Picoplankton Groups and Their Responses to Environmental Factors in Small Cascade Hydropower Stations
Abstract
1. Introduction
2. Materials and Methods
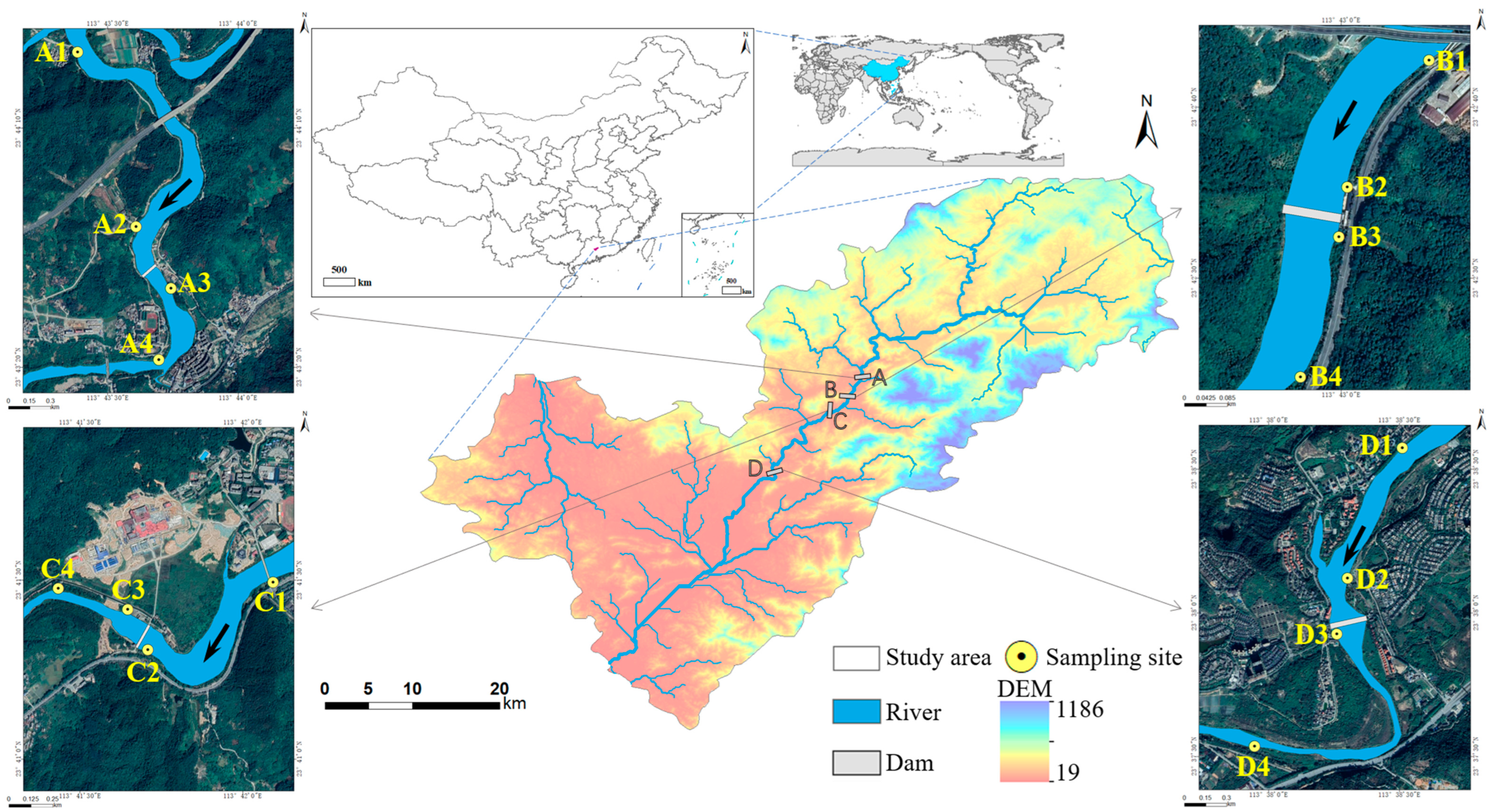
2.1. Study Area and Sampling
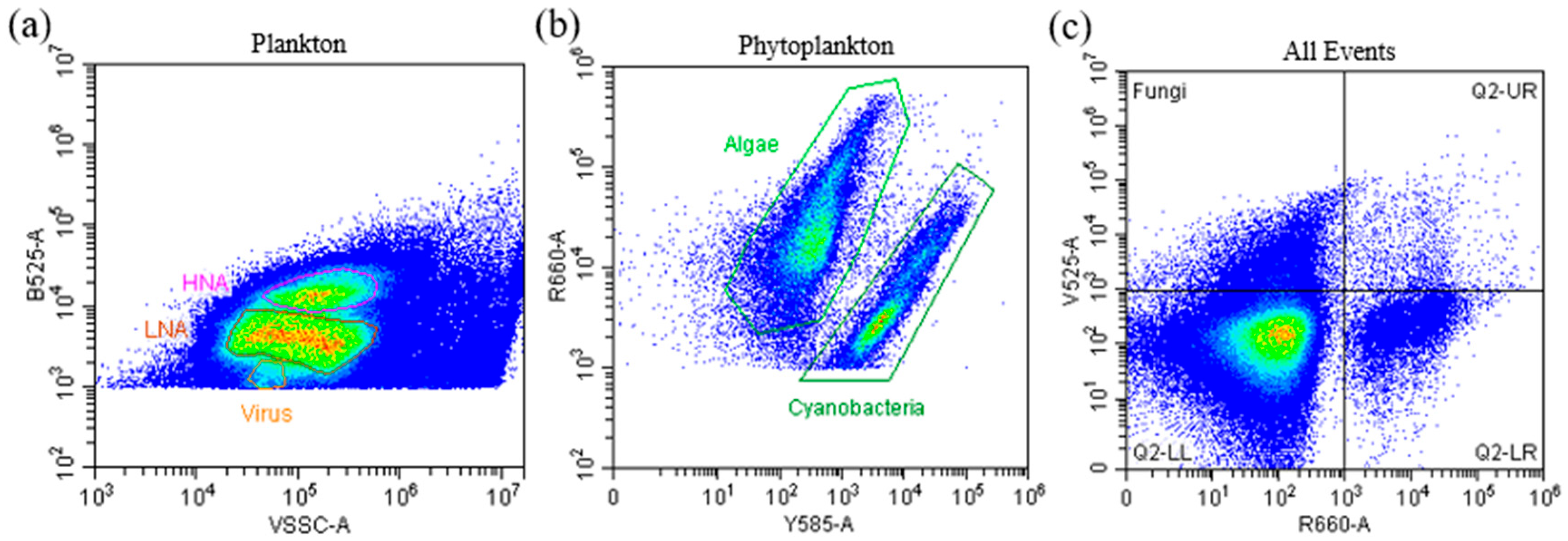
2.2. Sample Preparation and Flow Cytometry Analysis
2.3. Absolute Abundance of Picoplankton
2.4. Microbial Ecological Assessment Index
2.5. Data Processing and Statistical Analysis
