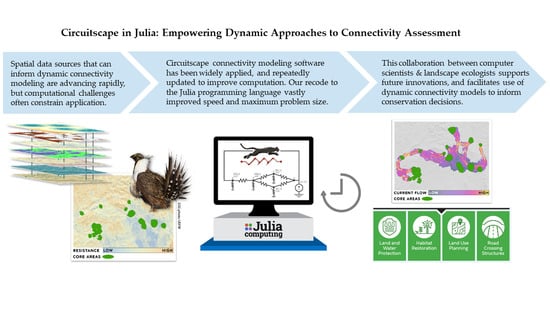
Circuitscape in Julia: Empowering Dynamic Approaches to Connectivity Assessment
Abstract
:1. Introduction
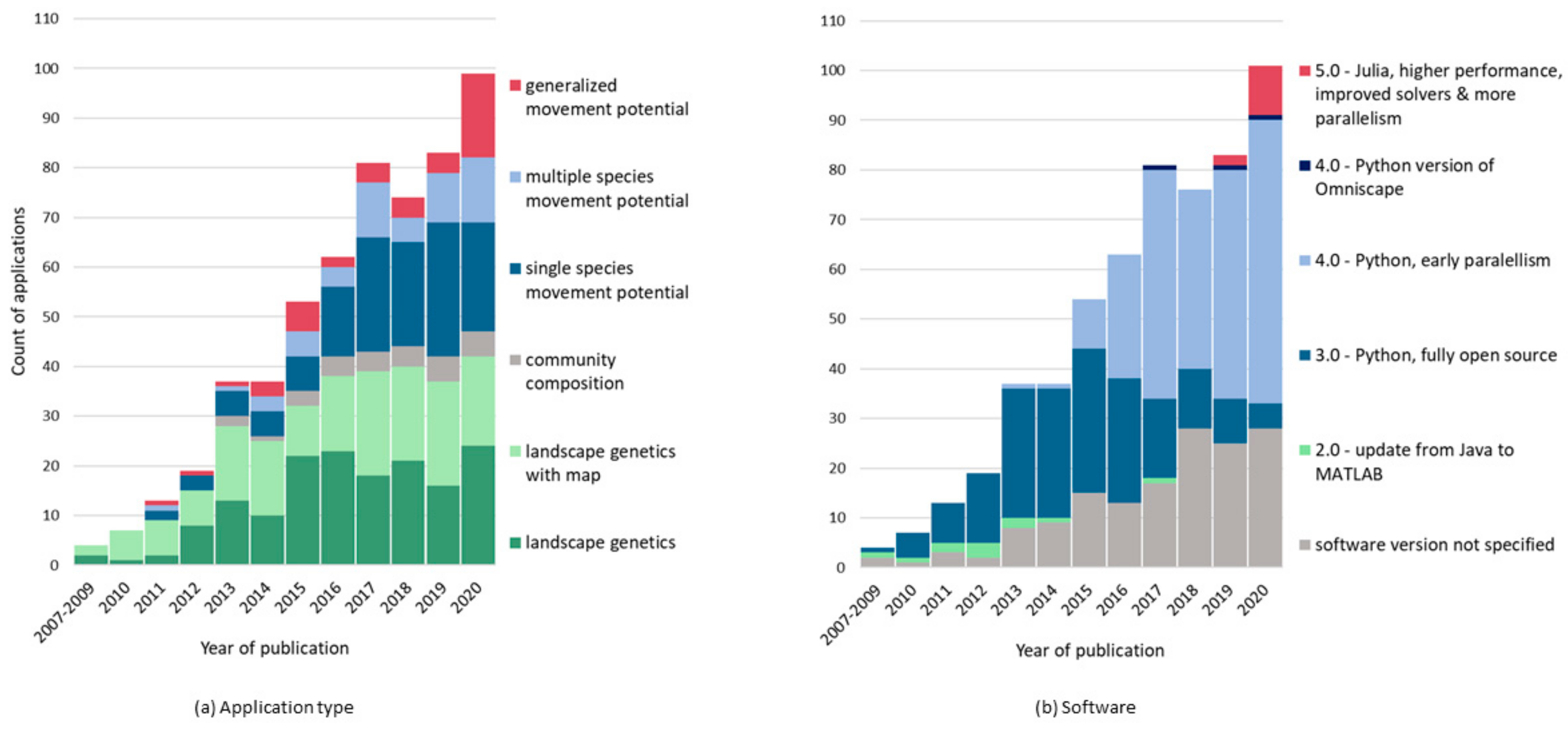
2. Dynamic Approaches to Connectivity: Circuitscape Applications as a Case Study
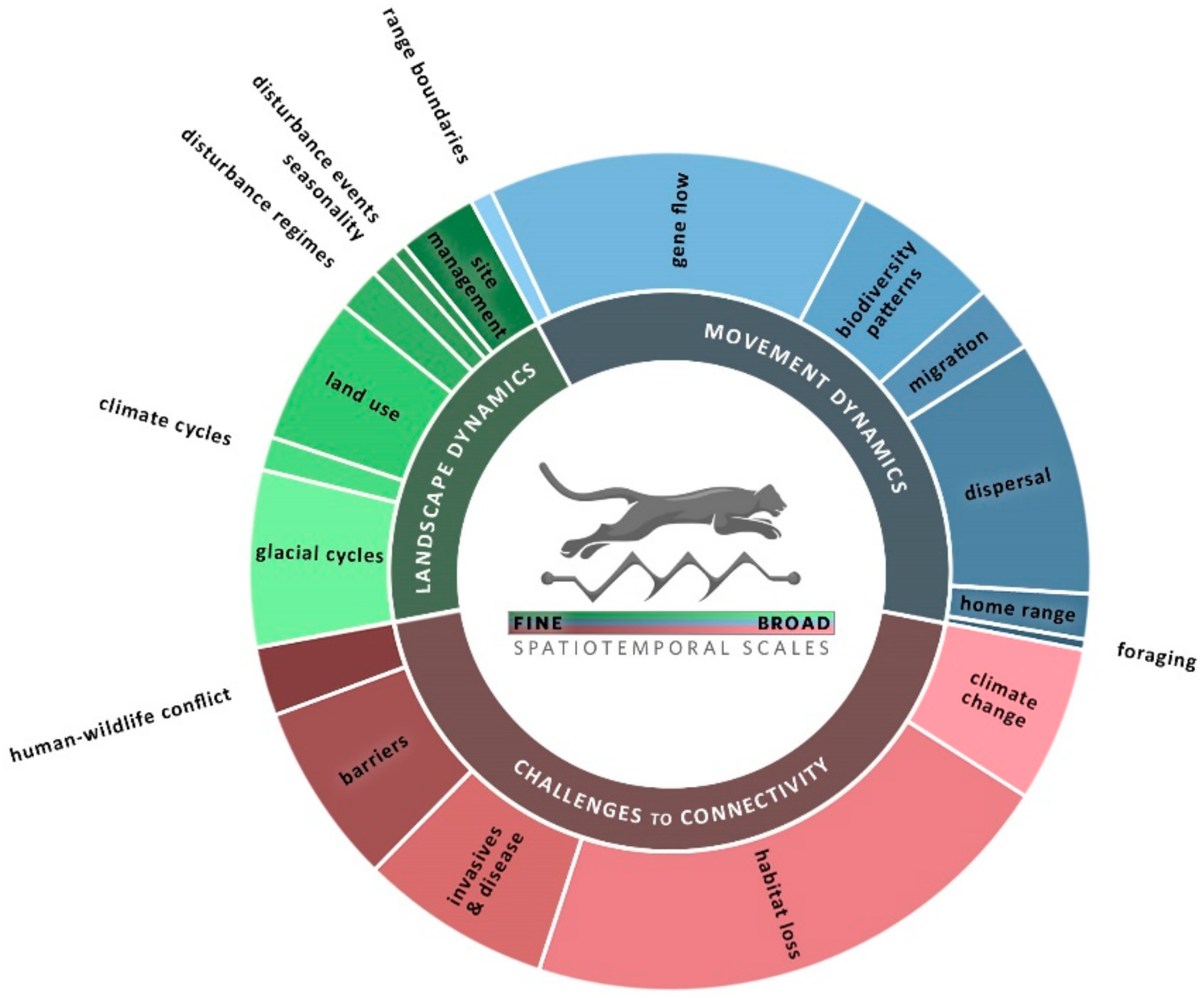
| Scale | Landscape Dynamics | Movement Dynamics | Challenges to Connectivity |
|---|---|---|---|
| Longer time & Larger extent | Glacial cycles Identify refugia & drivers of diversification [65,66,67,68]; detect a “glaciation signature” that provides context for current genetic variability [69,70,71] (LG). | Range boundaries Use multiple genetic markers to capture environmental constraints across generational times steps [72] (LG); test multiple factors to identify constraints [73] (MO). | Climate change Include projections of future climate [48,74,75,76,77] (ST & MO). Consider niche space under paleo, current, and future climates [78,79,80]; evaluate adaptive capacity [81,82] (LG). Integrate models of climate change & land use change [45,83,84,85,86] (ST, MO). |
| Climate cycles Evaluate the influence of climate variability on gene flow (“isolation by instability”) [87,88,89] (LG). Includes the influence of ocean currents [90] (ST). | Gene flow This group includes “snapshots” of genetic responses to heterogeneity. Also includes patterns in human language [91] & other distance-measurable traits. | ||
| Land use Evaluate broad-scale changes in land use in time series data [49] (ST); capture the genetic impact of forest cover change (pollen sources) by sampling trees of different ages [92] (LG). Model land use [93] (ST) or land function [94] (LG) scenarios, or the influence of land use policies [95] (ST). | Biodiversity patterns Identify changes in species assemblages & turnover as a function of resistance. Can incorporate genetics, morphology [96]. Consider multiple time steps [97] or assess many sites in different landscape contexts [98]; in freshwater, consider the role of different hydrologic periods or [99] precipitation events [100]. | Habitat loss/land use change Consider multiple habitat configurations or loss scenarios [101,102,103,104]; integrate multiple species [105,106]; evaluate changes in a protected area network [107,108,109] (LG, MO). Use multiple genetic methods to detect the influence of habitat loss relative to historical baselines [110,111,112,113,114,115,116] (LG). | |
| Disturbance regimes & succession Incorporate ocean currents & tidal influence [117,118,119] (LG & ST), fire regime [120], post-fire succession, [121] and dynamic (sand dunes) vs. static habitats [122] (LG). | Migration Incorporate seasonal patterns or variation in resources [27,123,124,125,126] (MO). Use multiple models to capture habitat choice and movement speed [42] (MO). | Invasive species & disease spread Use information from multiple regions or introduction events [127]; pair time series of high-resolution habitat data with on-the ground detections of invasive species [62]; evaluate barrier strategies [128] (MO). Use multiple waves to model viruses with wildlife vectors [129,130] (LG). Consider how dynamics of pathogens & hosts may interact [131,132,133], and be influenced by humans [134,135] (LG). | |
| Seasonality Change in habitat patch configuration due to seasonal flooding/drought (ST) [61,136,137,138]. (see migration for many seasonality examples with movement data). | Dispersal Recognize differences in home range use vs. dispersal [139,140,141,142] (MO); evaluate use of microclimates [40,143,144] (LG). Incorporate life history variation, e.g., by sex [145,146], seed dispersal type [147], or response to conspecifics [148] (LG & MO). | ||
