Application of Environmental DNA Technology in Fish Diversity Research in Dongting Lake, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Survey Area and Sampling Period
2.2. Traditional Fishing Gear
2.3. eDNA Collection from Water
2.4. DNA Extraction and Amplification from Filters
2.5. High-Throughput Sequencing
2.6. Cluster Analysis
2.7. Alpha Diversity Analysis
2.8. Beta Diversity Analysis
3. Results
3.1. Comparison of Traditional Fishing Gear and eDNA Monitoring Results
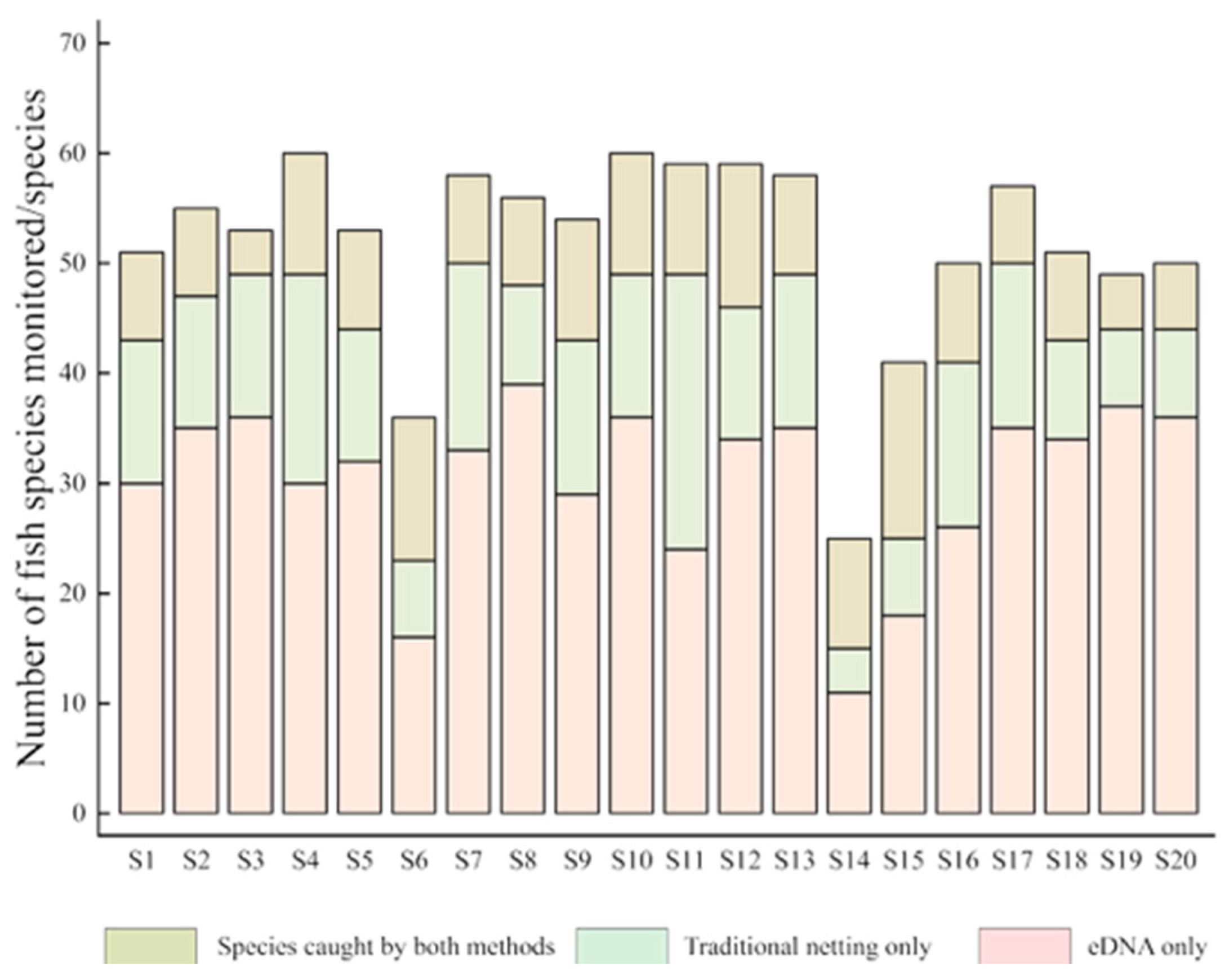
3.1.1. Comparison of Fish Species Detected
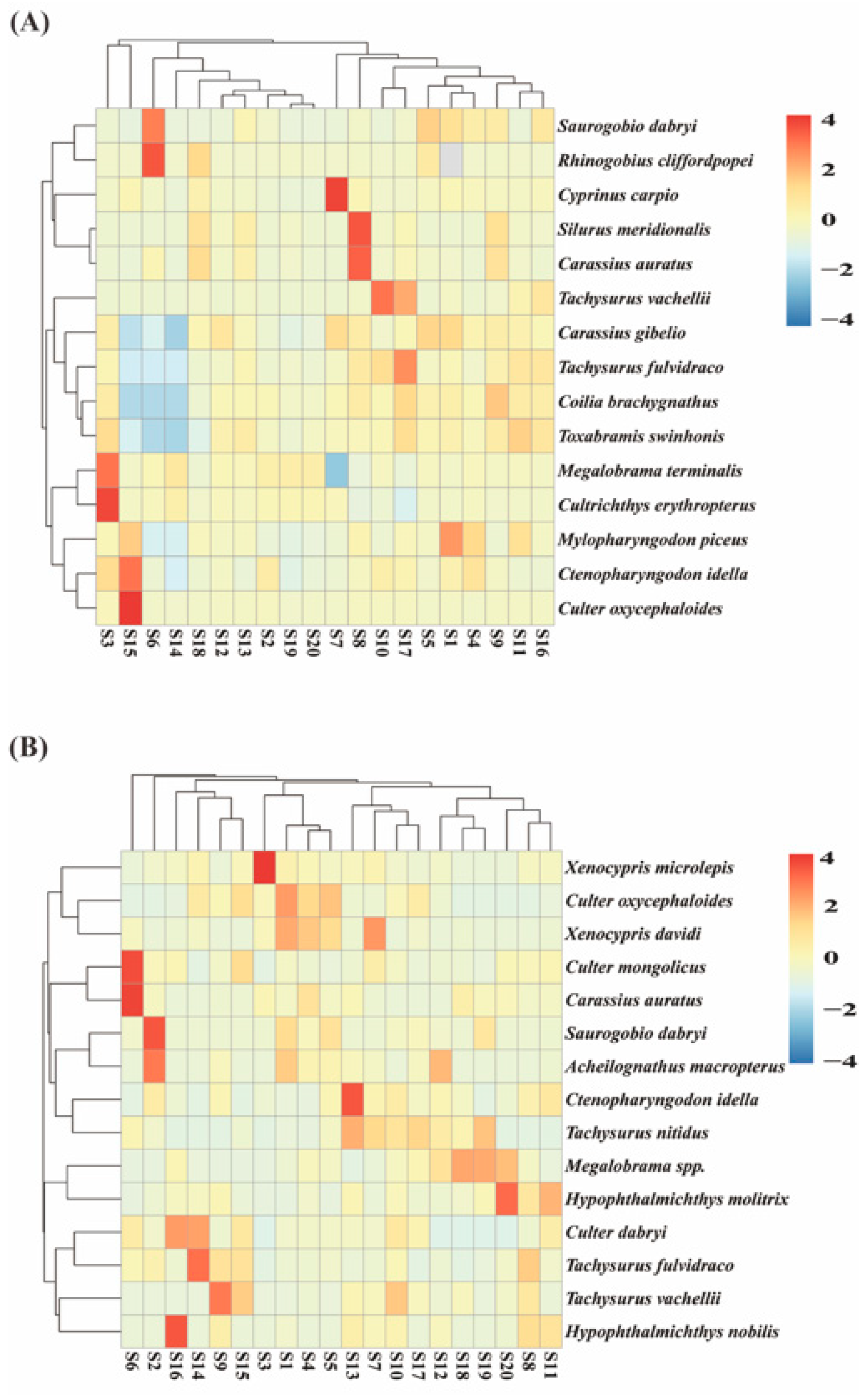
3.1.2. Comparison of Species Composition of Sampled Fish
3.2. Analysis of Fish Species Diversity
3.2.1. Alpha Diversity Analysis Based on eDNA and Traditional Fishing Gear
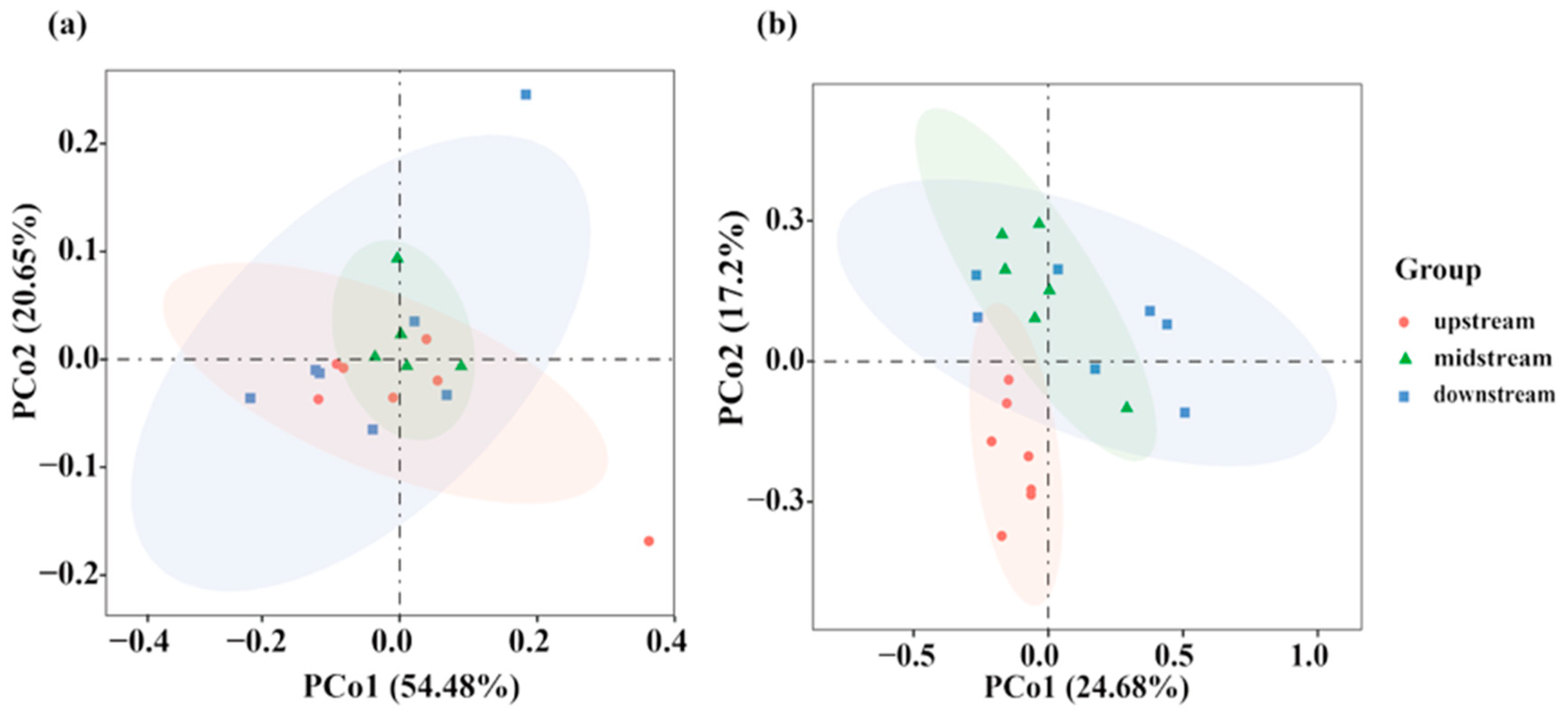
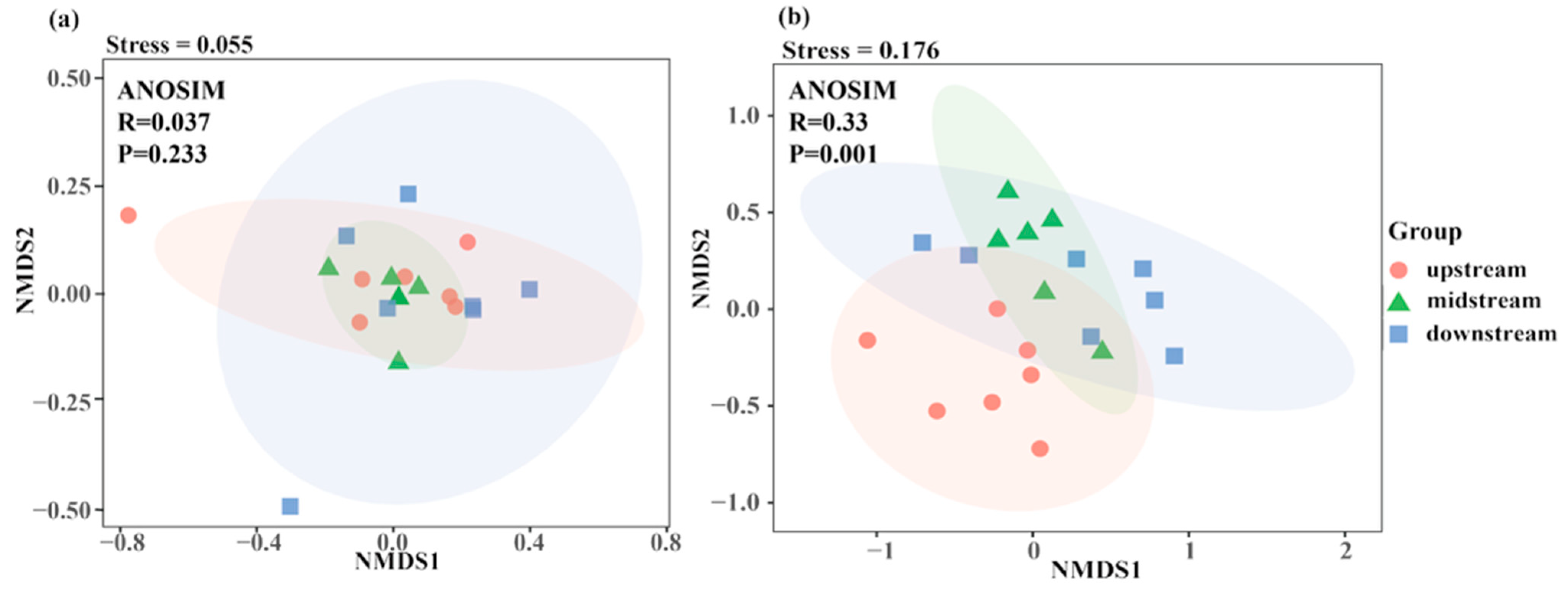
3.2.2. Beta Diversity Analysis Based on eDNA and Traditional Fishing Gear
4. Discussion
4.1. Complementary Benefits of eDNA Metabarcoding and Traditional Fishing Gear in Species Detection
4.2. Comparison of Fish Diversity Between Traditional Fishing Gear and eDNA Monitoring
4.3. Disparate Spatial Patterns of Fish Communities: A Methodological Perspective
4.4. Integrated Application of eDNA Technology and Traditional Fishing Gear
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zou, K.S.; Chen, J.W.; Ruan, H.T.; Li, Z.H.; Guo, W.J.; Li, M.; Liu, L. eDNA metabarcoding as a promising conservation tool for monitoring fish diversity in a coastal wetland of the Pearl River Estuary compared to bottom trawling. Sci. Total Environ. 2020, 702, 134704. [Google Scholar] [CrossRef]
- Turak, E.; Harrison, I.; Dudgeon, D.; Abell, R.; Bush, A.; Darwall, W.; Finlayson, C.M.; Ferrier, S.; Freyhof, J.; Hermoso, V.; et al. Essential Biodiversity Variables for measuring change in global freshwater biodiversity. Biol. Conserv. 2017, 213, 272–279. [Google Scholar] [CrossRef]