3. Results
3.1. Physicochemical Properties of Water Samples
3.2. Absolute Abundance of Picoplankton
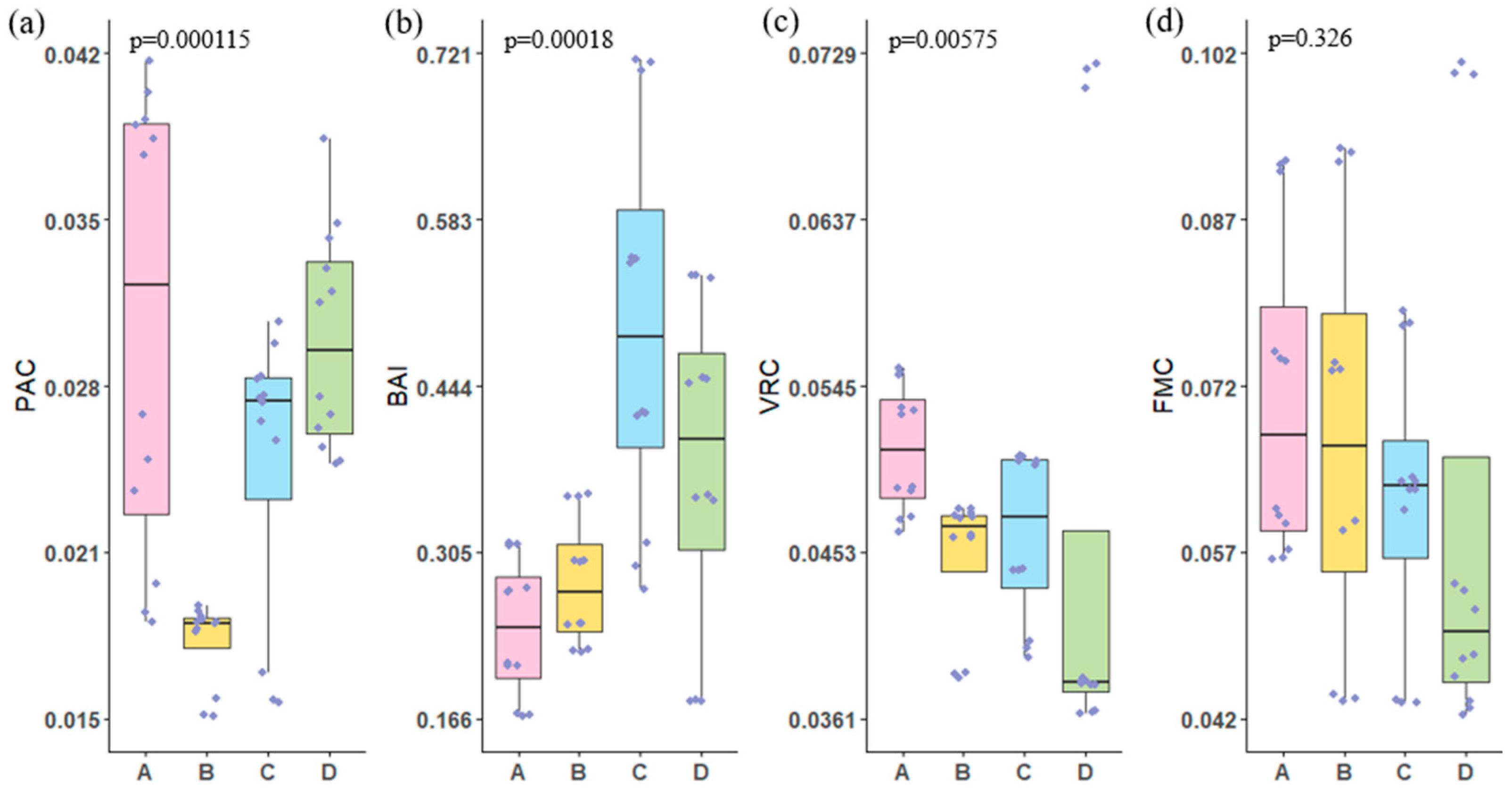
3.3. Microbial Ecological Assessment Index
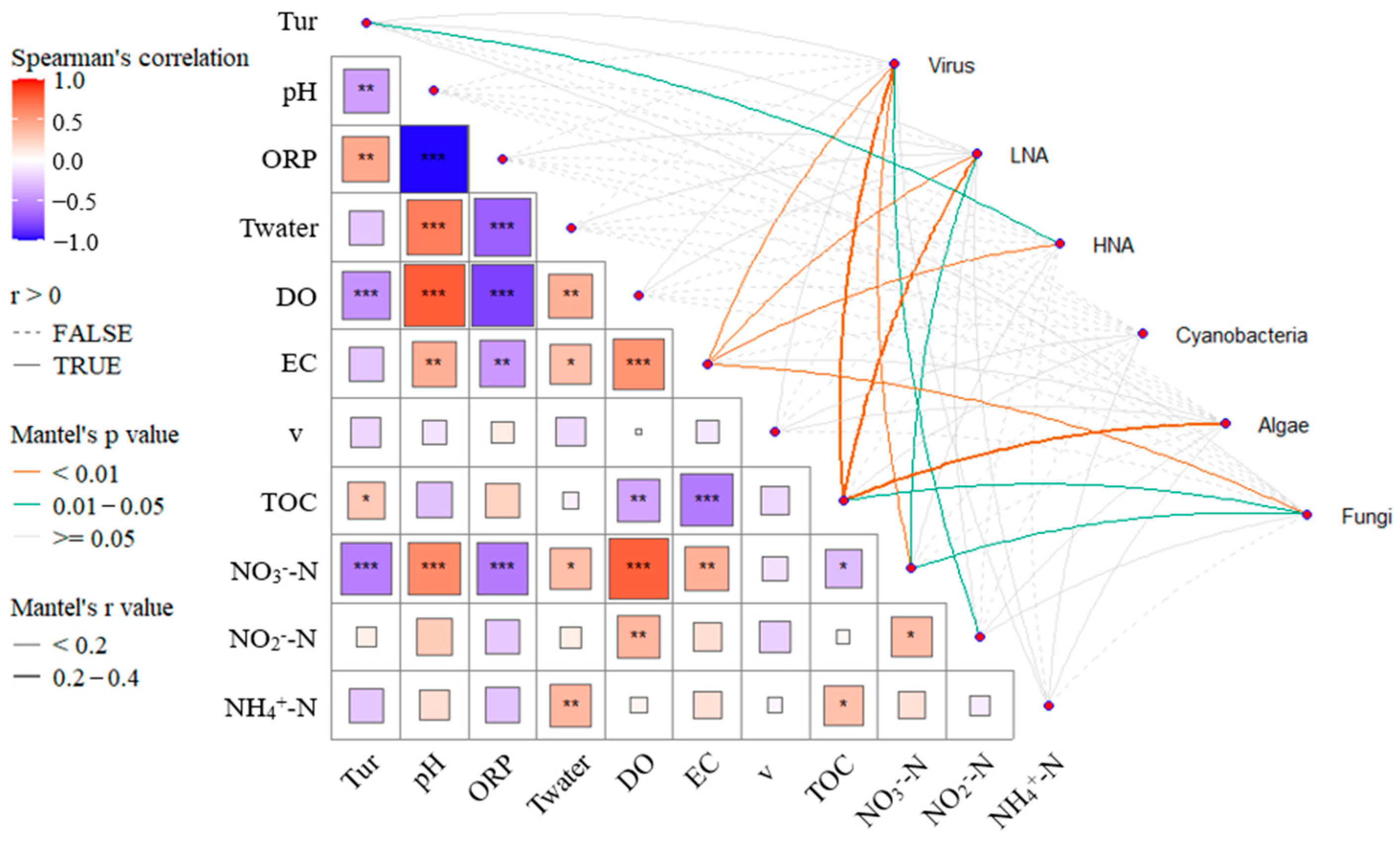
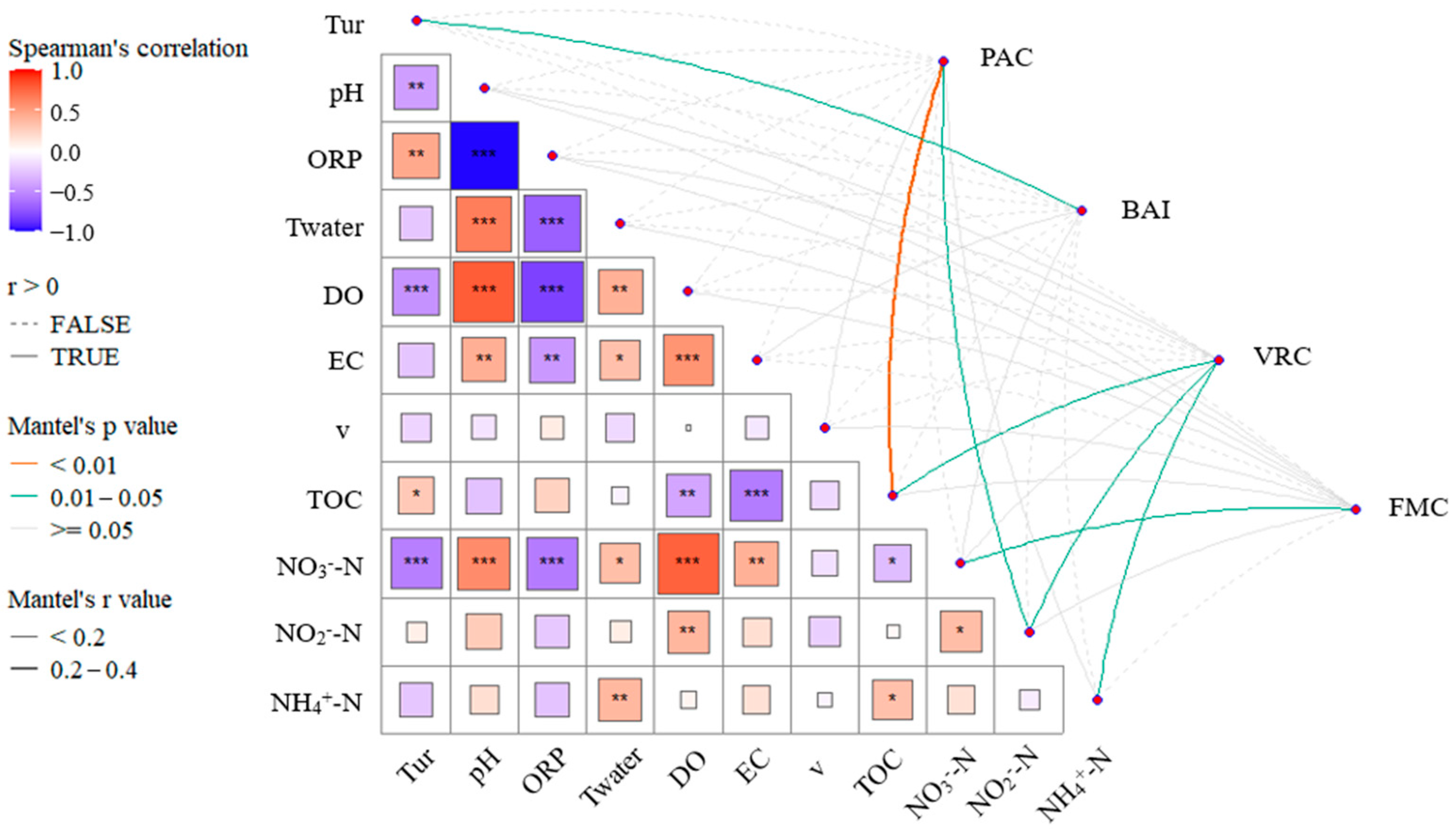
3.4. Analysis of Key Driving Factors
4. Discussion
4.1. Correlation Analysis Between Microbial Groups and Environmental Factors
4.2. Microbial Ecological Index for Assessing the Impact of Cascade Hydropower Stations
5. Conclusions
- Along the small cascade hydropower stations, DO and EC progressively increased from 5.71 mg/L and 49.87 μS/cm upstream to 6.80 mg/L and 56.18 μS/cm downstream, respectively. Meanwhile, the ORP and TOC concentrations decreased from 3.81 mV and 1.59 mg/L to −8.05 mV and 1.08 mg/L, respectively. No significant differences were observed in velocity and NO2−-N concentrations between the four cascade hydropower stations.
- Among the six microbial groups classified by flow cytometry, LNA had the highest abundance, with an average of 2551 cells/μL, and the abundance of Algae and Cyanobacteria was relatively low, averaging 47 cells/μL and 52 cells/μL, respectively. The abundance of Virus, LNA, and Fungi decreased by 30.9%, 30.5%, and 34.9%, respectively, along the cascade hydropower stations.