| Shorter time & Smaller extent | Disturbance events Disturbance events like fires can change dispersal patterns [149] (LG). Consider hurricane routes as a driver of species colonization [150] (LG). | Home range Consider variations by sex and/or seasons [151,152] (LG, MO), and the influence of social behavior [153] (MO). Home range may include multiple habitat types [154,155]; agriculture can be a key connector [156,157] (MO). | Barriers to migration Compare multiple sites [158,159] (LG); model current & future urbanization [160] or barrier removal [161] (MO), or passage structure locations [162] (MO). Consider demographic differences in response to barriers (LG) [163,164]. |
| Site restoration & management Use scenarios to evaluate the targeting of fire risk mitigation [165] (ST) & vegetation management [166,167] (MO). Integrate connectivity benefits & costs [168]; evaluate connectivity benefits of restoration [169] (MO). | Foraging Consider differences in foraging mode (active vs. passive) [170] (LG). Understanding reproductive timing may help scale the linkage between a dynamic resource and a focal species’ foraging range [171] (ST). | Human–wildlife conflict Incorporate season & time of day in studies of road crossing risk [43] (MO). Simulate changes in barriers [172]. Integrate social science to understand the human side of conflicts [173,174] (MO). |
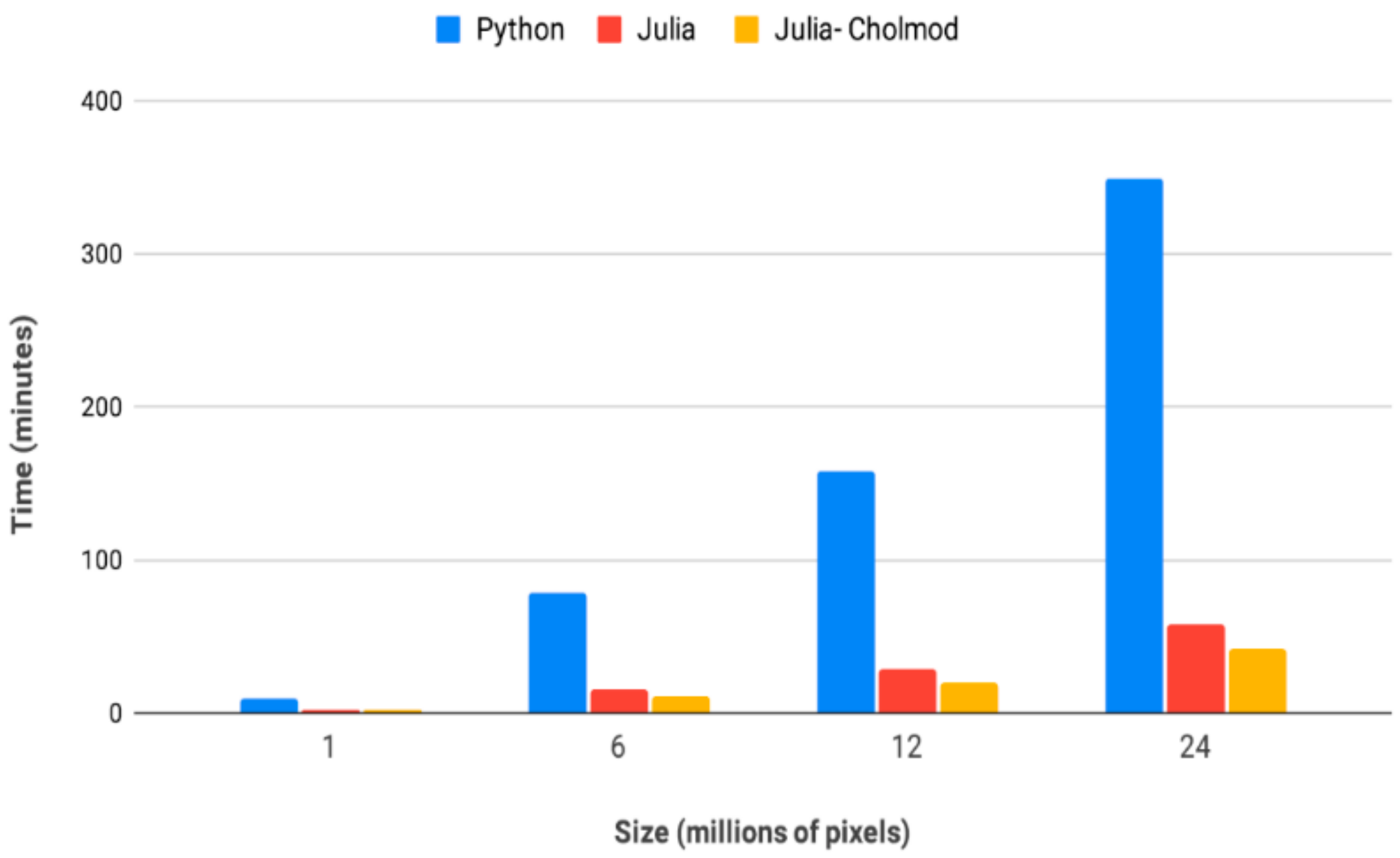
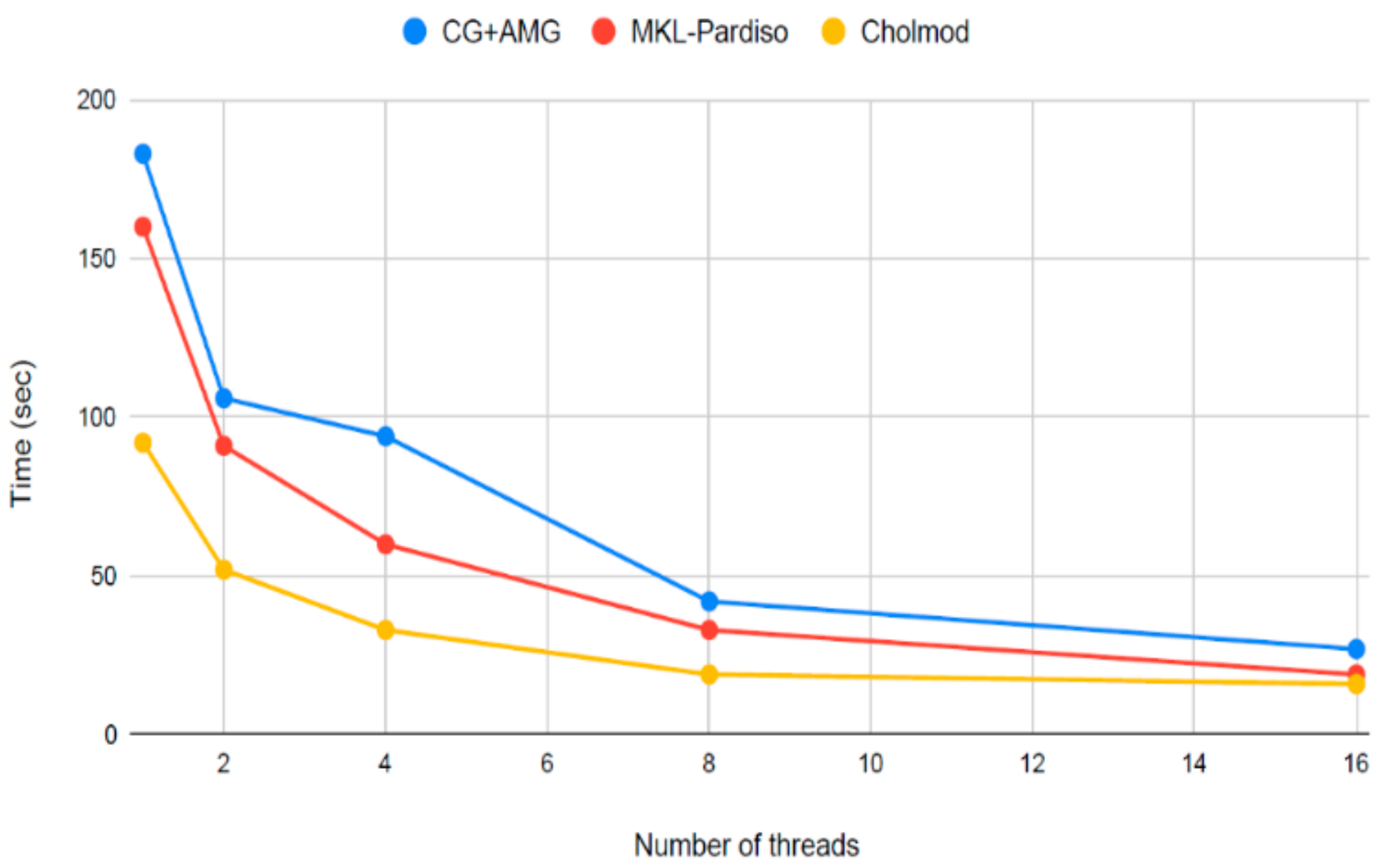
3. Computation as a Constraint to Dynamic Models and Driver of Innovation
4. An Introduction to the Julia Versions of Circuitscape and Omniscape
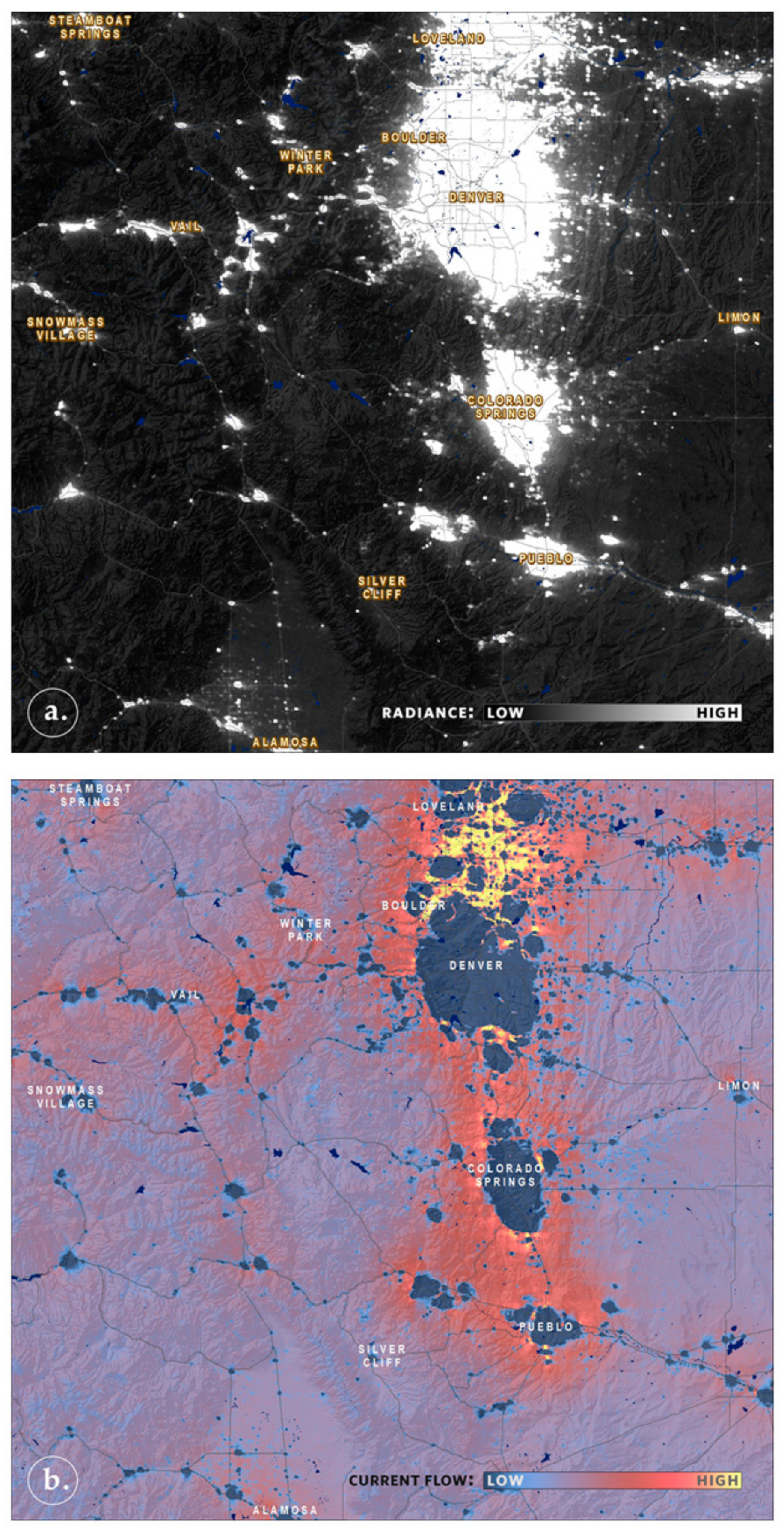
5. Faster Software Facilitates Data Exploration and Stakeholder Engagement
6. Dynamic Collaborations Empower Dynamic Approaches to Modeling
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B.; et al. Framing the concept of satellite remote sensing essential biodiversity variables: Challenges and future directions. Remote Sens. Ecol. Conserv. 2016, 2, 122–131. [Google Scholar] [CrossRef]
- Farley, S.S.; Dawson, A.; Goring, S.J.; Williams, J.W. Situating ecology as a big-data science: Current advances, challenges, and solutions. Bioscience 2018, 68, 563–576. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, R.; Fortin, M.-J. Spatial Ecology and Conservation Modeling; Applications with R; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Runting, R.K.; Phinn, S.; Xie, Z.; Venter, O.; Watson, J.E.M. Opportunities for big data in conservation and sustainability. Nat. Commun. 2020, 11, 2003. [Google Scholar] [CrossRef]
- Winsberg, E. Science in the Age of Computer Simulation; University of Chicago Press: Chicago, IL, USA, 2010; p. 168. [Google Scholar]
- Boyle, S.A.; Kennedy, C.M.; Torres, J.; Colman, K.; Perez-Estigarribia, P.E.; de la Sancha, N.U. High-resolution satellite imagery is an important yet underutilized resource in conservation biology. PLoS ONE 2014, 9, e86908. [Google Scholar] [CrossRef]
- Gomes, C.; Dietterich, T.; Barrett, C.; Conrad, J.; Dilkina, B.; Ermon, S.; Fang, F.; Farnsworth, A.; Fern, A.; Fern, X.; et al. Computational sustainability: Computing for a better world and a sustainable future. Commun. ACM 2019, 62, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Enquist, C.A.F.; Jackson, S.T.; Garfin, G.M.; Davis, F.W.; Gerber, L.R.; Littell, J.A.; Tank, J.L.; Terando, A.J.; Wall, T.U.; Halpern, B.; et al. Foundations of translational ecology. Front. Ecol. Environ. 2017, 15, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Wood, K.A.; Stillman, R.A.; Hilton, G.M. Conservation in a changing world needs predictive models. Anim. Conserv. 2018, 21, 87–88. [Google Scholar] [CrossRef] [Green Version]
