Characterization of Active MMP9 in Chronic Inflammatory Diseases Using a Novel Anti-MMP9 Antibody
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibody Generation and Characterization
2.2. In Vitro MMP9 Activation
2.3. Assessment of Active MMP9 in Human Tissues
- Deparaffinization with EZ Prep (Ventana Medical Systems; Cat #5279755001) for 12 min at 69 °C;
- Antigen retrieval with CC1 cell conditioning (Ventana Medical Systems; Cat #6414575001) for 64 min at 95 °C;
- ChromoMap Inhibitor (Ventana Medical Systems; Cat #5266645001) for 8 min at room temperature;
- Manual addition of 100 µL of AB006988 at a concentration of 5 µg/mL for 1 h at room temperature;
- Manual addition of 100 µL Mach 3 Anti-Rabbit Probe (Biocare Medical, Concord, CA, USA; Cat #RP531) for 30 min at room temperature;
- Anti-Mouse HQ (Ventana Medical Systems; Cat #7017782001) for 32 min at room temperature;
- Anti-HQ HRP (Ventana Medical Systems; Cat #7017936001) for 32 min at room temperature;
- ChromoMap DAB kit (Ventana Medical Systems; Cat #5266645001) for 8 min at room temperature;
- Hematoxylin II (Ventana Medical Systems; Cat #5277965001) for 4 min at room temperature.
3. Results
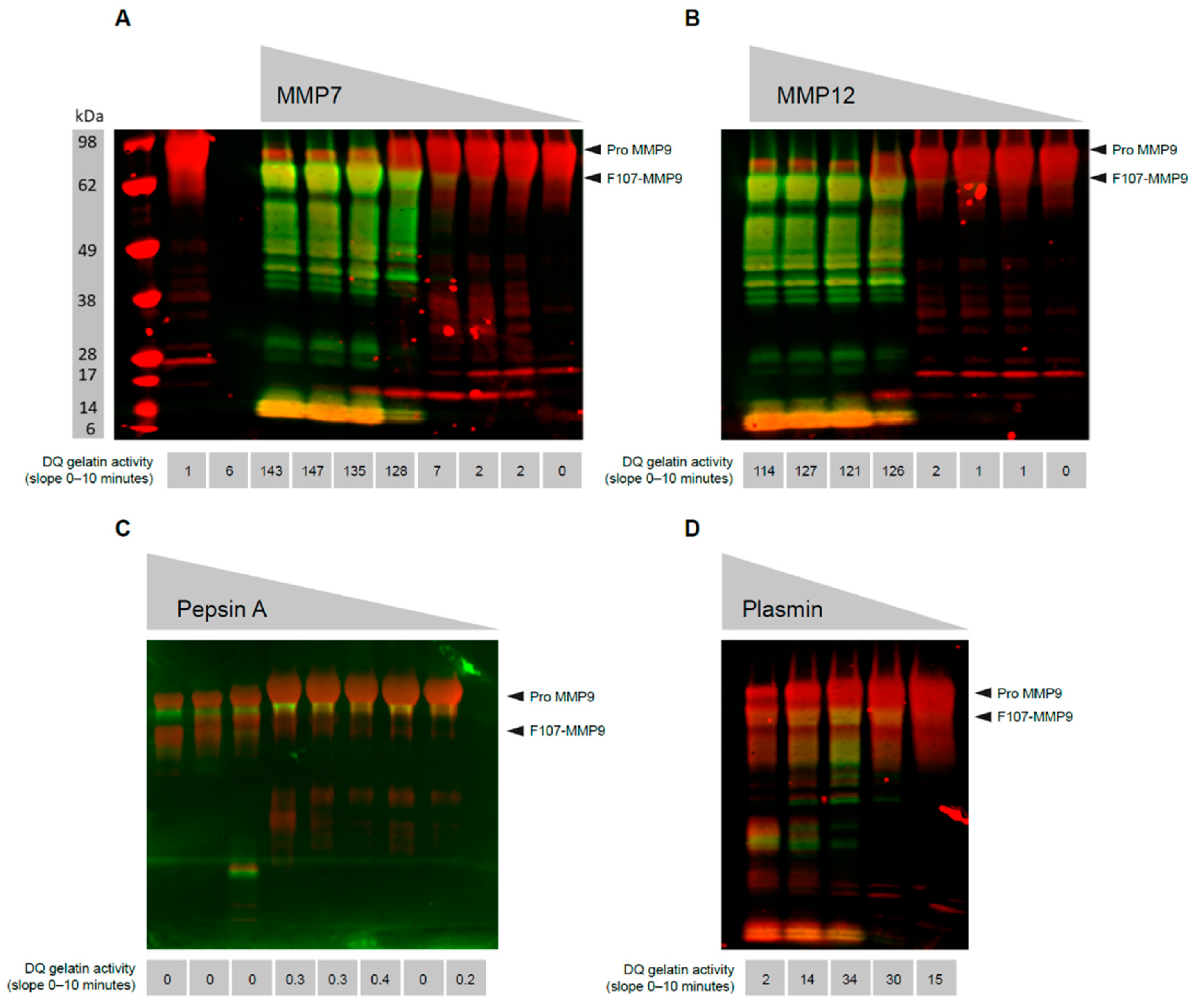
3.1. MMP9 Cleaved at Phe107 Is Proteolytically Active
3.2. Generation of a mAb That Recognizes the N-Terminus of Active MMP9
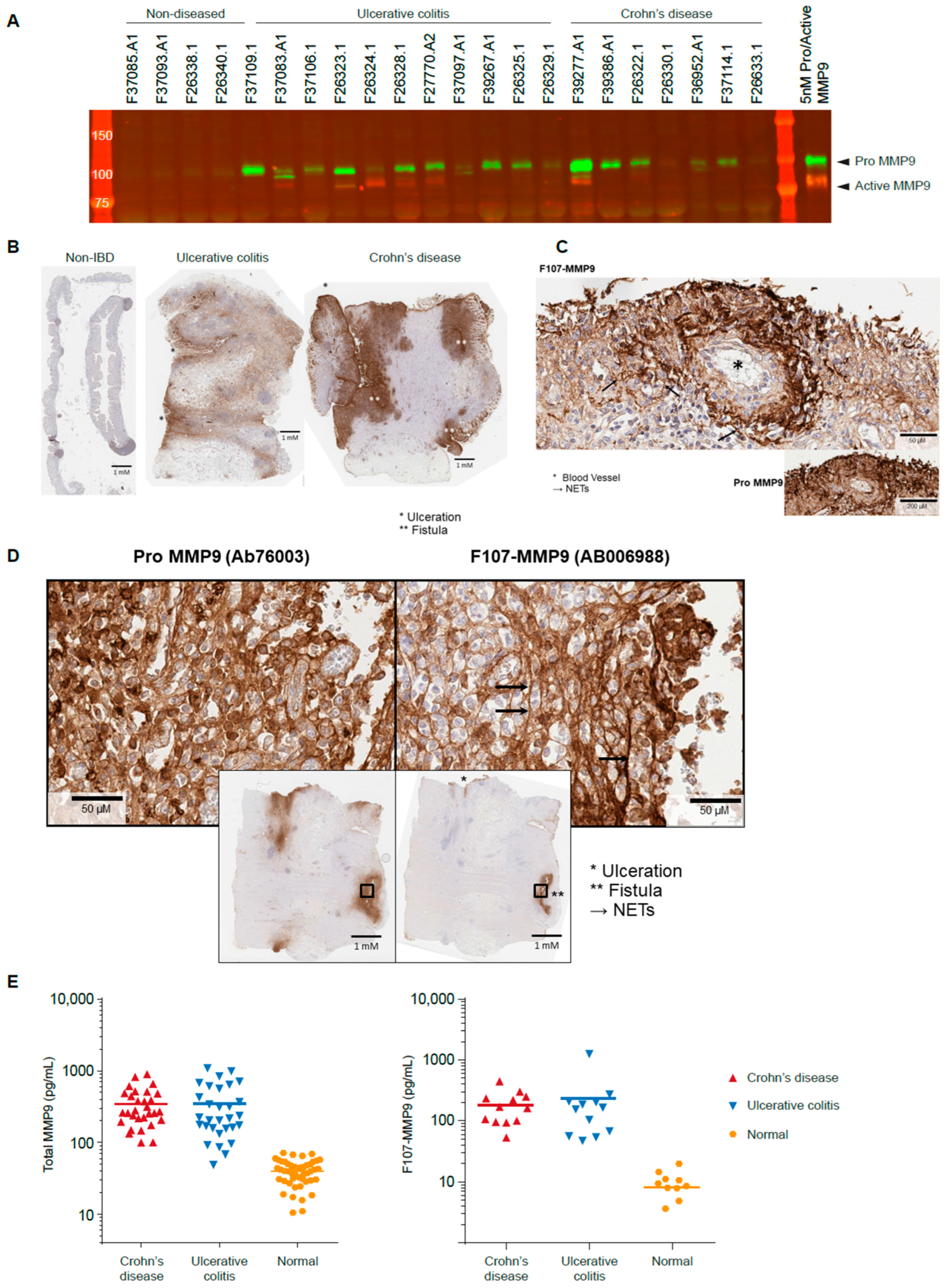
3.3. Active MMP9 Is Detectable in IBD
3.4. Active MMP9 Expression Is Pronounced in Inflamed Blood Vessels and Fistulae
3.5. Active MMP9 Is Observed in IBD Sera
3.6. Active MMP9 Expression in HS
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Transgene Amino Acid Sequences Used in This Report
- >MMP3G100L
- mkslpillllcvavcsaypldgaargedtsmnlvqkylenyydlkkdvkqfvrrkdsgpvvkkiremqkflglevtgkldsdtlevmrkprcLvpdvghfrtfpgipkwrkthltyrivnytpdlpkdavdsavekalkvweevtpltfsrlyegeadimisfavrehgdfypfdgpgnvlahayapgpgingdahfdddeqwtkdttgtnlflvaaheighslglfhsantealmyplyhsltdltrfrlsqddingiqslygpppdspetplvptepvppepgtpancdpalsfdavstlrgeilifkdrhfwrkslrklepelhlissfwpslpsgvdaayevtskdlvfifkgnqfwairgnevragyprgihtlgfpptvrkidaaisdkeknktyffvedkywrfdekrnsmepgfpkqiaedfpgidskidavfeefgffyfftgssqlefdpnakkvthtlksnswlnc