- Wang, J.M.; Tang, W.Q. Preliminary Investigation of Fish Diversity in Huayang River based on eDNA Technology. Int. J. Biol. Life Sci. 2024, 8, 22–27. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.K.; Bekkevold, D.; Clausen, L.W.; Nielsen, E.E. The sceptical optimist: Challenges and perspectives for the application of environmental DNA in marine fisheries. Fish Fish. 2018, 19, 751–768. [Google Scholar] [CrossRef]
- Thompson, L.R.; Thielen, P. Decoding dissolved information: Environmental DNA sequencing at global scale to monitor a changing ocean. Curr. Opin. Biotechnol. 2023, 81, 102936. [Google Scholar] [CrossRef]
- Shaw, J.L.A.; Clarke, L.J.; Wedderburn, S.D.; Barnes, T.C.; Weyrich, L.S.; Cooper, A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol. Conserv. 2016, 197, 131–138. [Google Scholar] [CrossRef]
- Padilla, D.K.; Williams, S.L. Beyond ballast water: Aquarium and ornamental trades as sources of invasive species in aquatic ecosystems. Front. Ecol. Environ. 2004, 2, 131–138. [Google Scholar] [CrossRef]
- Bertozzi, T.; Adams, M.A.; Walker, K.F. Species boundaries in carp gudgeons (Eleotrididae: Hypseleotris) from the River Murray, South Australia: Evidence for multiple species and extensive hybridization. Mar. Freshw. Res. 2000, 51, 805–815. [Google Scholar] [CrossRef]
- Shen, Y.J.; Zhou, X.X.; Zhang, Y.F.; Zhang, J.M.; Li, Q.H.; Chen, Q.L.; Liu, Z.H.; Li, Y.W.; Cheng, R.; Luo, Y. Important fish diversity maintenance status of the tributaries in a hotspot fish conservation area in the upper Yangtze River revealed by eDNA metabarcoding. Sci. Rep. 2024, 14, 24128. [Google Scholar] [CrossRef]
