Integrating Microbial Source Tracking to Unravel Impacts of Wastewater Discharge on Spatial Distribution of Riverine Microbial Community
Abstract
1. Introduction
2. Materials and Methods
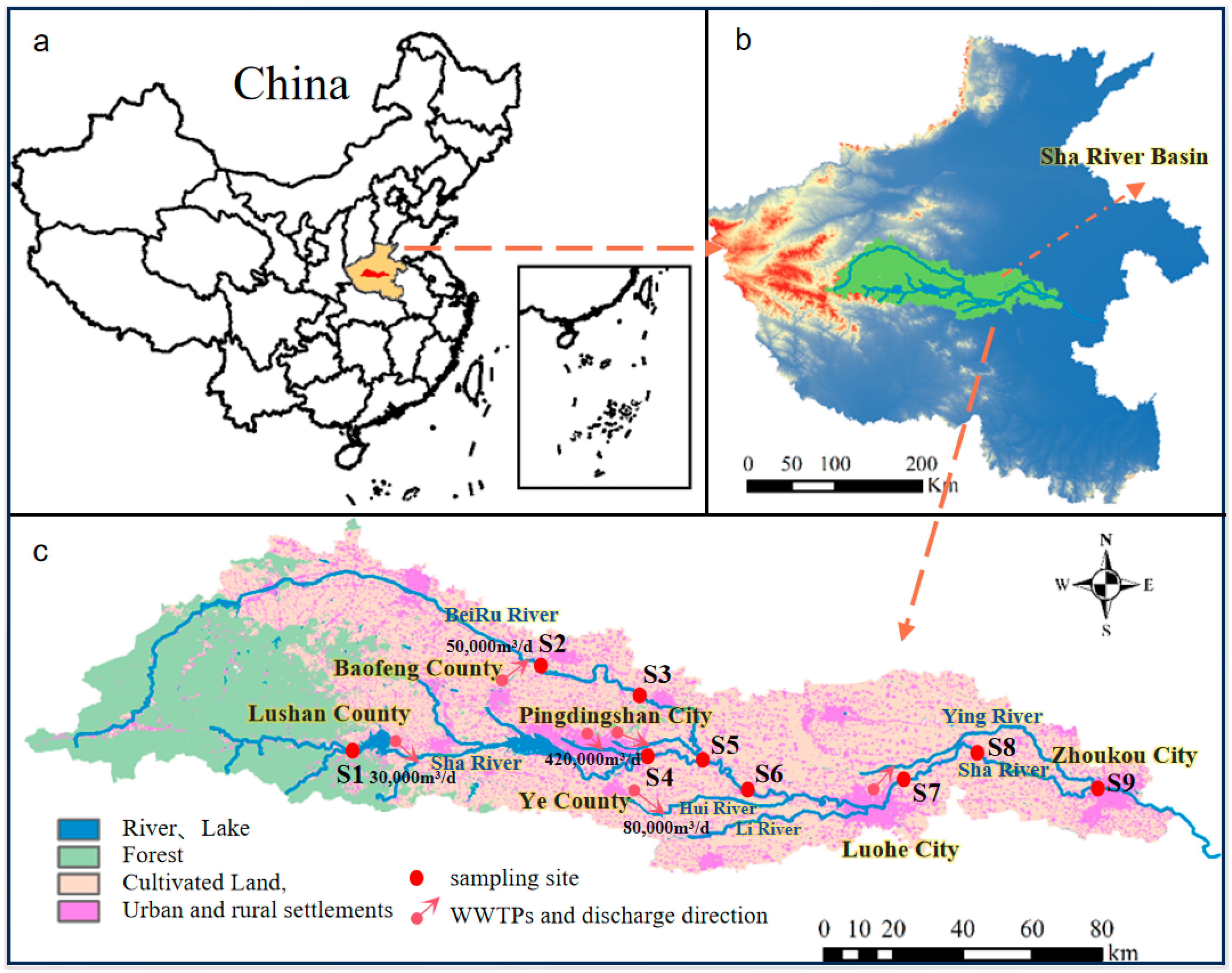
2.1. Collection of Samples
2.2. Laboratory Analyses
2.3. Source Tracker Model
3. Results and Discussion
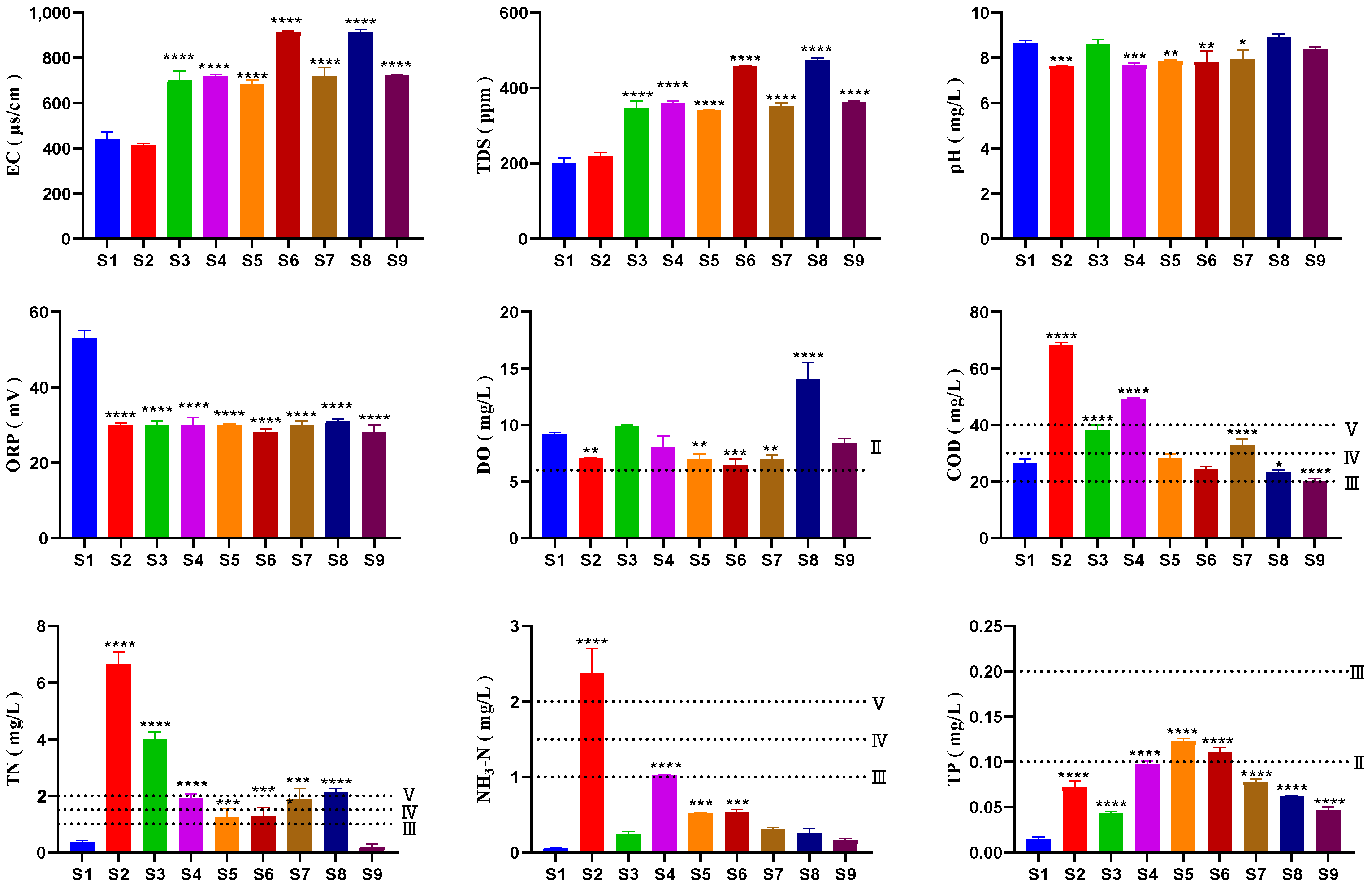
3.1. Physicochemical Properties
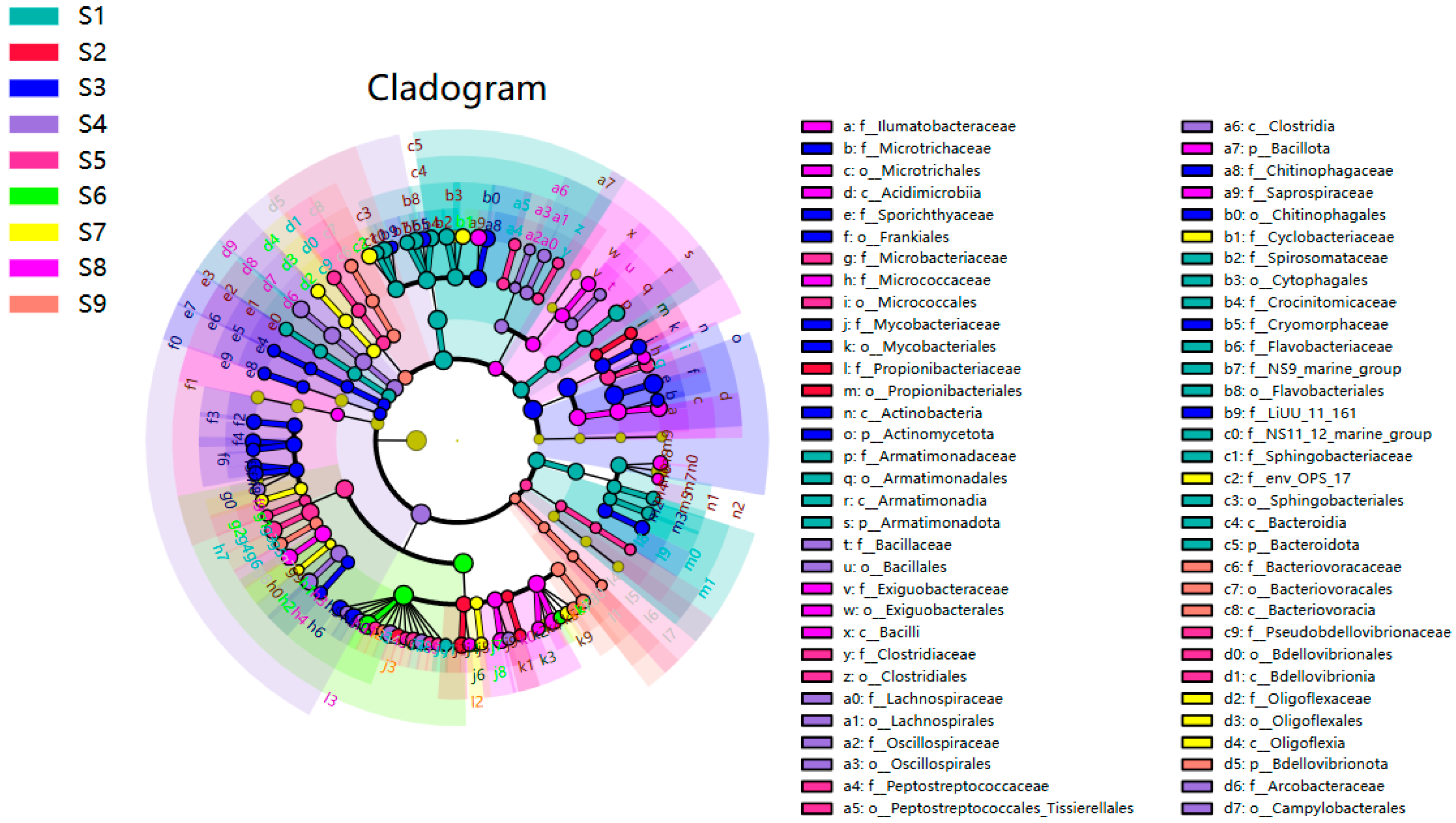
3.2. Microbial Community
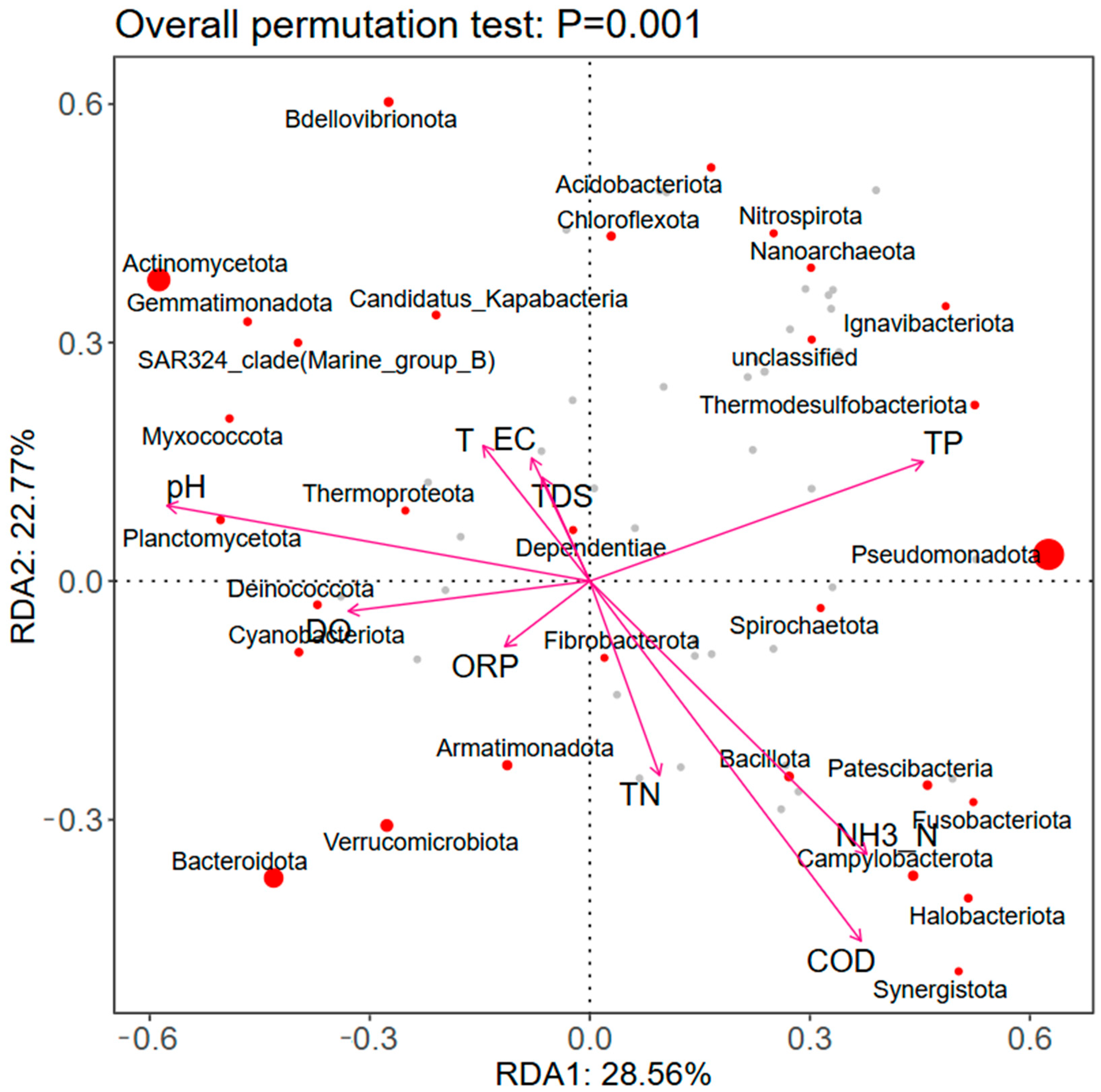
3.3. Factors Influencing Microbial Distribution
3.4. Source Apportionment of Downstream of the River Using the Source Tracker Model