- EC, TOC, and NO3−-N concentrations are significantly correlated with the abundance of Virus, LNA, and Fungi. The concentrations of carbon and nitrogen nutrients significantly influence the microbial ecological assessment indices. Cascade hydropower stations significantly impact the PAC, BAI, and VRC of the river’s microbial ecosystem, while their influence on the FMC is relatively small. The VRC shows a decreasing trend along the cascade hydropower stations, suggesting a possible weakening effect of the stations on VRC.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| qPCR | quantitative Polymerase Chain Reaction |
| OTUs | Operational Taxonomic Units |
| ASVs | Amplicon Sequence Variants |
| eDNA | environmental Deoxyribonucleic Acid |
| FCM | Flow cytometry |
| CFW | Calcofluor White |
| LNA | Low Nucleic Acid |
| HNA | High Nucleic Acid |
| DNA | Deoxyribonucleic Acid |
| PAC | Photosynthetic Autotrophic Capacity |
| BAI | Bacterial Activity Index |
| VRC | Virus Regulatory Capacity |
| FMC | Fungal Metabolic Capacity |
| ORP | Oxidation-Reduction Potential |
| DO | Dissolved Oxygen |
| EC | Electrical Conductivity |
| TOC | Total Organic Carbon |
References
- Sachdev, H.S.; Akella, A.K.; Kumar, N. Analysis and evaluation of small hydropower plants: A bibliographical survey. Renew. Sustain. Energy Rev. 2015, 51, 1013–1022. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Li, S.L.; Liang, X.; Li, X.D.; Wang, F.; Yang, M.; Liu, C.Q. Anthropogenic regulation governs nutrient cycling and biological succession in hydropower reservoirs. Sci. Total Environ. 2022, 834, 155392. [Google Scholar] [CrossRef]
- Moran, E.F.; Lopez, M.C.; Moore, N.; Müller, N.; Hyndman, D.W. Sustainable hydropower in the 21st century. Proc. Natl. Acad. Sci. USA 2018, 115, 11891–11898. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Chen, X.; Zhang, L. Evaluating the impacts of small cascade hydropower from a perspective of stream health that integrates eco-environmental and hydrological values. J. Environ. Manag. 2022, 305, 114366. [Google Scholar] [CrossRef]
- He, F.; Zarfl, C.; Tockner, K.; Olden, J.D.; Campos, Z.; Muniz, F.; Svenning, J.C.; Jähnig, S.C. Hydropower impacts on riverine biodiversity. Nat. Rev. Earth Environ. 2024, 5, 755–772. [Google Scholar] [CrossRef]
- Li, W.; Wang, B.; Liu, N.; Yang, M.; Liu, C.Q.; Xu, S. River damming enhances ecological functional stability of planktonic microorganisms. Front. Microbiol. 2022, 13, 1049120. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.Z.; Hu, L.X.; Liu, Y.S.; Qiao, L.K.; Chen, Z.Y.; Su, J.Q.; He, L.Y.; Bai, H.; Zhu, Y.G.; Ying, G.G. Unveiling the overlooked small-sized microbiome in river ecosystems. Water Res. 2024, 265, 122302. [Google Scholar] [CrossRef]
- Drummond, J.D.; Davies-Colley, R.J.; Stott, R.; Sukias, J.P.; Nagels, J.W.; Sharp, A.; Packman, A.I. Retention and remobilization dynamics of fine particles and microorganisms in pastoral streams. Water Res. 2014, 66, 459–472. [Google Scholar] [CrossRef]
- Li, J.; Dong, S.; Liu, S.; Yang, Z.; Peng, M.; Zhao, C. Effects of cascading hydropower dams on the composition, biomass and biological integrity of phytoplankton assemblages in the middle Lancang-Mekong River. Ecol. Eng. 2013, 60, 316–324. [Google Scholar] [CrossRef]
- Vaulot, D.; Eikrem, W.; Viprey, M.; Moreau, H. The diversity of small eukaryotic phytoplankton (≤3 μm) in marine ecosystems. FEMS Microbiol. Rev. 2008, 32, 795–820. [Google Scholar] [CrossRef]
- Dashkova, V.; Malashenkov, D.; Poulton, N.; Vorobjev, I.; Barteneva, N.S. Imaging flow cytometry for phytoplankton analysis. Methods 2017, 112, 188–200. [Google Scholar] [PubMed]
- Coggins, L.X.; Larma, I.; Hinchliffe, A.; Props, R.; Ghadouani, A. Flow cytometry for rapid characterisation of microbial community dynamics in waste stabilisation ponds. Water Res. 2020, 169, 115243. [Google Scholar]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [PubMed]
- Li, K.; Hu, J.; Li, T.; Liu, F.; Tao, J.; Liu, J.; Zhang, Z.; Luo, X.; Li, L.; Deng, Y.; et al. Microbial abundance and diversity investigations along rivers: Current knowledge and future directions. Wiley Interdiscip. Rev. Water 2021, 8, e1547. [Google Scholar]
- Roots, P.; Wang, Y.; Rosenthal, A.F.; Griffin, J.S.; Sabba, F.; Petrovich, M.; Yang, F.; Kozak, J.A.; Zhang, H.; Wells, G.F. Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor. Water Res. 2019, 157, 396–405. [Google Scholar] [PubMed]