- Sigsgaard, E.E.; Nielsen, I.B.; Carl, H.; Krag, M.A.; Knudsen, S.W.; Xing, Y.C.; Holm Hansen, T.H.; Møller, P.R.; Thomsen, P.F. Seawater environmental DNA reflects seasonality of a coastal fish community. Mar. Biol. 2017, 164, 128. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef]
- Munian, K.; Ramli, F.F.; Othman, N.; Mahyudin, N.A.A.; Sariyati, N.H.; Abdullah Fauzi, N.A.F.; Haris, H.; Ilham Norhakim, M.L.; Abdul Latiff, M.A.B. Environmental DNA metabarcoding of freshwater fish in Malaysian tropical rivers using short-read nanopore sequencing as a potential biomonitoring tool. Mol. Ecol. Resour. 2024, 24, e13936. [Google Scholar] [CrossRef]
- Cheng, R.L.; Luo, Y.; Li, Q.H.; Zhang, Y.F.; Liu, Z.H.; Chen, Q.L.; Li, Y.W.; Shen, Y.J. Application of eDNA metabarcoding for monitoring the fish diversity of the Jiangjin to Fuling section of the upper reaches of the Yangtze River. Hydrobiologia 2023, 850, 4067–4088. [Google Scholar] [CrossRef]
- Xu, N.; Chang, J. Preliminary Study on Fish Species Detection in the Middle and Lower Yangtze River Using Environmental DNA. J. Hydroecology 2016, 37, 49–55. [Google Scholar]
- Tong, Y.D.; Kuang, Z.; Liu, P.F.; Liang, Y.D.; Fan, Y.C.; Xu, D.P. Fish diversity in the Dongping Lake based on environmental DNA techniques. J. Fish. Sci. China 2023, 30, 1530–1542. [Google Scholar]
- Guo, N.; Shen, M.; Xiao, N.; Gao, X.; Guo, X.; Li, J. Distribution characteristics of autumn fish diversity in Chishui River based on environmental DNA metabarcoding technology. Acta Ecol. Sin. 2023, 43, 1676–1690. [Google Scholar] [CrossRef]
- Miya, M. Environmental DNA Metabarcoding: A Novel Method for Biodiversity Monitoring of Marine Fish Communities. Ann. Rev. Mar. Sci. 2022, 14, 161–185. [Google Scholar] [CrossRef]