- Mairota, P.; Cafarelli, B.; Didham, R.K.; Lovergine, F.P.; Lucas, R.M.; Nagendra, H.; Rocchini, D.; Tarantino, C. Challenges and opportunities in harnessing satellite remote-sensing for biodiversity monitoring. Ecol. Inform. 2015, 30, 207–214. [Google Scholar] [CrossRef]
- Neumann, W.; Martinuzzi, S.; Estes, A.B.; Pidgeon, A.M.; Dettki, H.; Ericsson, G.; Radeloff, V.C. Opportunities for the application of advanced remotely-sensed data in ecological studies of terrestrial animal movement. Mov. Ecol. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, B.L.; Wood, C.L.; Iliff, M.J.; Bonney, R.E.; Fink, D.; Kelling, S. eBird: A citizen-based bird observation network in the biological sciences. Biol. Conserv. 2009, 142, 2282–2292. [Google Scholar] [CrossRef]
- Kranstauber, B.; Cameron, A.; Weinzerl, R.; Fountain, T.; Tilak, S.; Wikelski, M.; Kays, R. The Movebank data model for animal tracking. Environ. Model. Software 2011, 26, 834–835. [Google Scholar] [CrossRef]
- Hilty, J.A.; Keeley, A.T.H.; Lidicker, W.Z., Jr.; Merenlender, A.M. Corridor Ecology: Linking Landscapes for Biodiversity Conservation and Climate Adaptation, 2nd ed.; Island Press: Washington, DC, USA, 2019. [Google Scholar]
- Wade, A.A.; McKelvey, K.S.; Schwartz, M.K. Resistance-Surface-Based Wildlife Conservation Connectivity Modeling: Summary of Efforts in the United States and Guide for Practitioners; Gen. Tech. Rep. RMRS-GTR-333; U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station: Fort Collins, CO, USA, 2015; p. 93.
- Dickson, B.G.; Albano, C.M.; Gray, M.E.; McClure, M.L.; Theobald, D.M.; Anantharaman, R.; Shah, V.B.; Beier, P.; Fargione, J.; Hall, K.R.; et al. Circuit-theory applications to connectivity science and conservation. Conserv. Biol. 2019, 33, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Diniz, M.F.; Cushman, S.A.; Machado, R.B.; De Marco, P. Landscape connectivity modeling from the perspective of animal dispersal. Landsc. Ecol. 2020, 35, 41–58. [Google Scholar] [CrossRef]
- Keeley, A.T.H.; Ackerly, D.D.; Cameron, D.R.; Heller, N.E.; Huber, P.R.; Schloss, C.A.; Thorne, J.H.; Merenlender, A.M. New concepts, models, and assessments of climate-wise connectivity. Environ. Res. Lett. 2018, 13, 073002. [Google Scholar] [CrossRef]
- Correa Ayram, C.A.; Mendoza, M.E.; Etter, A.; Salicrup, D.R.P. Habitat connectivity in biodiversity conservation: A review of recent studies and applications. Prog. Phys. Geogr.-Earth Environ. 2016, 40, 7–37. [Google Scholar] [CrossRef]
- Spear, S.F.; Balkenhol, N.; Fortin, M.J.; McRae, B.H.; Scribner, K. Use of resistance surfaces for landscape genetic studies: Considerations for parameterization and analysis. Mol. Ecol. 2010, 19, 3576–3591. [Google Scholar] [CrossRef]
- Zeller, K.A.; McGarigal, K.; Whiteley, A.R. Estimating landscape resistance to movement: A review. Landsc. Ecol. 2012, 27, 777–797. [Google Scholar] [CrossRef]
- McRae, B.H.; Dickson, B.G.; Keitt, T.H.; Shah, V.B. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 2008, 89, 2712–2724. [Google Scholar] [CrossRef]
- Baldwin, R.F.; Perkl, R.M.; Trombulak, S.C.; Burwell, W.B. Modeling Ecoregional Connectivity; Springer: New York, NY, USA, 2010; pp. 349–367. [Google Scholar]
- Spear, S.F.; Cushman, S.A.; McRae, B.H. Resistance surface modeling in landscape genetics. In Landscape Genetics: Concepts, Methods, Applications; Balkenhol, N., Cushman, S.A., Storfer, A.T., Waits, L.P., Eds.; Wiley and Sons: Oxford, UK, 2016; pp. 129–148. [Google Scholar]
- McRae, B.; Shah, V.; Mohapatra, T. Circuitscape 4 User Guide; The Nature Conservancy: Seattle, WA, USA, 2014; Available online: http://www.circuitscape.org (accessed on 8 September 2018).