- >MMP9 TM
- mslwqplvlvllvlgccfaaprqrqstlvlfpgdlrtnltdrqlaeeylyrygytrvaemrgeskslgpallllqkqlslpetgeldsatlkamrtprcgvpdlgrfqtfegdlkwhhhnitywiqnysedlpraviddafarafalwsavtpltftrvysrdadiviqfgvaehgdgypfdgkdgllahafppgpgiqgdahfdddelwslgkgvvvptrfgnadgaachfpfifegrsysacttdgrsdglpwcsttanydtddrfgfcpserlytrdgnadgkpcqfpfifqgqsysacttdgrsdgyrwcattanydrdklfgfcptradstvmggnsagelcvfpftflgkeystctsegrgdgrlwcattsnfdsdkkwgfcpdqgyslflvaahefghalgldhssvpealmypmyrftegpplhkddvngirhlygprpepeprppttttpqptapptvcptgpptvhpserptagptgppsagptgpptagpstattvplspvddacnvnifdaiaeignqlylfkdgkywrfsegrgsrpqgpfliadkwpalprkldsvfeeplskklfffsgrqvwvytgasvlgprrldklglgadvaqvtgalrsgrgkmllfsgrrlwrfdvkaqmvdprsasevdrmfpgvpldthdvfqyrekayfcqdrfywrvssrselnqvdqvgyvtydilqcpedGTALSIVLPIVLLVFLCLGVFLLWKNWRLKN
- >>MMP9_dead_PDGFR
- METDTLLLWVLLLWVPGSTGDfqtfegdlkwhhhnitywiqnysedlpraviddafarafalwsavtpltftrvysrdadiviqfgvaehgdgypfdgkdgllahafppgpgiqgdahfdddelwslgkgvvvptrfgnadgaachfpfifegrsysacttdgrsdglpwcsttanydtddrfgfcpserlytrdgnadgkpcqfpfifqgqsysacttdgrsdgyrwcattanydrdklfgfcptradstvmggnsagelcvfpftflgkeystctsegrgdgrlwcattsnfdsdkkwgfcpdqgyslflvaahAfghalgldhssvpealmypmyrftegpplhkddvngirhlygprpepeEQKLISEEDLNAVGQDTQEVIVVPHSLPFKVVVISAILALVVLTIISLIILIMLWQKKPR
- >>MMP9_dead_PDGFR_D20A
- METDTLLLWVLLLWVPGSTGAfqtfegdlkwhhhnitywiqnysedlpraviddafarafalwsavtpltftrvysrdadiviqfgvaehgdgypfdgkdgllahafppgpgiqgdahfdddelwslgkgvvvptrfgnadgaachfpfifegrsysacttdgrsdglpwcsttanydtddrfgfcpserlytrdgnadgkpcqfpfifqgqsysacttdgrsdgyrwcattanydrdklfgfcptradstvmggnsagelcvfpftflgkeystctsegrgdgrlwcattsnfdsdkkwgfcpdqgyslflvaahAfghalgldhssvpealmypmyrftegpplhkddvngirhlygprpepeEQKLISEEDLNAVGQDTQEVIVVPHSLPFKVVVISAILALVVLTIISLIILIMLWQKKPR
- >>MMP9_dead_PDGFR_ΔD20
- METDTLLLWVLLLWVPGSTGfqtfegdlkwhhhnitywiqnysedlpraviddafarafalwsavtpltftrvysrdadiviqfgvaehgdgypfdgkdgllahafppgpgiqgdahfdddelwslgkgvvvptrfgnadgaachfpfifegrsysacttdgrsdglpwcsttanydtddrfgfcpserlytrdgnadgkpcqfpfifqgqsysacttdgrsdgyrwcattanydrdklfgfcptradstvmggnsagelcvfpftflgkeystctsegrgdgrlwcattsnfdsdkkwgfcpdqgyslflvaahAfghalgldhssvpealmypmyrftegpplhkddvngirhlygprpepeEQKLISEEDLNAVGQDTQEVIVVPHSLPFKVVVISAILALVVLTIISLIILIMLWQKKPR
References