- Wang, S.P.; Yan, Z.G.; Hänfling, B.; Zheng, X.; Wang, P.Y.; Fan, J.T.; Li, J.L. Methodology of fish eDNA and its applications in ecology and environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Jin, S.G.; Wang, N.; Zhao, J.R.; Guo, H.W.; Pellikka, P. Total Phosphorus and Nitrogen Dynamics and Influencing Factors in Dongting Lake Using Landsat Data. Remote Sens. 2022, 14, 5648. [Google Scholar] [CrossRef]
- Fu, C.Z.; Wu, J.H.; Chen, J.K.; Wu, Q.H.; Lei, G.C. Freshwater fish biodiversity in the Yangtze River basin of China: Patterns, threats and conservation. Biodivers. Conserv. 2003, 12, 1649–1685. [Google Scholar] [CrossRef]
- Xu, N.; Xiong, M.H.; Shao, K.; Que, Y.F.; Li, J.Y. Preliminary Study on Environmental DNA Metabarcoding for Detecting Biodiversity in the Middle and Lower Reaches of the Yangtze River. Res. Environ. Sci. 2020, 33, 1187–1196. [Google Scholar]
- Bylemans, J.; Gleeson, D.M.; Hardy, C.M.; Furlan, E. Toward an ecoregion scale evaluation of eDNA metabarcoding primers: A case study for the freshwater fish biodiversity of the Murray–Darling Basin (Australia). Ecol. Evol. 2018, 8, 8697–8712. [Google Scholar] [CrossRef] [PubMed]
- Djurhuus, A.; Port, J.; Closek, C.J.; Yamahara, K.M.; Romero Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B.; et al. Evaluation of Filtration and DNA Extraction Methods for Environmental DNA Biodiversity Assessments across Multiple Trophic Levels. Front. Mar. Sci. 2017, 4, 00314. [Google Scholar] [CrossRef]
- Lyu, S.; Tong, J.F.; Wu, J.H.; Wang, X.F.; Geng, X.Y.; Gao, C.X.; Wang, Y. Towards a comprehensive assessment of ichthyofaunal diversity in the Yangtze River estuary: Leveraging environmental DNA technology and bottom trawl surveys. Heliyon 2024, 10, e34761. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Adolf, J.; Charlop Powers, Z.; Dunton, K.J.; Hinks, G.; VanMorter, S.M. Trawl and eDNA assessment of marine fish diversity, seasonality, and relative abundance in coastal New Jersey, USA. ICES J. Mar. Sci. 2020, 78, 293–304. [Google Scholar] [CrossRef]