- Koen, E.L.; Ellington, E.H.; Bowman, J. Mapping landscape connectivity for large spatial extents. Landsc. Ecol. 2019, 34, 2421–2433. [Google Scholar] [CrossRef]
- Burke, R.A.; Frey, J.K.; Ganguli, A.; Stoner, K.E. Species distribution modelling supports “nectar corridor” hypothesis for migratory nectarivorous bats and conservation of tropical dry forest. Divers. Distrib. 2019, 25, 1399–1415. [Google Scholar] [CrossRef]
- Draheim, H.M.; Moore, J.A.; Fortin, M.-J.; Scribner, K.T. Beyond the snapshot: Landscape genetic analysis of time series data reveal responses of American black bears to landscape change. Evol. Appl. 2018, 11, 1219–1230. [Google Scholar] [CrossRef]
- Huang, J.L.; Andrello, M.; Martensen, A.C.; Saura, S.; Liu, D.F.; He, J.H.; Fortin, M.J. Importance of spatio-temporal connectivity to maintain species experiencing range shifts. Ecography 2020, 43, 591–603. [Google Scholar] [CrossRef]
- Zeller, K.A.; Lewison, R.; Fletcher, R.J.; Tulbure, M.G.; Jennings, M.K. Understanding the importance of dynamic landscape connectivity. Land 2020, 9, 303. [Google Scholar] [CrossRef]
- Shah, V.; McRae, B.H. Circuitscape: A Tool for Landcape Ecology. In Proceedings of the 7th Python in Science Conference (SciPy 2008), Pasadena, CA, USA, 19–24 August 2008; Available online: http://conference.scipy.org/proceedings/scipy2008/SciPy2008_proceedings.pdf (accessed on 9 August 2020).
- Bezanson, J.; Chen, J.; Chung, B.; Karpinski, S.; Shah, V.B.; Vitek, J.; Zoubritzky, L. Julia: Dynamism and performance reconciled by design. Proc. ACM Program. Lang. 2018, 2, 120. [Google Scholar] [CrossRef] [Green Version]
- Bezanson, J.; Edelman, A.; Karpinski, S.; Shah, V.B. Julia: A fresh approach to numerical computing. SIAM Rev. 2017, 59, 65–98. [Google Scholar] [CrossRef] [Green Version]
- Anantharaman, R.; Hall, K.R.; Shah, V.B.; Edelman, A. Circuitscape in Julia: High performance connectivity modelling to support conservation decisions. JuliaCon Proc. 2020, 1, 58. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- McRae, B.H. Isolation by resistance. Evolution 2006, 60, 1551–1561. [Google Scholar] [CrossRef]
- Novembre, J.; Beier, P.; Fargione, J.; Lawler, J.; Selkoe, K. Brad McRae (1966–2017). Mol. Ecol. 2018, 27, 3035–3036. [Google Scholar] [CrossRef]
- Lawler, J.; Beier, P.; Dickson, B.; Fargione, J.; Novembre, J.; Theobald, D. A tribute to a true conservation innovator, Brad McRae, 1966–2017. Conserv. Biol. 2018. [Google Scholar] [CrossRef]
- McRae, B.H.; Beier, P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. USA 2007, 104, 19885–19890. [Google Scholar] [CrossRef] [Green Version]
- Peterman, W.E.; Connette, G.M.; Semlitsch, R.D.; Eggert, L.S. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol. Ecol. 2014, 23, 2402–2413. [Google Scholar] [CrossRef]
- Peterman, W.E.; Winiarski, K.J.; Moore, C.E.; Carvalho, C.D.; Gilbert, A.L.; Spear, S.F. A comparison of popular approaches to optimize landscape resistance surfaces. Landsc. Ecol. 2019, 34, 2197–2208. [Google Scholar] [CrossRef]
- Brennan, A.; Hanks, E.M.; Merkle, J.A.; Cole, E.K.; Dewey, S.R.; Courtemanch, A.B.; Cross, P.C. Examining speed versus selection in connectivity models using elk migration as an example. Landsc. Ecol. 2018, 33, 955–968. [Google Scholar] [CrossRef] [Green Version]
- Zeller, K.A.; Wattles, D.W.; Destefano, S. Evaluating methods for identifying large mammal road crossing locations: Black bears as a case study. Landsc. Ecol. 2020, 35, 1799–1808. [Google Scholar] [CrossRef]
- Crawford, J.A.; Peterman, W.E.; Kuhns, A.R.; Eggert, L.S. Altered functional connectivity and genetic diversity of a threatened salamander in an agroecosystem. Landsc. Ecol. 2016, 31, 2231–2244. [Google Scholar] [CrossRef]
- Dilts, T.E.; Weisberg, P.J.; Leitner, P.; Matocq, M.D.; Inman, R.D.; Nussear, K.E.; Esque, T.C. Multiscale connectivity and graph theory highlight critical areas for conservation under climate change. Ecol. Appl. 2016, 26, 1223–1237. [Google Scholar] [CrossRef]
- McRae, B.H.; Popper, K.; Jones, A.; Schindel, M.; Buttrick, S.; Hall, K.R.; Unnasch, R.S.; Platt, J. Conserving Nature’s Stage: Mapping Omnidirectional Connectivity for Resilient Terrestrial Landscapes in the Pacific Northwest; The Nature Conservancy: Portland, OR, USA, 2016; p. 47. Available online: http://nature.org/resilienceNW (accessed on 8 January 2017).
- Pelletier, D.; Clark, M.; Anderson, M.G.; Rayfield, B.; Wulder, M.A.; Cardille, J.A. Applying circuit theory for corridor expansion and management at regional scales: Tiling, pinch points, and omnidirectional connectivity. PLoS ONE 2014, 9, e84135. [Google Scholar] [CrossRef] [Green Version]
- Littlefield, C.E.; McRae, B.H.; Michalak, J.; Lawler, J.J.; Carroll, C. Connecting today’s climates to future analogs to facilitate species movement under climate change. Conserv. Biol. 2017, 31, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Baumann, M.; Kamp, J.; Potzschner, F.; Bleyhl, B.; Dara, A.; Hankerson, B.; Prishchepov, A.V.; Schierhorn, F.; Muller, D.; Holzel, N.; et al. Declining human pressure and opportunities for rewilding in the steppes of Eurasia. Divers. Distrib. 2020, 26, 1058–1070. [Google Scholar] [CrossRef]
- Koen, E.L.; Bowman, J.; Sadowski, C.; Walpole, A.A. Landscape connectivity for wildlife: Development and validation of multispecies linkage maps. Methods Ecol. Evol. 2014, 5, 626–633. [Google Scholar] [CrossRef]
- Landau, V.A.; Shah, V.B.; Anantharaman, R.; Hall, K.R. Omniscape.jl: Software to compute omnidirectional landscape connectivity. J. Open Source Softw. 2021, 6, 2829. [Google Scholar] [CrossRef]
- McRae, B.; Shah, V.B. Circuitscape User Guide. Version 3.5, Updated December 2011. Available online: https://www.researchgate.net/publication/265494222_Circuitscape_User_Guide (accessed on 30 October 2020).
- Yao, H.M.; Hsieh, Y.P.; Kong, J.; Hofmann, M. Modelling electrical conduction in nanostructure assemblies through complex networks. Nat. Mater. 2020, 19, 745–751. [Google Scholar] [CrossRef] [PubMed]
- McRae, B.H.; Kavanagh, D.M. Linkage Mapper Connectivity Analysis Software; The Nature Conservancy: Seattle, WA, USA, 2011; Available online: http://www.circuitscape.org/linkagemapper (accessed on 15 August 2018).