- Ardi, V.C.; Van den Steen, P.E.; Opdenakker, G.; Schweighofer, B.; Deryugina, E.I.; Quigley, J.P. Neutrophil MMP-9 Proenzyme, Unencumbered by TIMP-1, Undergoes Efficient Activation in Vivo and Catalytically Induces Angiogenesis via a Basic Fibroblast Growth Factor (FGF-2)/FGFR-2 Pathway. J. Biol. Chem. 2009, 284, 25854–25866. [Google Scholar] [CrossRef] [PubMed]
- Deryugina, E.I.; Quigley, J.P. Pleiotropic Roles of Matrix Metalloproteinases in Tumor Angiogenesis: Contrasting, Overlapping and Compensatory Functions. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 103–120. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Buhmeida, A.; Bendardaf, R.; Hilska, M.; Collan, Y.; Laato, M.; Syrjänen, S.; Syrjänen, K.; Pyrhönen, S. Prognostic Significance of Matrix Metalloproteinase-9 (MMP-9) in Stage II Colorectal Carcinoma. J. Gastrointest. Cancer 2009, 40, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Zhang, Z.; Li, Y.; Zheng, J.; Dong, G.; Wang, W.; Ji, G. Matrix Metalloproteinase-9 Is Associated with Disease-Free Survival and Overall Survival in Patients with Gastric Cancer. Int. J. Cancer 2010, 129, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Zhao, Z.; Zhou, Y.; Li, Y.; Li, J.; Zheng, J.; Zhao, Q.; Wang, W. Matrix Metalloproteinase-9 Is Associated with Relapse and Prognosis of Patients with Colorectal Cancer. Ann. Surg. Oncol. 2011, 19, 318–325. [Google Scholar] [CrossRef]
- Al-Batran, S.-E.; Pauligk, C.; Wirtz, R.; Werner, D.; Steinmetz, K.; Homann, N.; Schmalenberg, H.; Hofheinz, R.-D.; Hartmann, J.; Atmaca, A.; et al. The Validation of Matrix Metalloproteinase-9 mRNA Gene Expression as a Predictor of Outcome in Patients with Metastatic Gastric Cancer. Ann. Oncol. 2012, 23, 1699–1705. [Google Scholar] [CrossRef]
- Iniesta, P.; Morán, A.; De Juan, C.; Gómez, A.; Hernando, F.; García-Aranda, C.; Frías, C.; Díaz-López, A.; Rodríguez-Jiménez, F.-J.; Balibrea, J.-L.; et al. Biological and Clinical Significance of MMP-2, MMP-9, TIMP-1 and TIMP-2 in Non-Small Cell Lung Cancer. Oncol. Rep. 2007, 17, 217–223. [Google Scholar] [CrossRef]