- Somerfield, P.J.; Clarke, K.R.; Gorley, R.N. Analysis of similarities (ANOSIM) for 2-way layouts using a generalised ANOSIM statistic, with comparative notes on Permutational Multivariate Analysis of Variance (PERMANOVA). Austral Ecol. 2021, 46, 911–926. [Google Scholar] [CrossRef]
- Willis, A.D.; Martin, B.D. Estimating diversity in networked ecological communities. Biostatistics 2022, 23, 207–222. [Google Scholar] [CrossRef]
- Wu, Y.A.; Li, H.; Liao, F.; Yang, X.; Xie, Z.G. The Freshwater Fishes of Hunan Province; Science Press: Beijing, China, 2019; pp. 15–485. [Google Scholar]
- Macher, T.H.; Schütz, R.; Arle, J.; Beermann, A.J.; Koschorreck, J.; Leese, F. Beyond fish eDNA metabarcoding: Field replicates disproportionately improve the detection of stream associated vertebrate species. Metabarcoding Metagenom. 2021, 5. [Google Scholar] [CrossRef]
- Nakagawa, H.; Yamamoto, S.; Sato, Y.; Sado, T.; Minamoto, T.; Miya, M. Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshw. Biol. 2018, 63, 569–580. [Google Scholar] [CrossRef]
- Charan Dixon, H.; Lamker, P.S.; Arvin-Blaauw, A.S.; Nokise, F.; Brons, J.K.; Ziebell, A.C.; Maathuis, M.A.M.; Tulp, I.; Eriksson, B.K. The molecularly and visually identified prey of fish in Dutch salt marshes. J. Sea Res. 2025, 208, 102634. [Google Scholar] [CrossRef]
- Troth, C.R.; Sweet, M.J.; Nightingale, J.; Burian, A. Seasonality, DNA degradation and spatial heterogeneity as drivers of eDNA detection dynamics. Sci. Total Environ. 2021, 768, 144466. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yang, G.; Geng, Z.; Zhao, F.; Feng, X.; Zhang, T. Application of environmental DNA technology in fish diversity analysis in the yangtze river estuary. Acta Hydrobiol. Sin. 2023, 47, 365–375. [Google Scholar]