- Peterman, W.E. ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 2018, 9, 1638–1647. [Google Scholar] [CrossRef] [Green Version]
- Dellicour, S.; Rose, R.; Faria, N.R.; Lemey, P.; Pybus, O.G. SERAPHIM: Studying environmental rasters and phylogenetically informed movements. Bioinformatics 2016, 32, 3204–3206. [Google Scholar] [CrossRef]
- Leonard, P.B.; Duffy, E.B.; Baldwin, R.F.; McRae, B.H.; Shah, V.B.; Mohapatra, T.K. GFLOW: Software for modelling circuit theory-based connectivity at any scale. Methods Ecol. Evol. 2017, 8, 519–526. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudik, M.; Shapire, R.E. A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004; pp. 655–662. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, D.; Lapointe, M.E.; Wulder, M.A.; White, J.C.; Cardille, J.A. Forest connectivity regions of Canada using circuit theory and image analysis. PLoS ONE 2017, 12, e0169428. [Google Scholar] [CrossRef] [PubMed]
- Bishop-Taylor, R.; Tulbure, M.G.; Broich, M. Evaluating static and dynamic landscape connectivity modelling using a 25-year remote sensing time series. Landsc. Ecol. 2018, 33, 625–640. [Google Scholar] [CrossRef]
- Elmes, A.; Rogan, J.; Williams, C.; Ratick, S.; Nowak, D. Modeling the potential dispersal of Asian longhorned beetle using circuit theory. Prof. Geogr. 2019, 71, 580–594. [Google Scholar] [CrossRef]
- Filz, K.J.; Engler, J.O.; Stoffels, J.; Weitzel, M.; Schmitt, T. Missing the target? A critical view on butterfly conservation efforts on calcareous grasslands in south-western Germany. Biodivers. Conserv. 2013, 22, 2223–2241. [Google Scholar] [CrossRef]
- Brodie, J.F.; Newmark, W.D. Heterogeneous matrix habitat drives species occurrences in complex, fragmented landscapes. Am. Nat. 2019, 193, 748–754. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Knowles, L.L. Linking micro- and macroevolutionary perspectives to evaluate the role of Quaternary sea-level oscillations in island diversification. Evolution 2017, 71, 2901–2917. [Google Scholar] [CrossRef]
- He, Q.X.; Edwards, D.L.; Knowles, L.L. Integrative testing of how environments from the past to the present shape genetic structure across landscapes. Evolution 2013, 67, 3386–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, J.; Parra, J.L.; Goodman, S.M.; Raherilalao, M.J.; Vanderwal, J.; Bowie, R.C.K. Extending ecological niche models to the past 120000 years corroborates the lack of strong phylogeographic structure in the Crested Drongo (Dicrurus forficatus forficatus) on Madagascar. Biol. J. Linn. Soc. 2013, 108, 658–676. [Google Scholar] [CrossRef] [Green Version]
- Afroosheh, M.; Rodder, D.; Mikulicek, P.; Akmali, V.; Vaissi, S.; Fleck, J.; Schneider, W.; Sharifi, M. Mitochondrial DNA variation and Quaternary range dynamics in the endangered Yellow Spotted Mountain Newt, Neurergus derjugini (Caudata, Salamandridae). J. Zool. Syst. Evol. Res. 2019, 57, 580–590. [Google Scholar] [CrossRef]
- Yannic, G.; Pellissier, L.; Dubey, S.; Vega, R.; Basset, P.; Mazzotti, S.; Pecchioli, E.; Vernesi, C.; Hauffe, H.C.; Searle, J.B.; et al. Multiple refugia and barriers explain the phylogeography of the Valais shrew, Sorex antinorii (Mammalia: Soricomorpha). Biol. J. Linn. Soc. 2012, 105, 864–880. [Google Scholar] [CrossRef] [Green Version]
- Lanier, H.C.; Massatti, R.; He, Q.X.; Olson, L.E.; Knowles, L.L. Colonization from divergent ancestors: Glaciation signatures on contemporary patterns of genomic variation in Collared Pikas (Ochotona collaris). Mol. Ecol. 2015, 24, 3688–3705. [Google Scholar] [CrossRef] [Green Version]
- Koch, J.B.; Vandame, R.; Merida-Rivas, J.; Sagot, P.; Strange, J. Quaternary climate instability is correlated with patterns of population genetic variability in Bombus huntii. Ecol. Evol. 2018, 8, 7849–7864. [Google Scholar] [CrossRef] [Green Version]
- Zellmer, A.J.; Hanes, M.M.; Hird, S.M.; Carstens, B.C. Deep phylogeographic structure and environmental differentiation in the carnivorous plant Sarracenia alata. Syst. Biol. 2012, 61, 763–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walpole, A.A.; Bowman, J.; Murray, D.L.; Wilson, P.J. Functional connectivity of lynx at their southern range periphery in Ontario, Canada. Landsc. Ecol. 2012, 27, 761–773. [Google Scholar] [CrossRef]
- Malakoutikhah, S.; Fakheran, S.; Hemami, M.R.; Tarkesh, M.; Senn, J. Assessing future distribution, suitability of corridors and efficiency of protected areas to conserve vulnerable ungulates under climate change. Divers. Distrib. 2020, 26, 1383–1396. [Google Scholar] [CrossRef]
- Maher, S.P.; Morelli, T.L.; Hershey, M.; Flint, A.L.; Flint, L.E.; Moritz, C.; Beissinger, S.R. Erosion of refugia in the Sierra Nevada meadows network with climate change. Ecosphere 2017, 8, e01673. [Google Scholar] [CrossRef]
- Li, W.W.; Yu, Y.; Liu, P.; Tang, R.C.; Dai, Y.C.; Li, L.; Zhang, L. Identifying climate refugia and its potential impact on small population of Asian elephant (Elephas maximus) in China. Glob. Ecol. Conserv. 2019, 19, e00664. [Google Scholar] [CrossRef]
- Ashrafzadeh, M.R.; Naghipour, A.A.; Haidarian, M.; Kusza, S.; Pilliod, D.S. Effects of climate change on habitat and connectivity for populations of a vulnerable, endemic salamander in Iran. Glob. Ecol. Conserv. 2019, 19, e00637. [Google Scholar] [CrossRef]
- Barker, B.S.; Rodriguez-Robles, J.A.; Cook, J.A. Climate as a driver of tropical insular diversity: Comparative phylogeography of two ecologically distinctive frogs in Puerto Rico. Ecography 2015, 38, 769–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.S.; Gaddis, K.D.; Cairns, D.M.; Konganti, K.; Krutovsky, K.V. Landscape genomic insights into the historic migration of mountain hemlock in response to Holocene climate change. Am. J. Bot. 2017, 104, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Metzger, G.; Espindola, A.; Waits, L.P.; Sullivan, J. Genetic structure across broad spatial and temporal scales: Rocky Mountain tailed frogs (Ascaphus montanus; Anura: Ascaphidae) in the inland temperate rainforest. J. Hered. 2015, 106, 700–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razgour, O.; Forester, B.; Taggart, J.B.; Bekaert, M.; Juste, J.; Ibanez, C.; Puechmaille, S.J.; Novella-Fernandez, R.; Alberdi, A.; Manel, S. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Natl. Acad. Sci. USA 2019, 116, 10418–10423. [Google Scholar] [CrossRef] [Green Version]
- Razgour, O.; Taggart, J.B.; Manel, S.; Juste, J.; Ibanez, C.; Rebelo, H.; Alberdi, A.; Jones, G.; Park, K. An integrated framework to identify wildlife populations under threat from climate change. Molec. Ecol. Resour. 2018, 18, 18–31. [Google Scholar] [CrossRef] [PubMed]
- De Kort, H.; Prunier, J.G.; Tessier, M.; Turlure, C.; Baguette, M.; Stevens, V.M. Interacting grassland species under threat of multiple global change drivers. J. Biogeogr. 2018, 45, 2133–2145. [Google Scholar] [CrossRef]
- Hamilton, C.M.; Bateman, B.L.; Gorzo, J.M.; Reid, B.; Thogmartin, W.E.; Peery, M.Z.; Heglund, P.J.; Radeloff, V.C.; Pidgeon, A.M. Slow and steady wins the race? Future climate and land use change leaves the imperiled Blanding’s turtle (Emydoidea blandingii) behind. Biol. Conserv. 2018, 222, 75–85. [Google Scholar] [CrossRef]
- Jennings, M.K.; Haeuser, E.; Foote, D.; Lewison, R.L.; Conlisk, E. Planning for dynamic connectivity: Operationalizing robust decision-making and prioritization across landscapes experiencing climate and land-use change. Land 2020, 9, 341. [Google Scholar] [CrossRef]
- Leonard, P.B.; Sutherland, R.W.; Baldwin, R.F.; Fedak, D.A.; Carnes, R.G.; Montgomery, A.P. Landscape connectivity losses due to sea level rise and land use change. Anim. Conserv. 2017, 20, 80–90. [Google Scholar] [CrossRef]
- Bell, R.C.; Parra, J.L.; Tonione, M.; Hoskin, C.J.; Mackenzie, J.B.; Williams, S.E.; Moritz, C. Patterns of persistence and isolation indicate resilience to climate change in montane rainforest lizards. Mol. Ecol. 2010, 19, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.C.; Parra, J.L.; Badjedjea, G.; Barej, M.F.; Blackburn, D.C.; Burger, M.; Channing, A.; Dehling, J.M.; Greenbaum, E.; Gvozdik, V.; et al. Idiosyncratic responses to climate-driven forest fragmentation and marine incursions in reed frogs from Central Africa and the Gulf of Guinea Islands. Mol. Ecol. 2017, 26, 5223–5244. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Moran-Ordonez, A.; Garcia, J.T.; Calero-Riestra, M.; Alda, F.; Sanz, J.; Suarez-Seoane, S. Current landscape attributes and landscape stability in breeding grounds explain genetic differentiation in a long-distance migratory bird. Anim. Conserv. 2020. [Google Scholar] [CrossRef]
- Dambach, J.; Raupach, M.J.; Leese, F.; Schwarzer, J.; Engler, J.O. Ocean currents determine functional connectivity in an Antarctic deep-sea shrimp. Mar. Ecol. 2016, 37, 1336–1344. [Google Scholar] [CrossRef]
- Tarkhnishvili, D.; Gavashelishvili, A.; Murtskhvaladze, M.; Latsuzbaia, A. Landscape complexity in the Caucasus impedes genetic assimilation of human populations more effectively than language or ethnicity. Human Biol. 2016, 88, 287. [Google Scholar] [CrossRef] [PubMed]