4. Conclusions
- (1)
- The sequencing results of species at the phylum level revealed that the dominant microbial phyla in the Sha River were primarily Proteobacteria (55.4%), Actinobacteriota (24.0%), Bacteroidota (14.3%), and Verrucomicrobiota (2.6%). The most dominant phylum, Proteobacteria, exhibited varying abundances across different sampling sites in the Sha River basin, with the highest abundances observed at Sites S2, S4, S5 and S6. It can be seen from this that the discharge of wastewater from wastewater treatment plants has a significant impact on the distribution of microorganisms in rivers.
- (2)
- Correlation analysis of microbial communities and environmental factors in the Sha River shows that despite wastewater treatment plants discharging effluent meeting the Class 1 Grade A standard into the river, the water body still has relatively high concentrations of ammonia nitrogen, chemical oxygen demand, and total phosphorus. This not only significantly affects the structure of aquatic microbial communities, but also worsens water eutrophication.
- (3)
- The results of quantitative analysis showed that S2 (36.7%) and S4 (31.3%) in the upper reaches of the Sha River are the primary contributors to the microbial community in the downstream catchment area (S6). The study found that the impact of wastewater discharge on the microbial community in the downstream water body exhibits a “longitudinal persistence of microbial signatures”—even though the physicochemical pollution indicators of the water body have decreased. Unfortunately, a limitation of this study is that its hydrological data were derived solely from a single field sampling campaign. This sampling only captured the low-flow hydrological state of the study area, which did not fully account for how extreme hydrological events, such as monsoons or droughts, might alter the source–sink dynamics of microbial community.