- Perry, W.B.; Seymour, M.; Orsini, L.; Jâms, I.B.; Milner, N.; Edwards, F.; Harvey, R.; de Bruyn, M.; Bista, I.; Walsh, K.; et al. An integrated spatio-temporal view of riverine biodiversity using environmental DNA metabarcoding. Nat. Commun. 2024, 15, 4372. [Google Scholar] [PubMed]
- Marie, D.; Rigaut-Jalabert, F.; Vaulot, D. An improved protocol for flow cytometry analysis of phytoplankton cultures and natural samples. Cytom. Part A 2014, 85, 962–968. [Google Scholar]
- Zamorska, J.; Karwowska, E.; Przystaś, W. Assessment of microbiological quality of water using culture methods, flow cytometry and luminometry. Water 2023, 15, 4077. [Google Scholar] [CrossRef]
- Metz, S.; Lopes Dos Santos, A.; Berman, M.C.; Bigeard, E.; Licursi, M.; Not, F.; Lara, E.; Unrein, F. Diversity of photosynthetic picoeukaryotes in eutrophic shallow lakes as assessed by combining flow cytometry cell-sorting and high throughput sequencing. FEMS Microbiol. Ecol. 2019, 95, fiz038. [Google Scholar]
- Manohar, S.M.; Shah, P.; Nair, A. Flow cytometry: Principles, applications and recent advances. Bioanalysis 2021, 13, 181–198. [Google Scholar]
- Zhao, M.M.; Wang, S.M.; Chen, Y.P.; Wu, J.-H.; Xue, L.-G.; Fan, T.T. Pollution status of the Yellow River tributaries in middle and lower reaches. Sci. Total Environ. 2020, 722, 137861. [Google Scholar] [PubMed]
- Liu, Z.; Wang, X.; Jia, S.; Mao, B. Multi-methods to investigate spatiotemporal variations of nitrogen-nitrate and its risks to human health in China’s largest fresh water lake (Poyang Lake). Sci. Total Environ. 2023, 863, 160975. [Google Scholar]
- Prest, E.I.; Hammes, F.; Kötzsch, S.; van Loosdrecht, M.C.M.; Vrouwenvelder, J.S. Monitoring microbiological changes in drinking water systems using a fast and reproducible flow cytometric method. Water Res. 2013, 47, 7131–7142. [Google Scholar] [PubMed]
- Shen, C.F.; Meghrous, J.; Kamen, A. Quantitation of baculovirus particles by flow cytometry. J. Virol. Methods 2002, 105, 321–330. [Google Scholar] [PubMed]
- Lichius, A.; Zeilinger, S. Application of membrane and cell wall selective fluorescent dyes for live-cell imaging of filamentous fungi. J. Vis. Exp. 2019, 153, e60613. [Google Scholar]
- Hammes, F.; Egli, T. Cytometric methods for measuring bacteria in water: Advantages, pitfalls and applications. Anal. Bioanal. Chem. 2010, 397, 1083–1095. [Google Scholar]
- Song, Y.; Wang, Y.; Mao, G.; Gao, G.; Wang, Y. Impact of planktonic low nucleic acid-content bacteria to bacterial community structure and associated ecological functions in a shallow lake. Sci. Total Environ. 2019, 658, 868–878. [Google Scholar]
- García, F.C.; López-Urrutia, Á.; Morán, X.A.G. Automated clustering of heterotrophic bacterioplankton in flow cytometry data. Aquat. Microb. Ecol. 2014, 72, 175–185. [Google Scholar]
- Marie, D.; Shi, X.L.; Rigaut-Jalabert, F.; Vaulot, D. Use of flow cytometric sorting to better assess the diversity of small photosynthetic eukaryotes in the English Channel. FEMS Microbiol. Ecol. 2010, 72, 165–178. [Google Scholar]
- Thompson, A.W.; Foster, R.A.; Krupke, A.; Carter, B.J.; Musat, N.; Vaulot, D.; Kuypers, M.M.M.; Zehr, J.P. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 2012, 337, 1546–1550. [Google Scholar]
- Gasol, J.M.; Morán, X.A.G. Flow Cytometric Determination of Microbial Abundances and Its Use to Obtain Indices of Community Structure and Relative Activity. In Hydrocarbon and Lipid Microbiology Protocols: Single-Cell and Single-Molecule Methods; Springer: Heidelberg, Germany, 2016; pp. 159–187. [Google Scholar]
- Suparna, M.; Paul, R.; Daniel, R.; Tim, U.; Jack, G.; Folker, M.; Andreas, W.; Daniel, H. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinform. 2011, 12, S21. [Google Scholar]
- Clark, D.R.; Ferguson, R.M.W.; Harris, D.N.; Matthews Nicholass, K.J.; Prentice, H.J.; Randall, K.C.; Randell, L.; Warren, S.L.; Dumbrell, A.J. Streams of data from drops of water: 21st century molecular microbial ecology. WIREs Water 2018, 5, e1280. [Google Scholar]
- Gaïa, M.; Meng, L.; Pelletier, E.; Forterre, P.; Vanni, C.; Fernandez-Guerra, A.; Jaillon, O.; Wincker, P.; Ogata, H.; Krupovic, M.; et al. Mirusviruses link herpesviruses to giant viruses. Nature 2023, 616, 783–789. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, H.; Liang, X.; Li, X.; Wang, F. Cumulative effects of cascade dams on river water cycle: Evidence from hydrogen and oxygen isotopes. J. Hydrol. 2019, 568, 604–610. [Google Scholar]