- Oyama, K.; Herrera-Arroyo, M.L.; Rocha-Ramirez, V.; Benitez-Malvido, J.; Ruiz-Sanchez, E.; Gonzalez-Rodriguez, A. Gene flow interruption in a recently human-modified landscape: The value of isolated trees for the maintenance of genetic diversity in a Mexican endemic red oak. For. Ecol. Manage. 2017, 390, 27–35. [Google Scholar] [CrossRef]
- Goretskaia, M.I.; Beme, I.R.; Popova, D.V.; Amos, N.; Buchanan, K.L.; Sunnucks, P.; Pavlova, A. Song parameters of the fuscous honeyeater Lichenostomus fuscus correlate with habitat characteristics in fragmented landscapes. J. Avian Biol. 2018, 49, e01493. [Google Scholar] [CrossRef]
- Youngquist, M.B.; Inoue, K.; Berg, D.J.; Boone, M.D. Effects of land use on population presence and genetic structure of an amphibian in an agricultural landscape. Landsc. Ecol. 2017, 32, 147–162. [Google Scholar] [CrossRef]
- Liu, X.J.; Liu, D.F.; Zhao, H.Z.; He, J.H.; Liu, Y.L. Exploring the spatio-temporal impacts of farmland reforestation on ecological connectivity using circuit theory: A case study in the agro-pastoral ecotone of North China. J. Geogr. Sci. 2020, 30, 1419–1435. [Google Scholar] [CrossRef]
- Thomassen, H.A.; Fuller, T.; Buermann, W.; Mila, B.; Kieswetter, C.M.; Jarrin, P.; Cameron, S.E.; Mason, E.; Schweizer, R.; Schlunegger, J.; et al. Mapping evolutionary process: A multi-taxa approach to conservation prioritization. Evol. Appl. 2011, 4, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Palacio, R.D.; Kattan, G.H.; Pimm, S.L. Bird extirpations and community dynamics in an Andean cloud forest over 100 years of land-use change. Conserv. Biol. 2020, 34, 677–687. [Google Scholar] [CrossRef]
- Schirmel, J.; Thiele, J.; Entling, M.H.; Buchholz, S. Trait composition and functional diversity of spiders and carabids in linear landscape elements. Agric. Ecosyst. Environ. 2016, 235, 318–328. [Google Scholar] [CrossRef]
- Sarremejane, R.; Canedo-Arguelles, M.; Prat, N.; Mykra, H.; Muotka, T.; Bonada, N. Do metacommunities vary through time? Intermittent rivers as model systems. J. Biogeogr. 2017, 44, 2752–2763. [Google Scholar] [CrossRef]
- Razeng, E.; Moran-Ordonez, A.; Box, J.B.; Thompson, R.; Davis, J.; Sunnucks, P. A potential role for overland dispersal in shaping aquatic invertebrate communities in arid regions. Freshwat. Biol. 2016, 61, 745–757. [Google Scholar] [CrossRef]
- Grafius, D.R.; Corstanje, R.; Siriwardena, G.M.; Plummer, K.E.; Harris, J.A. A bird’s eye view: Using circuit theory to study urban landscape connectivity for birds. Landsc. Ecol. 2017, 32, 1771–1787. [Google Scholar] [CrossRef]
- Cameron, A.C.; Page, R.B.; Watling, J.I.; Hickerson, C.A.M.; Anthony, C.D. Using a comparative approach to investigate the relationship between landscape and genetic connectivity among woodland salamander populations. Conserv. Genet. 2019, 20, 1265–1280. [Google Scholar] [CrossRef]
- Correa Ayram, C.A.; Mendoza, M.E.; Salicrup, D.R.P.; Granados, E.L. Identifying potential conservation areas in the Cuitzeo Lake basin, Mexico by multitemporal analysis of landscape connectivity. J. Nat. Conserv. 2014, 22, 424–435. [Google Scholar] [CrossRef]
- Schaffer-Smith, D.; Swenson, J.J.; Boveda-Penalba, A.J. Rapid conservation assessment for endangered species using habitat connectivity models. Environ. Conserv. 2016, 43, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Fedorca, A.; Popa, M.; Jurj, R.; Ionescu, G.; Ionescu, O.; Fedorca, M. Assessing the regional landscape connectivity for multispecies to coordinate on-the-ground needs for mitigating linear infrastructure impact in Brasov—Prahova region. J. Nat. Conserv. 2020, 58, 11. [Google Scholar] [CrossRef]
- Meurant, M.; Gonzalez, A.; Doxa, A.; Albert, C.H. Selecting surrogate species for connectivity conservation. Biol. Conserv. 2018, 227, 326–334. [Google Scholar] [CrossRef]
- Jewitt, D.; Goodman, P.S.; Erasmus, B.F.N.; O’Connor, T.G.; Witkowski, E.T.F. Planning for the maintenance of floristic diversity in the face of land cover and climate change. Environ. Manag. 2017, 59, 792–806. [Google Scholar] [CrossRef]
- Castilho, C.S.; Hackbart, V.C.S.; Pivello, V.R.; dos Santos, R.F. Evaluating landscape connectivity for Puma concolor and Panthera onca among Atlantic forest protected areas. Environ. Manag. 2015, 55, 1377–1389. [Google Scholar] [CrossRef]
- Schindler, A.R.; Haukos, D.A.; Hagen, C.A.; Ross, B.E. A decision-support tool to prioritize candidate landscapes for lesser prairie-chicken conservation. Landsc. Ecol. 2020, 35, 1417–1434. [Google Scholar] [CrossRef]
- González-Serna, M.J.; Cordero, P.J.; Ortego, J. Using high-throughput sequencing to investigate the factors structuring genomic variation of a Mediterranean grasshopper of great conservation concern. Sci. Rep. 2018, 8, 13436. [Google Scholar] [CrossRef] [PubMed]
- Epps, C.W.; Wasser, S.K.; Keim, J.L.; Mutayoba, B.M.; Brashares, J.S. Quantifying past and present connectivity illuminates a rapidly changing landscape for the African elephant. Mol. Ecol. 2013, 22, 1574–1588. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.; Arcos, V.H.J.; de la Cruz, F.R.M.; Murphy, R.W. Landscape genetics of leaf-toed geckos in the tropical dry forest of Northern Mexico. PLoS ONE 2013, 8, e57433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chávez-Pesqueira, M.; Suarez-Montes, P.; Castillo, G.; Nunez-Farfan, J. Habitat fragmentation threatens wild populations of Carica papaya (Caricaceae) in a lowland rainforest. Am. J. Bot. 2014, 101, 1092–1101. [Google Scholar] [CrossRef] [Green Version]
- Landaverde-Gonzalez, P.; Baltz, L.M.; Escobedo-Kenefic, N.; Merida, J.; Paxton, R.J.; Husemann, M. Recent low levels of differentiation in the native Bombus ephippiatus (Hymenoptera: Apidae) along two Neotropical mountain-ranges in Guatemala. Biodivers. Conserv. 2018, 27, 3513–3531. [Google Scholar] [CrossRef]
- Landaverde-Gonzalez, P.; Enriquez, E.; Ariza, M.; Murray, T.; Paxton, R.J.; Husemann, M. Fragmentation in the clouds? The population genetics of the native bee Partamona bilineata (Hymenoptera: Apidae: Meliponini) in the cloud forests of Guatemala. Conserv. Genet. 2017, 18, 631–643. [Google Scholar] [CrossRef]
- Blair, C.; Arcos, V.H.J.; de la Cruz, F.R.M.; Murphy, R.W. Historical and contemporary demography of leaf-toed geckos (Phyllodactylidae: Phyllodactylus tuberculosus saxatilis) in the Mexican dry forest. Conserv. Genet. 2015, 16, 419–429. [Google Scholar] [CrossRef]
- Rouger, R.; Jump, A.S. A seascape genetic analysis reveals strong biogeographical structuring driven by contrasting processes in the polyploid saltmarsh species Puccinellia maritima and Triglochin maritima. Mol. Ecol. 2014, 23, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Seymour, M.; Rasanen, K.; Holderegger, R.; Kristjansson, B.K. Connectivity in a pond system influences migration and genetic structure in threespine stickleback. Ecol. Evol. 2013, 3, 492–502. [Google Scholar] [CrossRef]
- Afán, I.; Giménez, J.; Forero, M.G.; Ramirez, F. An adaptive method for identifying marine areas of high conservation priority. Conserv. Biol. 2018, 32, 1436–1447. [Google Scholar] [CrossRef]
- Ruiz-Lopez, M.J.; Barelli, C.; Rovero, F.; Hodges, K.; Roos, C.; Peterman, W.E.; Ting, N. A novel landscape genetic approach demonstrates the effects of human disturbance on the Udzungwa red colobus monkey (Procolobus gordonorum). Heredity 2016, 116, 167–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.L.; Landguth, E.L.; Bull, C.M.; Banks, S.C.; Gardner, M.G.; Driscoll, D.A. Dispersal responses override density effects on genetic diversity during post-disturbance succession. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152934. [Google Scholar] [CrossRef] [Green Version]
- Andrew, R.L.; Ostevik, K.L.; Ebert, D.P.; Rieseberg, L.H. Adaptation with gene flow across the landscape in a dune sunflower. Mol. Ecol. 2012, 21, 2078–2091. [Google Scholar] [CrossRef] [PubMed]
- Osipova, L.; Okello, M.M.; Njumbi, S.J.; Ngene, S.; Western, D.; Hayward, M.W.; Balkenhol, N. Using step-selection functions to model landscape connectivity for African elephants: Accounting for variability across individuals and seasons. Anim. Conserv. 2019, 22, 35–48. [Google Scholar] [CrossRef]
- Jahel, C.; Lenormand, M.; Seck, I.; Apolloni, A.; Toure, I.; Faye, C.; Sall, B.; Lo, M.; Diaw, C.S.; Lancelot, R.; et al. Mapping livestock movements in Sahelian Africa. Sci. Rep. 2020, 10, 8339. [Google Scholar] [CrossRef] [PubMed]
- McClure, M.L.; Hansen, A.J.; Inman, R.M. Connecting models to movements: Testing connectivity model predictions against empirical migration and dispersal data. Landsc. Ecol. 2016, 31, 1419–1432. [Google Scholar] [CrossRef]
- Poor, E.E.; Loucks, C.; Jakes, A.; Urban, D.L. Comparing habitat suitability and connectivity modeling methods for conserving pronghorn migrations. PLoS ONE 2012, 7, e49390. [Google Scholar] [CrossRef]
- Fischer, M.L.; Salgado, I.; Beninde, J.; Klein, R.; Frantz, A.C.; Heddergott, M.; Cullingham, C.I.; Kyle, C.J.; Hochkirch, A. Multiple founder effects are followed by range expansion and admixture during the invasion process of the raccoon (Procyon lotor) in Europe. Divers. Distrib. 2017, 23, 409–420. [Google Scholar] [CrossRef] [Green Version]
- Rees, E.E.; Pond, B.A.; Tinline, R.R.; Bélanger, D. Modelling the effect of landscape heterogeneity on the efficacy of vaccination for wildlife infectious disease control. J. Appl. Ecol. 2013, 50, 881–891. [Google Scholar] [CrossRef]