- (4)
- These findings of this study represent the application in microbial source tracking in the upstream and downstream sections of rivers, providing strong support for formulating more effective environmental protection strategies in the Sha River basin. Given that Site S2 (domestic wastewater with untreated aquaculture wastewater) and Site S4 (domestic wastewater discharge) contribute the highest proportion of microorganisms to the downstream confluence area (S6), it is necessary to strengthen the upgrading and transformation of wastewater treatment plants and reduce the direct discharge of aquaculture wastewater.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drury, B.; Rosi-Marshall, E.; Kelly, J.J. Wastewater Treatment Effluent Reduces the Abundance and Diversity of Benthic Bacterial Communities in Urban and Suburban Rivers. Appl. Environ. Microbiol. 2013, 79, 1897–1905. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A Review on the Occurrence of Micropollutants in the Aquatic Environment and Their Fate and Removal during Wastewater Treatment. Sci. Total Environ. 2014, 473–474, 619–641. [Google Scholar] [CrossRef]
- Lacaze, É.; Gendron, A.D.; Miller, J.L.; Colson, T.L.L.; Sherry, J.P.; Giraudo, M.; Marcogliese, D.J.; Houde, M. Cumulative Effects of Municipal Effluent and Parasite Infection in Yellow Perch: A Field Study Using High-Throughput RNA-Sequencing. Sci. Total Environ. 2019, 665, 797–809. [Google Scholar] [CrossRef]
- Wang, J.W.; Chen, Y.; Cai, P.G.; Gao, Q. Impacts of Municipal Wastewater Treatment Plant Discharge on Microbial Community Structure and Function of the Receiving River in Northwest Tibetan Plateau. J. Hazard. Mater. 2022, 423, 127170. [Google Scholar] [CrossRef]
- Pascual-Benito, M.; Ballesté, E.; Monleón-Getino, T.; Urmeneta, J.; Blanch, A.R.; García-Aljaro, C.; Lucena, F. Impact of Treated Sewage Effluent on the Bacterial Community Composition in an Intermittent Mediterranean Stream. Environ. Pollut. 2020, 266, 115254. [Google Scholar] [CrossRef]
- Olano, H.; Martigani, F.; Somma, A.; Aubriot, L. Wastewater Discharge with Phytoplankton May Favor Cyanobacterial Development in the Main Drinking Water Supply River in Uruguay. Environ. Monit. Assess. 2019, 191, 146. [Google Scholar] [CrossRef]
- Ruiz-González, C.; Niño-García, J.P.; Del Giorgio, P.A. Terrestrial Origin of Bacterial Communities in Complex Boreal Freshwater Networks. Ecol. Lett. 2015, 18, 1198–1206. [Google Scholar] [CrossRef]
- Begmatov, S.; Dorofeev, A.G.; Kadnikov, V.V.; Beletsky, A.V.; Pimenov, N.V.; Ravin, N.V.; Mardanov, A.V. The Structure of Microbial Communities of Activated Sludge of Large-Scale Wastewater Treatment Plants in the City of Moscow. Sci. Rep. 2022, 12, 3458. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.T.T.; Petrovich, M.L.; Chaudhary, A.; Wright, D.; Murphy, B.; Wells, G.; Poretsky, R. Metagenomics Reveals the Impact of Wastewater Treatment Plants on the Dispersal of Microorganisms and Genes in Aquatic Sediments. Appl. Environ. Microbiol. 2018, 84, e02168-17. [Google Scholar] [CrossRef] [PubMed]
- Abia, A.L.K.; Alisoltani, A.; Keshri, J.; Ubomba-Jaswa, E. Metagenomic Analysis of the Bacterial Communities and Their Functional Profiles in Water and Sediments of the Apies River, South Africa, as a Function of Land Use. Sci. Total Environ. 2018, 616–617, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J. Structure and Function of the Global Topsoil Microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, X.; Shu, H.Y.; Lin, X.R.; Zhou, Q.X.; Bramryd, T.; Shu, W.S.; Huang, L.N. Microbial Community Structure and Function in Sediments from E-Waste Contaminated Rivers at Guiyu Area of China. Environ. Pollut. 2018, 235, 171–179. [Google Scholar] [CrossRef]
- Azli, B.; Razak, M.N.; Omar, A.R.; Mohd Zain, N.A.; Abdul Razak, F.; Nurulfiza, I. Metagenomics Insights into the Microbial Diversity and Microbiome Network Analysis on the Heterogeneity of Influent to Effluent Water. Front. Microbiol. 2022, 13, 779196. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.L. Microorganisms and Their Roles in Fundamental Biogeochemical Cycles. Curr. Opin. Biotechnol. 2011, 22, 456–464. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Wang, P.F.; Zhang, H.; Lin, C. Response of Bacterial Community Compositions to Different Sources of Pollutants in Sediments of a Tributary of Taihu Lake, China. Environ. Sci. Pollut. Res. 2016, 23, 13886–13894. [Google Scholar] [CrossRef]
