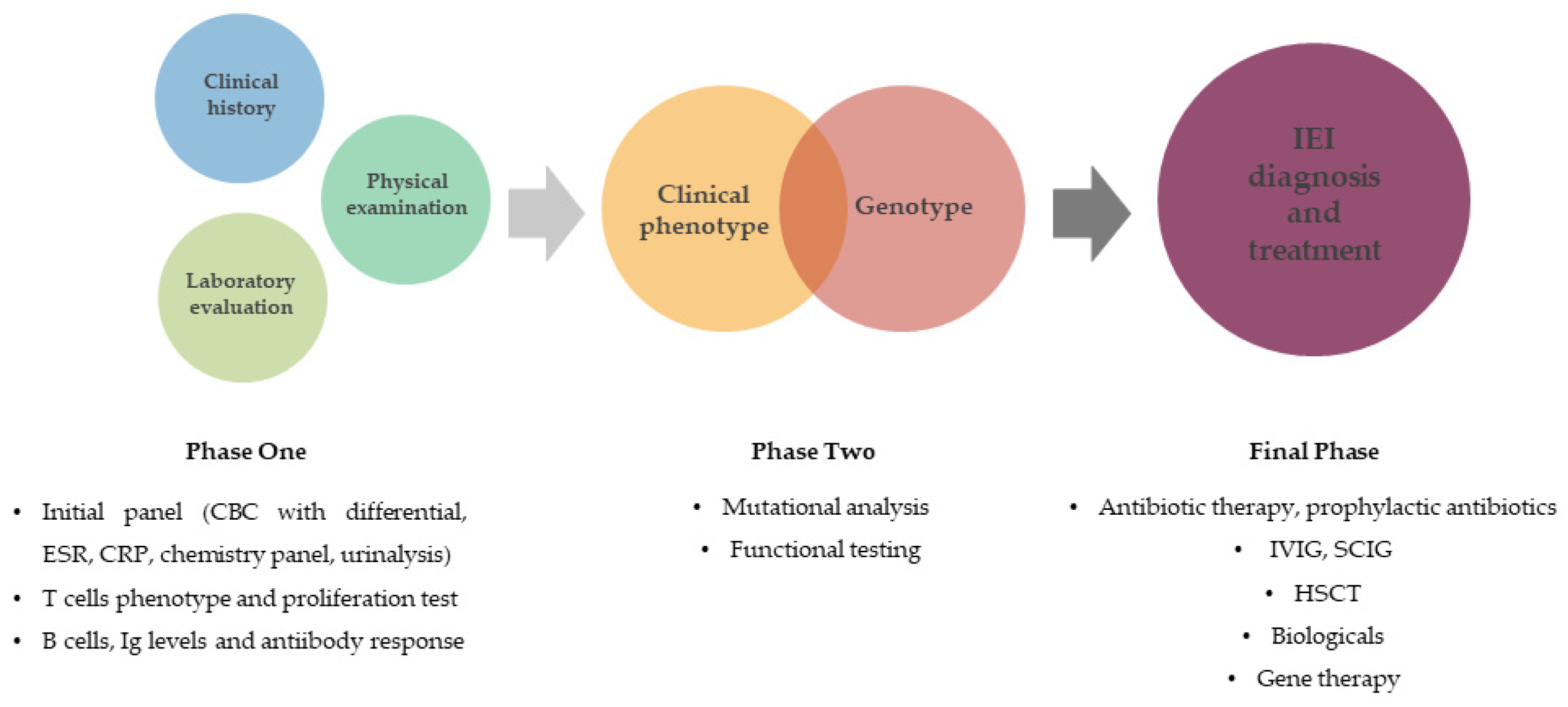
| Patient Results | |
|---|---|
| Mean age (±SD), years | 5.75 ± 3.847 |
| Sex (m:f) | 214:171 |
| Leading reasons for suspected IEI and number of patients referred for it | Recurrent respiratory infections—296 Chronic diarrhea and GIT infections—21 Recurrent fever of unknown origin—15 Recurrent skin infections—10 Unexplained anemia—4 Lymphoproliferative disease (unusual presentation or type for the age)—22 Immune thrombocytopenia—14 Tumor—3 |
| Comorbidities | No comorbidities—80 Allergic diseases (incl. asthma)—252 Congenital heart disease—6 Epilepsy—4 Diabetes—3 Anemia—4 Thrombocytopenia—14 Neoplasm—22 |
| Allergic diseases and HIES | Asthma—109 Allergic rhinitis—74 Atopic dermatitis—43 Drug allergy—26 HIES—3 No allergy—130 |
| Family history for atopy | No family history—106 Atopy—83 Autoimmunity—33 Autoimmunity and atopy—27 Neoplasm—9 Autoimmunity and neoplasm—1 Atopy and neoplasm—13 Autoimmunity and atopy and neoplasm—4 Bronchial asthma—62 Other chronic diseases (diabetes, arterial hypertension, epilepsy, etc.)—47 |
| Turbimetry | 0–6 Months | 6 Months–2 Years | 2–4 Years | 4–6 Years | 6–10 Years | 10–12 Years | 12–14 Years | Over 14 Years |
|---|---|---|---|---|---|---|---|---|
| IgA g/L | 0.2–1.0 | 0.2–1.0 | 0.2–1.0 | 0.3–1.95 | 0.5–2.5 | 0.5–2.0 | 0.6–3.6 | 0.6–3.5 |
| IgG g/L | 3.2–12.7 | 3.2–12.7 | 4.7–12.5 | 5.3–13.4 | 5.4–16.1 | 5.5–14.9 | 6.6–14.9 | 5.5–14.4 |
| IgM g/L | 0.2–1.0 | 0.4–2.2 | 0.3–2.0 | 0.3–2.0 | 0.5–1.8 | 0.5–1.8 | 0.35–2.42 | 0.35–2.42 |
| IgE IU/mL | <60 | <60 | <60 | <100 | <100 | <100 | <120 | <120 |
| IgG1 g/L | 1.3–6.33 | 3.1–9.4 | 3.1–9.4 | 2.88–9.18 | 4.3–10.6 | 3.4–11.5 | 3.15–8.55 | |
| IgG2 g/L | 0.3–1.29 | 0.3–2.2 | 0.6–3.4 | 0.44–3.75 | 0.4–3.5 | 1.0–4.5 | 0.64–4.95 | |