- Laenen, L.; Dellicour, S.; Vergote, V.; Nauwelaers, I.; De Coster, S.; Verbeeck, I.; Vanmechelen, B.; Lemey, P.; Maes, P. Spatio-temporal analysis of Nova virus, a divergent hantavirus circulating in the European mole in Belgium. Mol. Ecol. 2016, 25, 5994–6008. [Google Scholar] [CrossRef]
- Laenen, L.; Vergote, V.; Vanmechelen, B.; Tersago, K.; Baele, G.; Lemey, P.; Leirs, H.; Dellicour, S.; Vrancken, B.; Maes, P. Identifying the patterns and drivers of Puumala hantavirus enzootic dynamics using reservoir sampling. Virus Evol. 2019, 5, vez009. [Google Scholar] [CrossRef] [Green Version]
- Gryseels, S.; Baird, S.J.E.; Borremans, B.; Makundi, R.; Leirs, H.; de Bellocq, J.G. When viruses don’t go viral: The importance of host phylogeographic structure in the spatial spread of arenaviruses. PLoS Pathog. 2017, 13, e1006073. [Google Scholar] [CrossRef] [Green Version]
- Kozakiewicz, C.P.; Ricci, L.; Patton, A.H.; Stahlke, A.R.; Hendricks, S.A.; Margres, M.J.; Ruiz-Aravena, M.; Hamilton, D.G.; Hamede, R.; McCallum, H.; et al. Comparative landscape genetics reveals differential effects of environment on host and pathogen genetic structure in Tasmanian devils (Sarcophilus harrisii) and their transmissible tumour. Mol. Ecol. 2020, 29, 3217–3233. [Google Scholar] [CrossRef]
- Trovao, N.S.; Baele, G.; Vrancken, B.; Bielejec, F.; Suchard, M.A.; Fargette, D.; Lemey, P. Host ecology determines the dispersal patterns of a plant virus. Virus Evol. 2015, 1, vev016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talbi, C.; Lemey, P.; Suchard, M.A.; Abdelatif, E.; Elharrak, M.; Jalal, N.; Faouzi, A.; Echevarria, J.E.; Moron, S.V.; Rambaut, A.; et al. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog. 2010, 6, e1001166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, S.G.; Carrel, M.; Kitchen, A.; Malanson, G.P.; Tamerius, J.; Ali, M.; Kayali, G. How’s the flu getting through? Landscape genetics suggests both humans and birds spread H5N1 in Egypt. Infect. Genet. Evol. 2017, 49, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Bishop-Taylor, R.; Tulbure, M.G.; Broich, M. Surface-water dynamics and land use influence landscape connectivity across a major dryland region. Ecol. Appl. 2017, 27, 1124–1137. [Google Scholar] [CrossRef]
- Bishop-Taylor, R.; Tulbure, M.G.; Broich, M. Surface water network structure, landscape resistance to movement and flooding vital for maintaining ecological connectivity across Australia’s largest river basin. Landsc. Ecol. 2015, 30, 2045–2065. [Google Scholar] [CrossRef]
- Bishop-Taylor, R.; Tulbure, M.G.; Broich, M. Impact of hydroclimatic variability on regional-scale landscape connectivity across a dynamic dryland region. Ecol. Indic. 2018, 94, 142–150. [Google Scholar] [CrossRef]
- Jackson, C.R.; Marnewick, K.; Lindsey, P.A.; Roskaft, E.; Robertson, M.P. Evaluating habitat connectivity methodologies: A case study with endangered African wild dogs in South Africa. Landsc. Ecol. 2016, 31, 1433–1447. [Google Scholar] [CrossRef]
- Maiorano, L.; Chiaverini, L.; Falco, M.; Ciucci, P. Combining multi-state species distribution models, mortality estimates, and landscape connectivity to model potential species distribution for endangered species in human dominated landscapes. Biol. Conserv. 2019, 237, 19–27. [Google Scholar] [CrossRef]
- Rio-Maior, H.; Nakamura, M.; Alvares, F.; Beja, P. Designing the landscape of coexistence: Integrating risk avoidance, habitat selection and functional connectivity to inform large carnivore conservation. Biol. Conserv. 2019, 235, 178–188. [Google Scholar] [CrossRef]
- Keeley, A.T.H.; Beier, P.; Keeley, B.W.; Fagan, M.E. Habitat suitability is a poor proxy for landscape connectivity during dispersal and mating movements. Landsc. Urban Plan. 2017, 161, 90–102. [Google Scholar] [CrossRef]
- Mulder, K.P.; Cortes-Rodriguez, N.; Grant, E.H.C.; Brand, A.; Fleischer, R.C. North-facing slopes and elevation shape asymmetric genetic structure in the range-restricted salamander Plethodon shenandoah. Ecol. Evol. 2019, 9, 5094–5105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, J.A.; Epps, C.W.; Jeffress, M.R.; Ray, C.; Rodhouse, T.J.; Schwalm, D. Replicated landscape genetic and network analyses reveal wide variation in functional connectivity for American pikas. Ecol. Appl. 2016, 26, 1660–1676. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Garduno, T.; Tellez-Valdes, O.; Manel, S.; Vazquez-Dominguez, E. Role of habitat heterogeneity and landscape connectivity in shaping gene flow and spatial population structure of a dominant rodent species in a tropical dry forest. J. Zool. 2016, 298, 293–302. [Google Scholar] [CrossRef]
- Maiorano, L.; Boitani, L.; Chiaverini, L.; Ciucci, P. Uncertainties in the identification of potential dispersal corridors: The importance of behaviour, sex, and algorithm. Basic Appl. Ecol. 2017, 21, 66–75. [Google Scholar] [CrossRef]
- Grasty, M.R.; Thompson, P.G.; Hendrickson, E.C.; Pheil, A.E.; Cruzan, M.B. Fine-scale habitat heterogeneity and vole runways influence seed dispersal in Plagiobothrys nothofulvus. Am. J. Bot. 2020, 107, 413–422. [Google Scholar] [CrossRef]
- DiLeo, M.F.; Husby, A.; Saastamoinen, M. Landscape permeability and individual variation in a dispersal-linked gene jointly determine genetic structure in the Glanville fritillary butterfly. Evol. Lett. 2018, 2, 544–556. [Google Scholar] [CrossRef] [Green Version]
- Pereoglou, F.; Lindenmayer, D.B.; Macgregor, C.; Ford, F.; Wood, J.; Banks, S.C. Landscape genetics of an early successional specialist in a disturbance-prone environment. Mol. Ecol. 2013, 22, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Andraca-Gomez, G.; Lombaert, E.; Ordano, M.; Perez-Ishiwara, R.; Boege, K.; Dominguez, C.A.; Fornoni, J. Local dispersal pathways during the invasion of the cactus moth, Cactoblastis cactorum, within North America and the Caribbean. Sci. Rep. 2020, 10, 10. [Google Scholar] [CrossRef]
- Shafer, A.B.A.; Northrup, J.M.; White, K.S.; Boyce, M.S.; Cote, S.D.; Coltman, D.W. Habitat selection predicts genetic relatedness in an alpine ungulate. Ecology 2012, 93, 1317–1329. [Google Scholar] [CrossRef]
- Mui, A.B.; Caverhill, B.; Johnson, B.; Fortin, M.J.; He, Y.H. Using multiple metrics to estimate seasonal landscape connectivity for Blanding’s turtles (Emydoidea blandingii) in a fragmented landscape. Landsc. Ecol. 2017, 32, 531–546. [Google Scholar] [CrossRef]
- Koen, E.L.; Tosa, M.I.; Nielsen, C.K.; Schauber, E.M. Does landscape connectivity shape local and global social network structure in white-tailed deer? PLoS ONE 2017, 12, e0173570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, S.J.; Watson, D.M.; Luck, G.W.; Spooner, P.G. Effects of landscape composition and connectivity on the distribution of an endangered parrot in agricultural landscapes. Landsc. Ecol. 2014, 29, 1249–1259. [Google Scholar] [CrossRef]
- Haase, C.G.; Fletcher, R.J.; Slone, D.H.; Reid, J.P.; Butler, S.M. Landscape complementation revealed through bipartite networks: An example with the Florida manatee. Landsc. Ecol. 2017, 32, 1999–2014. [Google Scholar] [CrossRef]
- Nogeire, T.M.; Davis, F.W.; Crooks, K.R.; McRae, B.H.; Lyren, L.M.; Boydston, E.E. Can orchards help connect Mediterranean ecosystems? Animal movement data alter conservation priorities. Am. Midl. Nat. 2015, 174, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Luck, G.W.; Spooner, P.G.; Watson, D.M.; Watson, S.J.; Saunders, M.E. Interactions between almond plantations and native ecosystems: Lessons learned from north-western Victoria. Ecol. Manag. Restor. 2014, 15, 4–15. [Google Scholar] [CrossRef]
- DiLeo, M.F.; Rouse, J.D.; Davila, J.A.; Lougheed, S.C. The influence of landscape on gene flow in the eastern massasauga rattlesnake (Sistrurus c. catenatus): Insight from computer simulations. Mol. Ecol. 2013, 22, 4483–4498. [Google Scholar] [CrossRef]
- Balbi, M.; Ernoult, A.; Poli, P.; Madec, L.; Guiller, A.; Martin, M.C.; Nabucet, J.; Beaujouan, V.; Petit, E.J. Functional connectivity in replicated urban landscapes in the land snail (Cornu aspersum). Mol. Ecol. 2018, 27, 1357–1370. [Google Scholar] [CrossRef]
- McClure, M.L.; Dickson, B.G.; Nicholson, K.L. Modeling connectivity to identify current and future anthropogenic barriers to movement of large carnivores: A case study in the American Southwest. Ecol. Evol. 2017, 7, 3762–3772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, T.; Sharma, S.; DeFries, R. Targeting restoration sites to improve connectivity in a tiger conservation landscape in India. PeerJ 2018, 6, e5587. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.J.; Huang, Q.Y.; Stabach, J.; Buho, H.; Leimgruber, P. Railway underpass location affects migration distance in Tibetan antelope (Pantholops hodgsonii). PLoS ONE 2019, 14, e0211798. [Google Scholar] [CrossRef] [Green Version]
- Chiappero, M.B.; Sommaro, L.V.; Priotto, J.W.; Wiernes, M.P.; Steinmann, A.R.; Gardenal, C.N. Spatio-temporal genetic structure of the rodent Calomys venustus in linear, fragmented habitats. J. Mammal. 2016, 97, 424–435. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Espona, S.; McLeod, J.E.; Franks, N.R. Landscape genetics of a top neotropical predator. Mol. Ecol. 2012, 21, 5969–5985. [Google Scholar] [CrossRef]