- McLellan, S.L.; Eren, A.M. Discovering New Indicators of Fecal Pollution. Trends Microbiol. 2014, 22, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Li, L.G.; Huang, Q.; Yin, X.; Zhang, T. Source Tracking of Antibiotic Resistance Genes in the Environment—Challenges, Progress, and Prospects. Water Res. 2020, 185, 116127. [Google Scholar] [CrossRef]
- Schiaffino, F.; Pisanic, N.; Colston, J.M.; Rengifo, D.; Paredes Olortegui, M.; Shapiama, V.; Peñataro Yori, P.; Heaney, C.D.; Davis, M.F.; Kosek, M.N. Validation of Microbial Source Tracking Markers for the Attribution of Fecal Contamination in Indoor-Household Environments of the Peruvian Amazon. Sci. Total Environ. 2020, 743, 140531. [Google Scholar] [CrossRef]
- Duarte, A.S.R.; Röder, T.; Van Gompel, L.; Petersen, T.N.; Hansen, R.B.; Hansen, I.M.; Bossers, A.; Aarestrup, F.M.; Wagenaar, J.A.; Hald, T. Metagenomics-Based Approach to Source-Attribution of Antimicrobial Resistance Determinants–Identification of Reservoir Resistome Signatures. Front. Microbiol. 2021, 12, 3447. [Google Scholar] [CrossRef]
- Chen, J.; Chen, H.; Liu, C.; Huan, H.; Teng, Y. Evaluation of FEAST for Metagenomics-Based Source Tracking of Antibiotic Resistance Genes. J. Hazard. Mater. 2023, 442, 130116. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, L.; Wang, X.; Li, L.; Wang, Z.; Lin, K. Source Apportionment Based on EEM-PARAFAC Combined with Microbial Tracing Model and Its Implication in Complex Pollution Area, Wujin District, China. Environ. Pollut. 2024, 346, 123596. [Google Scholar] [CrossRef] [PubMed]
- Shenhav, L.; Thompson, M.; Joseph, T.A.; Briscoe, L.; Furman, O.; Bogumil, D.; Mizrahi, I.; Pe Er, I.; Halperin, E. FEAST: Fast expectation-maximization for microbial source tracking. Nat. Methods 2019, 16, 627–632. [Google Scholar] [CrossRef]
- Hao, M.; Zuo, Q.; Li, J.; Shi, S.; Li, B.; Zhao, X. A Comprehensive Exploration on Distribution, Risk Assessment, and Source Quantification of Heavy Metals in the Multi-Media Environment from Shaying River Basin, China. Ecotoxicol. Environ. Saf. 2022, 231, 113190. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yuan, X.; van der Hoek, J.P. Residues of Organochlorine Pesticides (OCPs) in Aquatic Environment and Risk Assessment along Shaying River, China. Environ. Geochem. Health 2018, 40, 2525–2538. [Google Scholar] [CrossRef] [PubMed]
- He, B.; He, J.; Wang, L.; Zhang, X.; Bi, E. Effect of Hydrogeological Conditions and Surface Loads on Shallow Groundwater Nitrate Pollution in the Shaying River Basin: Based on Least Squares Surface Fitting Model. Water Res. 2019, 163, 114880. [Google Scholar] [CrossRef]
- GB18918-2002; Discharge Standard of Pollutants for Municipal Wastewater Treatment Plants. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2003.
- Wang, J.; Liu, T.; Sun, W.; Chen, Q. Bioavailable Metal(loid)s and Physicochemical Features Co-Meditating Microbial Communities at Combined Metal(loid) Pollution Sites. Chemosphere 2020, 260, 127619. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Ram, R.J.; Matthew, D.; Evan, B.; Rob, K. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Segata, N.; Abubucker, S.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Waldron, L.; Zucker, J.; Henrissat, B.; et al. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P. vegan: Community Ecology Package; R Package Version 2.2-1; CRAN: Vienna, Austria, 2015. [Google Scholar]
- Yoshiki, V.; Pirrung, M.; Gonzalez, A.; Knight, R. Emperor: A tool for visualizing high-throughput microbial community data. Giga Sci. 2013, 2, 16. [Google Scholar]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian Community-Wide Culture-Independent Microbial Source Tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, G.; Jiang, S.; Liu, Y.X. Wekemo Bioincloud: A User-Friendly Platform for Meta-Omics Data Analyses. iMeta 2024, 3, e175. [Google Scholar] [CrossRef]
- GB3838-2002; Environmental Quality Standards for Surface Water of China. Ministry of Environmental Protection of China: Beijing, China, 2002.