- Ogata, Y.; Matono, K.; Sasatomi, T.; Ishibashi, N.; Ohkita, A.; Mizobe, T.; Ogo, S.; Ikeda, S.; Ozasa, H.; Shirouzu, K. The MMP-9 Expression Determined the Efficacy of Postoperative Adjuvant Chemotherapy Using Oral Fluoropyrimidines in Stage II or III Colorectal Cancer. Cancer Chemother. Pharmacol. 2005, 57, 577–583. [Google Scholar] [CrossRef]
- Roy, R.; Yang, J.; Moses, M.A. Matrix Metalloproteinases As Novel Biomarkers and Potential Therapeutic Targets in Human Cancer. J. Clin. Oncol. 2009, 27, 5287–5297. [Google Scholar] [CrossRef]
- Shao, W.; Wang, W.; Xiong, X.-G.; Cao, C.; Yan, T.D.; Chen, G.; Chen, H.; Yin, W.; Liu, J.; Gu, Y.; et al. Prognostic Impact of MMP-2 and MMP-9 Expression in Pathologic Stage IA Non-Small Cell Lung Cancer. J. Surg. Oncol. 2011, 104, 841–846. [Google Scholar] [CrossRef]
- Svagzdys, S.; Lesauskaite, V.; Pangonyte, D.; Saladzinskas, Z.; Tamelis, A.; Pavalkis, D. Matrix Metalloproteinase-9 Is a Prognostic Marker to Predict Survival of Patients Who Underwent Surgery Due to Rectal Carcinoma. Tohoku J. Exp. Med. 2011, 223, 67–73. [Google Scholar] [CrossRef]
- Springman, E.B.; Angleton, E.L.; Birkedal-Hansen, H.; Van Wart, H.E. Multiple Modes of Activation of Latent Human Fibroblast Collagenase: Evidence for the Role of a Cys73 Active-Site Zinc Complex in Latency and a “Cysteine Switch” Mechanism for Activation. Proc. Natl. Acad. Sci. USA 1990, 87, 364–368. [Google Scholar] [CrossRef]
- Toth, M.; Chvyrkova, I.; Bernardo, M.; Hernandez-Barrantes, S.; Fridman, R. Pro-MMP-9 Activation by the MT1-MMP/MMP-2 Axis and MMP-3: Role of TIMP-2 and Plasma Membranes. Biochem. Biophys. Res. Commun. 2003, 308, 386–395. [Google Scholar] [CrossRef]
- Vandooren, J.; Geurts, N.; Martens, E.; Van den Steen, P.E.; Opdenakker, G. Zymography Methods for Visualizing Hydrolytic Enzymes. Nat. Methods 2013, 10, 211–220. [Google Scholar] [CrossRef]
- Frankowski, H.; Gu, Y.H.; Heo, J.H.; Milner, R.; Del Zoppo, G.J. Use of Gel Zymography to Examine Matrix Metalloproteinase (Gelatinase) Expression in Brain Tissue or in Primary Glial Cultures. Methods Mol. Biol. 2012, 814, 221–233. [Google Scholar] [CrossRef]
- Jones, G.T. Matrix Metalloproteinases in Biologic Samples. Adv. Clin. Chem. 2014, 65, 199–219. [Google Scholar] [CrossRef]