- Sun, M.; Huang, K.; Shao, J.; Wu, W.; Liang, X. Effects of mountain rivers cascade hydropower stations on water ecosystems. Res. Ecol. 2022, 4, 17–26. [Google Scholar]
- Wang, Z.; Ma, J.; Yu, S.; Xu, Y.; Tao, Z.; Zhang, J.; Xiao, R.; Wei, H.; Liu, D. Analysis of water temperature variations in the Yangtze River’s upper and middle reaches in the context of cascade hydropower development. Water 2024, 16, 1669. [Google Scholar] [CrossRef]
- Chang, E.; Zhao, Y.; Wei, Q.; Shi, S.; Jiang, Z. Isolation of high-quality RNA from Platycladus orientalis and other Cupressaceae plants. Electron. J. Biotechnol. 2016, 23, 21–27. [Google Scholar]
- Dembowska, E.A. The use of phytoplankton in the assessment of water quality in the lower section of Poland’s largest river. Water 2021, 13, 3471. [Google Scholar] [CrossRef]
- Ly, Q.V.; Nguyen, X.C.; Lê, N.C.; Truong, T.-D.; Hoang, T.-H.T.; Park, T.J.; Maqbool, T.; Pyo, J.; Cho, K.H.; Lee, K.-S.; et al. Application of Machine Learning for eutrophication analysis and algal bloom prediction in an urban river: A 10-year study of the Han River, South Korea. Sci. Total Environ. 2021, 797, 149040. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, L.; Xu, K.; Wang, X.C. Characteristics of external carbon uptake by microalgae growth and associated effects on algal biomass composition. Bioresour. Technol. 2019, 292, 121887. [Google Scholar] [CrossRef]
- Wang, X.; Wang, C.; Wang, P.; Chen, J.; Miao, L.; Feng, T.; Yuan, Q.; Liu, S. How bacterioplankton community can go with cascade damming in the highly regulated Lancang-Mekong River Basin. Mol. Ecol. 2018, 27, 4444–4458. [Google Scholar] [PubMed]
- Liu, L.; Yang, J.; Yu, X.; Chen, G.; Yu, Z. Patterns in the composition of microbial communities from a subtropical river: Effects of environmental, spatial and temporal factors. PLoS ONE 2013, 8, e81232. [Google Scholar]
- Qi, H.; Lv, J.; Liao, J.; Jin, J.; Ren, Y.; Tao, Y.; Wang, D.; Alvarez, P.J.; Yu, P. Metagenomic insights into microalgae-bacterium-virus interactions and viral functions in phycosphere facing environmental fluctuations. Water Res. 2025, 268, 122676. [Google Scholar]
- Liu, X.; Hu, S.; Sun, R.; Wu, Y. Dissolved oxygen disturbs nitrate transformation by modifying microbial community, co-occurrence networks, and functional genes during aerobic-anoxic transition. Sci. Total Environ. 2021, 790, 148245. [Google Scholar]
- Wang, P.; Zhao, J.; Xiao, H.; Yang, W.; Yu, X. Bacterial community composition shaped by water chemistry and geographic distance in an anthropogenically disturbed river. Sci. Total Environ. 2019, 655, 61–69. [Google Scholar] [CrossRef]
- Liu, L.; Wu, Y.; Yin, M.; Ma, X.; Yu, X.; Guo, X.; Du, N.; Eller, F.; Guo, W. Soil salinity, not plant genotype or geographical distance, shapes soil microbial community of a reed wetland at a fine scale in the Yellow River Delta. Sci. Total Environ. 2023, 856, 159136. [Google Scholar]
- Bailey, V.L.; Smith, J.L.; Bolton, H., Jr. Fungal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol. Biochem. 2002, 34, 997–1007. [Google Scholar]
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar]
- Yu, Z.; Gan, Z.; Tawfik, A.; Meng, F. Exploring interspecific interaction variability in microbiota: A review. Eng. Microbiol. 2024, 4, 100178. [Google Scholar]
- Luo, Z.; Li, S.; Hou, K.; Ji, G. Spatial and seasonal bacterioplankton community dynamics in the main channel of the Middle Route of South-to-North Water Diversion Project. Res. Microbiol. 2019, 170, 24–34. [Google Scholar]
- Pang, Y.; Ji, G. Biotic factors drive distinct DNRA potential rates and contributions in typical Chinese shallow lake sediments. Environ. Pollut. 2019, 254, 112903. [Google Scholar] [CrossRef] [PubMed]
- Zhikharev, V.; Vodeneeva, E.; Kudrin, I.; Gavrilko, D.; Startseva, N.; Kulizin, P.; Erina, O.; Tereshina, M.; Okhapkin, A.; Shurganova, G. The species structure of plankton communities as a response to changes in the trophic gradient of the mouth areas of large tributaries to a lowland reservoir. Water 2022, 15, 74. [Google Scholar] [CrossRef]
- Fan, H.; He, D.; Wang, H. Environmental consequences of damming the mainstream Lancang-Mekong River: A review. Earth-Sci. Rev. 2015, 146, 77–91. [Google Scholar] [CrossRef]
- Yang, N.; Li, Y.; Zhang, W.; Lin, L.; Qian, B.; Wang, L.; Niu, L.; Zhang, H. Cascade dam impoundments restrain the trophic transfer efficiencies in benthic microbial food web. Water Res. 2020, 170, 115351. [Google Scholar]