- Wardi, M.; Slimani, N.; Ait Alla, A.; Dahchour, A.; El Mourabit, F.; Zekhnini, A.; Aziz, F. First Study of the Effect of Wastewater Treatment on Microbial Biodiversity at Three Wastewater Treatment Plants in Agadir, Morocco, Using 16S rRNA Sequencing. Environ. Pollut. 2023, 337, 122528. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Gao, Y.C.; Huang, X.N.; Li, J.; Lin, X.G. Adaptive Shifts of Bacterioplankton Communities in Response to Nitrogen Enrichment in a Highly Polluted River. Environ. Pollut. 2019, 245, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Yu, Z.; Ji, J. Microbial community structure and function indicate the severity of chromium contamination of the Yellow River. Front. Microbiol. 2018, 9, 38. [Google Scholar] [CrossRef]
- Chen, X.; Ji, J.; Li, P.; Peng, X.; Wang, J.; Fu, C.; Zong, Y. Characterization of microbial communities in the upstream water body of Linzhi City section of Yarlung Tsangpo River and changes in nitrogen cycle-related microbiota. J. Ecol. Rural Environ. 2025, 1, 1–24. (In Chinese) [Google Scholar]
- Chou, J.H.; Cho, N.T.; Arun, A.B. Nocardioides fonticola sp. nov, a novel actinomycete isolated from spring water. Int. J Syst. Evol. Microbiol. 2008, 58, 18641868. [Google Scholar] [CrossRef]
- Dang, H.; Li, J.; Chen, R.; Wang, L.; Guo, L.; Zhang, Z.; Klotz, M.G. Diversity, abundance, and spatial distribution of sediment ammonia oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl. Environ. Microbiol. 2010, 76, 4691–4702. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, B.; Li, J.; Gu, Z.; Liu, X. Wildfire Impacts Pinus tabulaeformis Forests on Soil Properties, Actinobacteriota, and Enzyme Activity in Northern China: Direct Effects or Mutual Interactions? Forests 2025, 16, 344. [Google Scholar] [CrossRef]
- Qin, B.Q.; Zhou, J.; Elser, J.J.; Gardner, W.S.; Deng, J.M.; Brookes, J.D. Water Depth Underpins the Relative Roles and Fates of Nitrogen and Phosphorus in Lakes. Environ. Sci. Technol. 2020, 54, 3191–3198. [Google Scholar] [CrossRef]
- Zhu, Z.; Ma, Y.; Huang, L.; Zhang, W. Homogeneous selection is not always important in bacterial community in the eutrophic enclosed bay. Ecol. Process. 2022, 11, 27. [Google Scholar] [CrossRef]
- Francis, C.; Beman, M.; Kuypers, M. New processes and players in the nitrogen cycle: The microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007, 1, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, M.; Liu, L.; Wang, Z.; Lin, K. Using EEM-PARAFAC to Identify and Trace the Pollution Sources of Surface Water with Receptor Models in Taihu Lake Basin, China. J. Environ. Manag. 2022, 321, 115925. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.Y.; Chen, L.; Dong, Y.W. Unraveling dissolved organic matter in drinking water through integrated ozonation/ceramic membrane and biological activated carbon process using FT-ICR MS. Water Res. 2022, 222, 118881. [Google Scholar] [CrossRef]
- Kong, Z.; Li, L.; Feng, C.; Dong, S.; Chen, N. Comparative investigation on integrated vertical-flow biofilters applying sulfur-based and pyrite-based autotrophic denitrification for domestic wastewater treatment. Bioresour. Technol. 2016, 211, 125–135. [Google Scholar] [CrossRef]