- Koivunen, E.; Arap, W.; Valtanen, H.; Rainisalo, A.; Medina, O.P.; Heikkilä, P.; Kantor, C.; Gahmberg, C.G.; Salo, T.; Konttinen, Y.T.; et al. Tumor Targeting With a Selective Gelatinase Inhibitor. Nat. Biotechnol. 1999, 17, 768–774. [Google Scholar] [CrossRef]
- Krüger, A.; Arlt, M.J.; Gerg, M.; Kopitz, C.; Bernardo, M.M.; Chang, M.; Mobashery, S.; Fridman, R. Antimetastatic Activity of a Novel Mechanism-Based Gelatinase Inhibitor. Cancer Res. 2005, 65, 3523–3526. [Google Scholar] [CrossRef]
- Vempati, P.; Karagiannis, E.D.; Popel, A.S. A Biochemical Model of Matrix Metalloproteinase 9 Activation and Inhibition. J. Biol. Chem. 2007, 282, 37585–37596. [Google Scholar] [CrossRef]
- Fisher, K.E.; Fei, Q.; Laird, E.R.; Stock, J.L.; Allen, M.R.; Sahagan, B.G.; Strick, C.A. Engineering Autoactivating Forms of Matrix Metalloproteinase-9 and Expression of the Active Enzyme in Cultured Cells and Transgenic Mouse Brain. Biochemistry 2002, 41, 8289–8297. [Google Scholar] [CrossRef]
- Ramos-DeSimone, N.; Hahn-Dantona, E.; Sipley, J.; Nagase, H.; French, D.L.; Quigley, J.P. Activation of Matrix Metalloproteinase-9 (MMP-9) via a Converging Plasmin/Stromelysin-1 Cascade Enhances Tumor Cell Invasion. J. Biol. Chem. 1999, 274, 13066–13076. [Google Scholar] [CrossRef]
- Krüger, A.; Kates, R.E.; Edwards, D.R. Avoiding Spam in the Proteolytic Internet: Future Strategies for Anti-Metastatic MMP Inhibition. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 95–102. [Google Scholar] [CrossRef]
- Hu, J.; Van den Steen, P.; Sang, Q.-X.A.; Opdenakker, G. Matrix Metalloproteinase Inhibitors as Therapy for Inflammatory and Vascular Diseases. Nat. Rev. Drug Discov. 2007, 6, 480–498. [Google Scholar] [CrossRef]
- Vandenbroucke, R.; Libert, C. Is There New Hope for Therapeutic Matrix Metalloproteinase Inhibition? Nat. Rev. Drug Discov. 2014, 13, 904–927. [Google Scholar] [CrossRef]
- Ren, Z.; Chen, J.; Khalil, R.A. Zymography as a Research Tool in the Study of Matrix Metalloproteinase Inhibitors. Methods Mol. Biol. 2017, 1626, 79–102. [Google Scholar] [CrossRef]