- Gray, M.E.; Dickson, B.G. Applying fire connectivity and centrality measures to mitigate the cheatgrass-fire cycle in the arid West, USA. Landsc. Ecol. 2016, 31, 1681–1696. [Google Scholar] [CrossRef]
- Estrada-Carmona, N.; Martínez-Salinas, A.; DeClerck, F.A.J.; Vílchez-Mendoza, S.; Garbach, K. Managing the farmscape for connectivity increases conservation value for tropical bird species with different forest-dependencies. J. Environ. Manag. 2019, 250, 109504. [Google Scholar] [CrossRef]
- Reinhardt, J.R.; Naugle, D.E.; Maestas, J.D.; Allred, B.; Evans, J.; Falkowski, M. Next-generation restoration for sage-grouse: A framework for visualizing local conifer cuts within a landscape context. Ecosphere 2017, 8, e01888. [Google Scholar] [CrossRef] [Green Version]
- Drake, J.C.; Griffis-Kyle, K.; McIntyre, N.E. Using nested connectivity models to resolve management conflicts of isolated water networks in the Sonoran Desert. Ecosphere 2017, 8, e01647. [Google Scholar] [CrossRef]
- Fagan, M.E.; DeFries, R.S.; Sesnie, S.E.; Arroyo-Mora, J.P.; Chazdon, R.L. Targeted reforestation could reverse declines in connectivity for understory birds in a tropical habitat corridor. Ecol. Appl. 2016, 26, 1456–1474. [Google Scholar] [CrossRef]
- De Fraga, R.; Lima, A.P.; Magnusson, W.E.; Ferrao, M.; Stow, A.J. Contrasting patterns of gene flow for Amazonian snakes that actively forage and those that wait in ambush. J. Hered. 2017, 108, 524–534. [Google Scholar] [CrossRef]
- Afán, I.; Chiaradia, A.; Forero, M.G.; Dann, P.; Ramirez, F. A novel spatio-temporal scale based on ocean currents unravels environmental drivers of reproductive timing in a marine predator. Proc. R. Soc. B Biol. Sci. 2015, 282. [Google Scholar] [CrossRef]
- Osipova, L.; Okello, M.M.; Njumbi, S.J.; Ngene, S.; Western, D.; Hayward, M.W.; Balkenhol, N. Fencing solves human-wildlife conflict locally but shifts problems elsewhere: A case study using functional connectivity modelling of the African elephant. J. Appl. Ecol. 2018, 55, 2673–2684. [Google Scholar] [CrossRef]
- Struebig, M.J.; Linkie, M.; Deere, N.J.; Martyr, D.J.; Millyanawati, B.; Faulkner, S.C.; Le Comber, S.C.; Mangunjaya, F.M.; Leader-Williams, N.; McKay, J.E.; et al. Addressing human-tiger conflict using socio-ecological information on tolerance and risk. Nat. Commun. 2018, 9, 3455. [Google Scholar] [CrossRef] [PubMed]
- Zarrate-Charry, D.A.; Massey, A.L.; Gonzalez-Maya, J.F.; Betts, M.G. Multi-criteria spatial identification of carnivore conservation areas under data scarcity and conflict: A jaguar case study in Sierra Nevada de Santa Marta, Colombia. Biodivers. Conserv. 2018, 27, 3373–3392. [Google Scholar] [CrossRef]
- Rakotomalala, M.; Vrancken, B.; Pinel-Galzi, A.; Ramavovololona, P.; Hebrard, E.; Randrianangaly, J.S.; Dellicour, S.; Lemey, P.; Fargette, D. Comparing patterns and scales of plant virus phylogeography: Rice yellow mottle virus in Madagascar and in continental Africa. Virus Evol. 2019, 5, vez023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howey, M.C.L. Multiple pathways across past landscapes: Circuit theory as a complementary geospatial method to least cost path for modeling past movement. J. Archaeol. Sci. 2011, 38, 2523–2535. [Google Scholar] [CrossRef]
- Tatem, A.J.; Hemelaar, J.; Gray, R.R.; Salemi, M. Spatial accessibility and the spread of HIV-1 subtypes and recombinants. AIDS 2012, 26, 2351–2360. [Google Scholar] [CrossRef]
- Gray, M.E.; Dickson, B.G. A new model of landscape-scale fire connectivity applied to resource and fire management in the Sonoran Desert, USA. Ecol. Appl. 2015, 25, 1099–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, M.E.; Dickson, B.G.; Nussear, K.E.; Esque, T.C.; Chang, T. A range-wide model of contemporary, omnidirectional connectivity for the threatened Mojave desert tortoise. Ecosphere 2019, 10, e02847. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.G.; Barnett, A.; Clark, M.; Olivero Sheldon, A.; Prince, J. Resilient and Connected Landscapes for Conservation across Eastern North America; The Nature Conservancy, Eastern Conservation Science: Boston, MA, USA, 2016; 161p, Available online: http://www.nature.ly/TNCResilience (accessed on 7 March 2017).
- Wiens, J. Spatial scaling in ecology. Funct. Ecol. 1989, 3, 385–397. [Google Scholar] [CrossRef]
- Van der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy array: A structure for efficient numerical computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef] [Green Version]
- Bell, W.; Olson, L.; Schroder, J. PyAMG: Algebraic Multigrid Solvers in Python v3. 0, 2015. Release 3. Available online: http://www.pyamg.org (accessed on 12 November 2020).
- Nissen, J.N. What Scientists Must Know about Hardware to Write Fast Code (Blog Post, April 19, 2020). 2020. Available online: https://biojulia.net/post/hardware/ (accessed on 12 November 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Wilk, A.J.; Donlon, K.C.; Peterman, W.E. Effects of habitat fragment size and isolation on the density and genetics of urban red-backed salamanders (Plethodon cinereus). Urban Ecosyst. 2020, 23, 761–773. [Google Scholar] [CrossRef]
- Wang, E.; Zhang, Q.; Shen, B.; Zhang, G.; Lu, X.; Wu, Q.; Wang, Y. Intel Math Kernel Library. In High Performance Computing on the Intel® Xeon Phi™; Springer: Cham, Switzerland, 2014; pp. 167–188. [Google Scholar] [CrossRef]
- Convention on Biological Diversity. Zero Draft of the Post-2020 Global Biodiversity Framework. 2020. Available online: https://www.cbd.int/doc/c/efb0/1f84/a892b98d2982a829962b6371/wg2020-02-03-en.pdf (accessed on 1 November 2020).
- Jennings, M.K.; Zeller, K.A.; Lewison, R.L. Supporting adaptive connectivity in dynamic landscapes. Land 2020, 9, 295. [Google Scholar] [CrossRef]
- Fleishman, E.; Anderson, J.; Dickson, B.G. Single-species and multiple-species connectivity models for large mammals on the Navajo Nation. West. N. Am. Nat. 2017, 77, 237–251. [Google Scholar] [CrossRef] [Green Version]
- Harihar, A.; Ghosh-Harihar, M.; MacMillan, D.C. Losing time for the tiger Panthera tigris: Delayed action puts a globally threatened species at risk of local extinction. Oryx 2018, 52, 78–88. [Google Scholar] [CrossRef] [Green Version]
- Mills, S.; Weiss, S.; Liang, C. VIIRS day/night band (DNB) stray light characterization and correction. Proc. SPIE 2013, 8866. [Google Scholar] [CrossRef]
- Elvidge, C.D.; Baugh, K.; Zhizhin, M.; Hsu, F.C.; Ghosh, T. VIIRS night-time lights. Int. J. Remote Sens. 2017, 38, 5860–5879. [Google Scholar] [CrossRef]
- NOAA. Visible Infrared Imaging Radiometer Suite (VIIRS) Day-Night Band (DNB) Nightly Mosaic Data. 2019. Available online: https://www.ngdc.noaa.gov/eog/viirs.html (accessed on 8 February 2020).
- Wall, T.U.; McNie, E.; Garfin, G.M. Use-inspired science: Making science usable by and useful to decision makers. Front. Ecol. Environ. 2017, 15, 551–559. [Google Scholar] [CrossRef] [Green Version]
- Voinov, A.; Jenni, K.; Gray, S.; Kolagani, N.; Glynn, P.D.; Bommel, P.; Prell, C.; Zellner, M.; Paolisso, M.; Jordan, R.; et al. Tools and methods in participatory modeling: Selecting the right tool for the job. Environ. Model. Softw. 2018, 109, 232–255. [Google Scholar] [CrossRef] [Green Version]
- Majka, D.R. Migrations in Motion. 2016. Available online: https://maps.tnc.org/migrations-in-motion (accessed on 20 May 2019).
- Lawler, J.J.; Ruesch, A.S.; Olden, J.D.; McRae, B.H. Projected climate-driven faunal movement routes. Ecol. Lett. 2013, 16, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Demšar, U.; Buchin, K.; Cagnacci, F.; Safi, K.; Speckmann, B.; Van de Weghe, N.; Weiskopf, D.; Weibel, R. Analysis and visualisation of movement: An interdisciplinary review. Mov. Ecol. 2015, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Perkel, J. Julia: Come for the syntax, stay for the speed. Nature 2019, 572, 141–142. [Google Scholar] [CrossRef] [PubMed]
- Poisot, T. Julia in Ecology: Why Multiple Dispatch is Good. 2017. Available online: https://armchairecology.blog/julia-in-ecology-why-multiple-dispatch-is-good/ (accessed on 1 December 2020).
- Poisot, T.; Bélisle, Z.; Hoebeke, L.; Stock, M.; Szefer, P. EcologicalNetworks.jl: Analysing ecological networks of species interactions. Ecography 2019, 42, 1850–1861. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hall, K.R.; Anantharaman, R.; Landau, V.A.; Clark, M.; Dickson, B.G.; Jones, A.; Platt, J.; Edelman, A.; Shah, V.B. Circuitscape in Julia: Empowering Dynamic Approaches to Connectivity Assessment. Land 2021, 10, 301. https://doi.org/10.3390/land10030301
Hall KR, Anantharaman R, Landau VA, Clark M, Dickson BG, Jones A, Platt J, Edelman A, Shah VB. Circuitscape in Julia: Empowering Dynamic Approaches to Connectivity Assessment. Land. 2021; 10(3):301. https://doi.org/10.3390/land10030301
Chicago/Turabian StyleHall, Kimberly R., Ranjan Anantharaman, Vincent A. Landau, Melissa Clark, Brett G. Dickson, Aaron Jones, Jim Platt, Alan Edelman, and Viral B. Shah. 2021. "Circuitscape in Julia: Empowering Dynamic Approaches to Connectivity Assessment" Land 10, no. 3: 301. https://doi.org/10.3390/land10030301