| Parameter | Mean | Min | Max | SD | CV |
|---|---|---|---|---|---|
| T (°C) | 21.59 | 19.20 | 23.20 | 1.28 | 0.06 |
| EC (µS/cm) | 690.78 | 414.00 | 913.00 | 163.38 | 0.24 |
| TDS (mg/L) | 345.56 | 201.00 | 474.00 | 85.73 | 0.25 |
| pH | 8.16 | 7.63 | 8.90 | 0.45 | 0.05 |
| ORP (mV) | 32.33 | 28.00 | 53.00 | 7.38 | 0.23 |
| DO (mg/L) | 8.60 | 6.49 | 14.04 | 2.18 | 0.25 |
| COD (mg/L) | 34.63 | 20.08 | 68.34 | 14.56 | 0.42 |
| TN (mg/L) | 2.22 | 0.21 | 6.80 | 1.94 | 0.87 |
| NH3-N (mg/L) | 0.61 | 0.06 | 2.39 | 0.68 | 1.12 |
| TP (mg/L) | 0.07 | 0.01 | 0.12 | 0.03 | 0.46 |
| Samples | Input | Filtered | Chao | Shannon | Simpson | Pielou Evenness | Coverage (%) |
|---|---|---|---|---|---|---|---|
| S1 | 18,217 | 164,760 | 1389.74 | 7.37 | 0.99 | 0.72 | 0.98 |
| S2 | 186,090 | 169,945 | 1472.93 | 7.32 | 0.98 | 0.70 | 0.98 |
| S3 | 187,824 | 172,317 | 593.68 | 6.45 | 0.97 | 0.75 | 0.97 |
| S4 | 185,918 | 169,179 | 1834.80 | 8.09 | 0.99 | 0.75 | 0.99 |
| S5 | 182,571 | 165,717 | 1451.91 | 7.16 | 0.98 | 0.69 | 0.98 |
| S6 | 185,307 | 169,706 | 1082.82 | 6.72 | 0.97 | 0.66 | 0.97 |
| S7 | 185,215 | 168,494 | 1307.15 | 7.09 | 0.98 | 0.68 | 0.98 |
| S8 | 183,904 | 165,052 | 1641.75 | 7.95 | 0.99 | 0.73 | 0.99 |
| S9 | 186,525 | 169,687 | 1162.36 | 7.06 | 0.98 | 0.68 | 0.98 |
| Rarefaction Depth | S1 | S2 | S3 | S4 |
|---|---|---|---|---|
| 1000 | 9.88% ± 0.69% | 31.28% ± 1.25% | 13.55% ± 1.84% | 36.75% ± 2.37% |
| 5000 | 8.25% ± 0.62% | 31.73% ± 0.78% | 14.72% ± 1.15% | 41.27% ± 0.94% |
| 10,000 | 8.61% ± 0.69% | 31.22% ± 0.22% | 15.23% ± 1.41% | 41.20% ± 1.36% |
| 20,000 | 9.33% ± 1.39% | 30.45% ± 0.69% | 13.69% ± 1.13% | 43.47% ± 0.91% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Gao, H.; Jiang, Z.; Lv, Y.; Guo, X.; Zhu, X.; Wu, J.; Li, Y.; Yu, W.; Li, Q.; et al. Integrating Microbial Source Tracking to Unravel Impacts of Wastewater Discharge on Spatial Distribution of Riverine Microbial Community. Water 2025, 17, 2753. https://doi.org/10.3390/w17182753
Fan Y, Gao H, Jiang Z, Lv Y, Guo X, Zhu X, Wu J, Li Y, Yu W, Li Q, et al. Integrating Microbial Source Tracking to Unravel Impacts of Wastewater Discharge on Spatial Distribution of Riverine Microbial Community. Water. 2025; 17(18):2753. https://doi.org/10.3390/w17182753
Chicago/Turabian StyleFan, Yanru, Hongbin Gao, Zhongfeng Jiang, Yuran Lv, Xiang Guo, Xinfeng Zhu, Junfeng Wu, Yizhe Li, Wenxiang Yu, Qi Li, and et al. 2025. "Integrating Microbial Source Tracking to Unravel Impacts of Wastewater Discharge on Spatial Distribution of Riverine Microbial Community" Water 17, no. 18: 2753. https://doi.org/10.3390/w17182753
APA StyleFan, Y., Gao, H., Jiang, Z., Lv, Y., Guo, X., Zhu, X., Wu, J., Li, Y., Yu, W., Li, Q., & Yuan, K. (2025). Integrating Microbial Source Tracking to Unravel Impacts of Wastewater Discharge on Spatial Distribution of Riverine Microbial Community. Water, 17(18), 2753. https://doi.org/10.3390/w17182753