- Marshall, D.C.; Lyman, S.K.; McCauley, S.; Kovalenko, M.; Spangler, R.; Liu, C.; Lee, M.; O’Sullivan, C.; Barry-Hamilton, V.; Ghermazien, H.; et al. Selective Allosteric Inhibition of MMP9 Is Efficacious in Preclinical Models of Ulcerative Colitis and Colorectal Cancer. PLoS ONE 2015, 10, e0127063. [Google Scholar] [CrossRef]
- Appleby, T.C.; Greenstein, A.; Hung, M.; Liclican, A.; Velasquez, M.; Villaseñor, A.G.; Wang, R.; Wong, M.H.; Liu, X.; Papalia, G.A.; et al. Biochemical Characterization and Structure Determination of a Potent, Selective Antibody Inhibitor of Human MMP9. J. Biol. Chem. 2017, 292, 6810–6820. [Google Scholar] [CrossRef]
- Gossage, D.L.; Cieslarová, B.; Ap, S.; Zheng, H.; Xin, Y.; Lal, P.; Chen, G.; Smith, V.; Sundy, J.S. Phase 1b Study of the Safety, Pharmacokinetics, and Disease-related Outcomes of the Matrix Metalloproteinase-9 Inhibitor Andecaliximab in Patients With Rheumatoid Arthritis. Clin. Ther. 2018, 40, 156–165.e5. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Bhandari, B.R.; Fogel, R.; Onken, J.; Yen, E.; Zhao, X.; Jiang, Z.; Ge, D.; Xin, Y.; Ye, Z.; et al. Randomised Clinical Trial: A Phase 1, Dose-Ranging Study of the Anti-Matrix Metalloproteinase-9 Monoclonal Antibody GS-5745 Versus Placebo for Ulcerative Colitis. Aliment. Pharmacol. Ther. 2016, 44, 157–169. [Google Scholar] [CrossRef]
- Beppu, M.; Ikebe, T.; Shirasuna, K. The Inhibitory Effects of Immunosuppressive Factors, Dexamethasone and Interleukin-4, on NF-KappaB-Mediated Protease Production by Oral Cancer. Biochim. Biophys. Acta 2002, 1586, 11–22. [Google Scholar] [CrossRef]
- Ogata, Y.; Enghild, J.; Nagase, H. Matrix Metalloproteinase 3 (Stromelysin) Activates the Precursor for the Human Matrix Metalloproteinase 9. J. Biol. Chem. 1992, 267, 3581–3584. [Google Scholar] [CrossRef]
- Imai, K.; Yokohama, Y.; Nakanishi, I.; Ohuchi, E.; Fujii, Y.; Nakai, N.; Okada, Y. Matrix Metalloproteinase 7 (Matrilysin) From Human Rectal Carcinoma Cells. Activation of the Precursor, Interaction With Other Matrix Metalloproteinases and Enzymic Properties. J. Biol. Chem. 1995, 270, 6691–6697. [Google Scholar] [CrossRef]
- Pittayapruek, P.; Meephansan, J.; Prapapan, O.; Komine, M.; Ohtsuki, M. Role of Matrix Metalloproteinases in Photoaging and Photocarcinogenesis. Int. J. Mol. Sci. 2016, 17, 868. [Google Scholar] [CrossRef]