- Li, Z.; Lu, L.; Guo, J.; Yang, J.; Zhang, J.; He, B.; Xu, L. Responses of spatial-temporal dynamics of bacterioplankton community to large-scale reservoir operation: A case study in the Three Gorges Reservoir, China. Sci. Rep. 2017, 7, 42469. [Google Scholar]
- Yang, N.; Li, Y.; Lin, L.; Zhang, W.; Wang, L.; Niu, L.; Zhang, H. Dam-induced flow velocity decrease leads to the transition from heterotrophic to autotrophic system through modifying microbial food web dynamics. Environ. Res. 2022, 212, 113568. [Google Scholar] [PubMed]
- Ramanan, R.; Kim, B.H.; Cho, D.H.; Oh, H.M.; Kim, H.S. Algae–bacteria interactions: Evolution, ecology and emerging applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar]
- Liu, Y.; Li, C.; Jian, S.; Miao, S.; Li, K.; Guan, H.; Mao, Y.; Wang, Z.; Li, C. Hydrodynamics regulate longitudinal plankton community structure in an alpine cascade reservoir system. Front. Microbiol. 2021, 12, 749888. [Google Scholar] [CrossRef]
- Santos, M.; Oliveira, H.; Pereira, J.L.; Pereira, M.J.; Gonçalves, F.J.; Vidal, T. Flow cytometry analysis of low/high DNA content (LNA/HNA) bacteria as bioindicator of water quality evaluation. Ecol. Indic. 2019, 103, 774–781. [Google Scholar]
- Johannessen, T.V.; Larsen, A.; Bratbak, G.; Pagarete, A.; Edvardsen, B.; Egge, E.D.; Sandaa, R.-A. Seasonal dynamics of haptophytes and dsDNA algal viruses suggest complex virus-host relationship. Viruses 2017, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lu, Y.; Dungait, J.A.J.; Liu, J.; Lin, S.; Jia, J.; Yu, G. The “regulator” function of viruses on ecosystem carbon cycling in the anthropocene. Front. Public Health 2022, 10, 858615. [Google Scholar]
- Martínez, A.; Larrañaga, A.; Pérez, J.; Descals, E.; Pozo, J. Temperature affects leaf litter decomposition in low-order forest streams: Field and microcosm approaches. FEMS Microbiol. Ecol. 2014, 87, 257–267. [Google Scholar] [CrossRef]
- Shishir, T.; Mahbub, N.; Kamal, N. Review on bioremediation: A tool to resurrect the polluted rivers. Pollution 2019, 5, 555–568. [Google Scholar]
- Hua, R.; Zhang, Y. Assessment of water quality improvements using the hydrodynamic simulation approach in regulated cascade reservoirs: A case study of drinking water sources of Shenzhen, China. Water 2017, 9, 825. [Google Scholar] [CrossRef]
- Wang, X.; Wang, P.; Wang, C.; Chen, J.; Hu, B.; Yuan, Q.; Du, C.; Xing, X. Cascade damming impacts on microbial mediated nitrogen cycling in rivers. Sci. Total Environ. 2023, 903, 166533. [Google Scholar]
- Bao, Y.; Wang, Y.; Hu, M.; Hu, P.; Wu, N.; Qu, X.; Liu, X.; Huang, W.; Wen, J.; Li, S. Deciphering the impact of cascade reservoirs on nitrogen transport and nitrate transformation: Insights from multiple isotope analysis and machine learning. Water Res. 2024, 268, 122638. [Google Scholar] [PubMed]
- Chen, Q.; Chen, Y.; Lin, Y.; Zhang, J.; Ni, J.; Xia, J.; Xiao, L.; Feng, T.; Ma, H. Does a hydropower reservoir cascade really harm downstream nutrient regimes. Sci. Bull. 2024, 69, 661–670. [Google Scholar]
- Jia, Y.; Hu, X.; Kang, W.; Dong, X. Unveiling microbial nitrogen metabolism in rivers using a machine learning approach. Environ. Sci. Technol. 2024, 58, 6605–6615. [Google Scholar] [CrossRef]
- Battin, T.J.; Lauerwald, R.; Bernhardt, E.S.; Bertuzzo, E.; Gener, L.G.; Hall, R.O., Jr.; Hotchkiss, E.R.; Maavara, T.; Pavelsky, T.M.; Ran, L. River ecosystem metabolism and carbon biogeochemistry in a changing world. Nature 2023, 613, 449–459. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Luo, Z.; Zhu, X.; Dang, Z.; Zhang, D.; Sui, X. Picoplankton Groups and Their Responses to Environmental Factors in Small Cascade Hydropower Stations. Water 2025, 17, 903. https://doi.org/10.3390/w17060903
Li P, Luo Z, Zhu X, Dang Z, Zhang D, Sui X. Picoplankton Groups and Their Responses to Environmental Factors in Small Cascade Hydropower Stations. Water. 2025; 17(6):903. https://doi.org/10.3390/w17060903
Chicago/Turabian StyleLi, Peiquan, Zhongxin Luo, Xianfang Zhu, Zhengzhu Dang, Daxin Zhang, and Xin Sui. 2025. "Picoplankton Groups and Their Responses to Environmental Factors in Small Cascade Hydropower Stations" Water 17, no. 6: 903. https://doi.org/10.3390/w17060903
APA StyleLi, P., Luo, Z., Zhu, X., Dang, Z., Zhang, D., & Sui, X. (2025). Picoplankton Groups and Their Responses to Environmental Factors in Small Cascade Hydropower Stations. Water, 17(6), 903. https://doi.org/10.3390/w17060903