- V. Wintzingerode, F.; Göbel, U.B.; Stackebrandt, E. Determination of microbial diversity in environmental samples: Pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 1997, 21, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.T.; Giovannoni, S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996, 62, 625–630. [Google Scholar] [CrossRef]
- Polz, M.F.; Cavanaugh, C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 1998, 64, 3724–3730. [Google Scholar] [CrossRef]
- Kontanis, E.J.; Reed, F.A. Evaluation of real-time PCR amplification efficiencies to detect PCR inhibitors. J. Forensic Sci. 2006, 51, 795–804. [Google Scholar] [CrossRef]
- Liu, W.H.; Saint, D.A. A New Quantitative Method of Real Time Reverse Transcription Polymerase Chain Reaction Assay Based on Simulation of Polymerase Chain Reaction Kinetics. Anal. Biochem. 2002, 302, 52–59. [Google Scholar] [CrossRef]
- Furlan, E.M.; Gleeson, D.; Hardy, C.M.; Duncan, R.P. A framework for estimating the sensitivity of eDNA surveys. Mol. Ecol. Resour. 2015, 16, 641–654. [Google Scholar] [CrossRef]
- Coble, A.A.; Flinders, C.A.; Homyack, J.A.; Penaluna, B.E.; Cronn, R.C.; Weitemier, K. eDNA as a tool for identifying freshwater species in sustainable forestry: A critical review and potential future applications. Sci. Total Environ. 2019, 649, 1157–1170. [Google Scholar] [CrossRef]
- Jeunen, G.J.; Knapp, M.; Spencer, H.G.; Lamare, M.D.; Taylor, H.R.; Stat, M.; Bunce, M.; Gemmell, N.J. Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 2019, 19, 426–438. [Google Scholar] [CrossRef]
- Li, Z.Y.; Jiang, P.W.; Wang, L.X.; Liu, L.; Li, M.; Zou, K.S. A comparison of seasonal composition and structure of fish community between environmental DNA technology and gillnetting in the Pearl River Estuary, China. Ecol. Indic. 2023, 147, 109915. [Google Scholar] [CrossRef]