- Greenfield, E.A. Standard Immunization of Rabbits. Cold Spring Harb. Protoc. 2020, 2020, 100305. [Google Scholar] [CrossRef]
- Seomun, Y.; Kim, J.-T.; Joo, C.-K. MMP-14 Mediated MMP-9 Expression is Involved in TGF-Beta1-Induced Keratinocyte Migration. J. Cell. Biochem. 2008, 104, 934–941. [Google Scholar] [CrossRef]
- Rasmussen, H.S.; McCann, P.P. Matrix Metalloproteinase Inhibition as a Novel Anticancer Strategy: A Review with Special Focus on Batimastat and Marimastat. Pharmacol. Ther. 1997, 75, 69–75. [Google Scholar] [CrossRef]
- Margesson, L.J.; Danby, F.W. Hidradenitis Suppurativa. Best Pract. Res. Clin. Obstet. Gynaecol. 2014, 28, 1013–1027. [Google Scholar] [CrossRef]
- Xu, D.; Suenaga, N.; Edelmann, M.J.; Fridman, R.; Muschel, R.J.; Kessler, B.M. Novel MMP-9 Substrates in Cancer Cells Revealed by a Label-free Quantitative Proteomics Approach. Mol. Cell. Proteom. 2008, 7, 2215–2228. [Google Scholar] [CrossRef]
- Van den Steen, P.E.; Husson, S.J.; Proost, P.; Van Damme, J.; Gopdenakker, G. Carboxyterminal Cleavage of the Chemokines MIG and IP-10 by Gelatinase B and Neutrophil Collagenase. Biochem. Biophys. Res. Commun. 2003, 310, 889–896. [Google Scholar] [CrossRef]
- Chua, P.K.; Melish, M.E.; Yu, Q.; Yanagihara, R.; Yamamoto, K.S.; Nerurkar, V.R. Elevated Levels of Matrix Metalloproteinase 9 and Tissue Inhibitor of Metalloproteinase 1 during the Acute Phase of Kawasaki Disease. Clin. Vaccine Immunol. 2003, 10, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, V.; van Putten, M.; Chaouch, A.; Garrood, P.; Straub, V.; Lochmüller, H.; Ginjaar, H.; Aartsma-Rus, A.; van Ommen, G.; Dunnen, J.D.; et al. Serum Matrix Metalloproteinase-9 (MMP-9) as a Biomarker for Monitoring Disease Progression in Duchenne Muscular Dystrophy (DMD). Neuromuscul. Disord. 2011, 21, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Stefanidakis, M.; Ruohtula, T.; Borregaard, N.; Gahmberg, C.G.; Koivunen, E. Intracellular and Cell Surface Localization of a Complex Between AlphaMbeta2 Integrin and Promatrix Metalloproteinase-9 Progelatinase in Neutrophils. J. Immunol. 2004, 172, 7060–7068. [Google Scholar] [CrossRef]