| IgG3 g/L | 0.1–0.6 | 0.1–0.6 | 0.1–1.2 | 0.15–1.5 | 0.1–1.7 | 0.3–1.2 | 0.23–1.96 | |
| IgG4 g/L | 0.01–0.2 | 0.01–0.5 | 0.02–1.1 | 0.01–1.1 | 0.01–1.1 | 0.04–1.3 | 0.11–1.57 |
| Immunoglobulins | IgA | IgM | IgG | IgE |
|---|---|---|---|---|
| Low | sIgAD—10 CVID—5 XLA—3 TrN—2 AT—2 CD—1 | CVID—6 XLA—3 CD—1 TrN—1 TCID—1 | CVID—6 XLA—3 Di George—1 TrN—1 TCID—1 CD—1 | CVID—3 XLA—2 sIgAD—2 TrN—1 AT—1 IgG2 deficiency—1 |
| High | LAD deficiency—2 SAVI—1 | AT—2 LAD deficiency—2 SAVI—1 sIgAD—1 HIES—1 | LAD deficiency—2 sIgAD—2 SAVI—1 HIES—1 TCID—1 | HIES—3 sIgAD—2 TCID—2 SAVI—1 Netherton—1 CARD11—1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostova, P.; Papochieva, V.; Miteva, D.; Georgieva, B.; Mileva, S.; Shahid, M.; Lukanov, T.; Petrova, G. Elevated IgE Levels—An Allergy or an Underlying Inborn Error of Immunity in Children with Recurrent Infections? Antibodies 2023, 12, 70. https://doi.org/10.3390/antib12040070
Kostova P, Papochieva V, Miteva D, Georgieva B, Mileva S, Shahid M, Lukanov T, Petrova G. Elevated IgE Levels—An Allergy or an Underlying Inborn Error of Immunity in Children with Recurrent Infections? Antibodies. 2023; 12(4):70. https://doi.org/10.3390/antib12040070
Chicago/Turabian StyleKostova, Polina, Vera Papochieva, Dimitrinka Miteva, Bilyana Georgieva, Sirma Mileva, Martin Shahid, Tsvetelin Lukanov, and Guergana Petrova. 2023. "Elevated IgE Levels—An Allergy or an Underlying Inborn Error of Immunity in Children with Recurrent Infections?" Antibodies 12, no. 4: 70. https://doi.org/10.3390/antib12040070
APA StyleKostova, P., Papochieva, V., Miteva, D., Georgieva, B., Mileva, S., Shahid, M., Lukanov, T., & Petrova, G. (2023). Elevated IgE Levels—An Allergy or an Underlying Inborn Error of Immunity in Children with Recurrent Infections? Antibodies, 12(4), 70. https://doi.org/10.3390/antib12040070