- Yuan, X.P.; Yang, X.; Ge, H.Z.; Li, H.; Deng, D.Q. Temporal Distribution of Fish Community Structure in Dongting Lake Estuary. Agric. Sci. 2019, 10, 294–301. [Google Scholar] [CrossRef]
- Cai, L.; Shi, L.L.; Guan, L.; Zhou, Y.; Jing, L.; Lei, G.C. Potential Impacts of Proposed Chenglingji Hydraulic Project on Wetlands in Dongting Lake. Wetl. Sci. 2018, 16, 377–392. [Google Scholar]
- Ji, F.F.; Shen, J.Z.; Zhang, W.Q.; Yan, S.H.; Shan, B.Q.; Zha, J.M. Discriminating spatiotemporal heterogeneity and environmental drivers of fish assemblages using environmental DNA metabarcoding in mosaic habitat ecosystems. Sci. Rep. 2025, 15, 15705. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Hinlo, R.; Furlan, E.; Suitor, L.; Gleeson, D. Environmental DNA monitoring and management of invasive fish: Comparison of eDNA and fyke netting. Manag. Biol. Invasions 2017, 8, 89–100. [Google Scholar] [CrossRef]
- Naputo, C.F.P.; Isowa, Y.; Gerona-Daga, M.E.; Artigas, M.D.; Kajita, T.; Salmo Iii, S.G. Application of eDNA metabarcoding in the assessment of fish biodiversity in Philippine mangroves: Challenges and opportunities. Reg. Stud. Mar. Sci. 2024, 77, 103642. [Google Scholar] [CrossRef]
- Qin, C.X.; Zuo, T.; Yu, G.; Zhou, W.l.; Li, C.H. Advances in research of environmental DNA (eDNA) in biomass assessment of aquatic ecosystems. South China Fish. Sci. 2020, 16, 123–128. [Google Scholar]
- Yang, C. Fish Community Pattern and Influencing Factors of the Yangtse River Basin; Xi’an University of Technology: Xi’an, China, 2021. [Google Scholar]
- Lacoursière Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef]
- Hänfling, B.; Lawson Handley, L.; Read, D.S.; Hahn, C.; Li, J.; Nichols, P.; Blackman, R.C.; Oliver, A.; Winfield, I.J. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 2016, 25, 3101–3119. [Google Scholar] [CrossRef]