- Christensen, J.; Shastri, V.P. Matrix-Metalloproteinase-9 is Cleaved and Activated by Cathepsin K. BMC Res. Notes 2015, 8, 322. [Google Scholar] [CrossRef]
- Demestre, M.; Parkin-Smith, G.; Petzold, A.; Pullen, A. The Pro and the Active Form of Matrix Metalloproteinase-9 is Increased in Serum of Patients With Amyotrophic Lateral Sclerosis. J. Neuroimmunol. 2005, 159, 146–154. [Google Scholar] [CrossRef]
- Shao, L.; Zhang, B.; Wang, L.; Wu, L.; Kan, Q.; Fan, K. MMP-9-Cleaved Osteopontin Isoform Mediates Tumor Immune Escape by Inducing Expansion of Myeloid-Derived Suppressor Cells. Biochem. Biophys. Res. Commun. 2017, 493, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Melani, C.; Sangaletti, S.; Barazzetta, F.M.; Werb, Z.; Colombo, M.P. Amino-Biphosphonate–Mediated MMP-9 Inhibition Breaks the Tumor-Bone Marrow Axis Responsible for Myeloid-Derived Suppressor Cell Expansion and Macrophage Infiltration in Tumor Stroma. Cancer Res 2007, 67, 11438–11446. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velasquez, M.; O’Sullivan, C.; Brockett, R.; Mikels-Vigdal, A.; Mikaelian, I.; Smith, V.; Greenstein, A.E. Characterization of Active MMP9 in Chronic Inflammatory Diseases Using a Novel Anti-MMP9 Antibody. Antibodies 2023, 12, 9. https://doi.org/10.3390/antib12010009
Velasquez M, O’Sullivan C, Brockett R, Mikels-Vigdal A, Mikaelian I, Smith V, Greenstein AE. Characterization of Active MMP9 in Chronic Inflammatory Diseases Using a Novel Anti-MMP9 Antibody. Antibodies. 2023; 12(1):9. https://doi.org/10.3390/antib12010009
Chicago/Turabian StyleVelasquez, Maile, Chris O’Sullivan, Robert Brockett, Amanda Mikels-Vigdal, Igor Mikaelian, Victoria Smith, and Andrew E. Greenstein. 2023. "Characterization of Active MMP9 in Chronic Inflammatory Diseases Using a Novel Anti-MMP9 Antibody" Antibodies 12, no. 1: 9. https://doi.org/10.3390/antib12010009
APA StyleVelasquez, M., O’Sullivan, C., Brockett, R., Mikels-Vigdal, A., Mikaelian, I., Smith, V., & Greenstein, A. E. (2023). Characterization of Active MMP9 in Chronic Inflammatory Diseases Using a Novel Anti-MMP9 Antibody. Antibodies, 12(1), 9. https://doi.org/10.3390/antib12010009