- Wang, S.Y.; Xiong, Y.; Zhang, H.S.; Song, D.D.; Wang, Y.P.; Ge, H.; Zhang, C.B.; Liang, L.; Zhong, X.M. Recovery of Coilia nasus resources after implementation of the 10-year fishing ban in the Yangtze River: Implied from the Yangtze River Estuary and its adjacent sea areas. Front. Mar. Sci. 2024, 11, 1474996. [Google Scholar] [CrossRef]


| Statistical Values | ||
|---|---|---|
| R2 | p | |
| Upstream-midstream | 0.0918 | 0.313 |
| Upstream-downstream | 0.0579 | 0.568 |
| Upstream-downstream | 0.0603 | 0.595 |
| Statistical Values | ||
|---|---|---|
| R2 | p | |
| Upstream-midstream | 0.2187 | 0.005 |
| Upstream-downstream | 0.2014 | 0.001 |
| Upstream-downstream | 0.1373 | 0.109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Hu, Y.; Liu, X.; Suo, W.; Liu, D.; Ku, Q.; Yuan, X.; Yang, X.; Yang, G.; Zhang, H.; et al. Application of Environmental DNA Technology in Fish Diversity Research in Dongting Lake, China. Water 2025, 17, 3282. https://doi.org/10.3390/w17223282
Wang C, Hu Y, Liu X, Suo W, Liu D, Ku Q, Yuan X, Yang X, Yang G, Zhang H, et al. Application of Environmental DNA Technology in Fish Diversity Research in Dongting Lake, China. Water. 2025; 17(22):3282. https://doi.org/10.3390/w17223282
Chicago/Turabian StyleWang, Chongrui, Yufeng Hu, Xiangrong Liu, Wenwen Suo, Dong Liu, Qianqian Ku, Xiping Yuan, Xin Yang, Guang Yang, Hui Zhang, and et al. 2025. "Application of Environmental DNA Technology in Fish Diversity Research in Dongting Lake, China" Water 17, no. 22: 3282. https://doi.org/10.3390/w17223282
APA StyleWang, C., Hu, Y., Liu, X., Suo, W., Liu, D., Ku, Q., Yuan, X., Yang, X., Yang, G., Zhang, H., & Ou, D. (2025). Application of Environmental DNA Technology in Fish Diversity Research in Dongting Lake, China. Water, 17(22), 3282. https://doi.org/10.3390/w17223282






