Microbial DNA-Based Monitoring of Underground Crude Oil Storage Bases Using Water-Sealed Rock-Cavern Tanks
Abstract
1. Introduction
2. Materials and Methods
2.1. General Description of the Sampling Sites
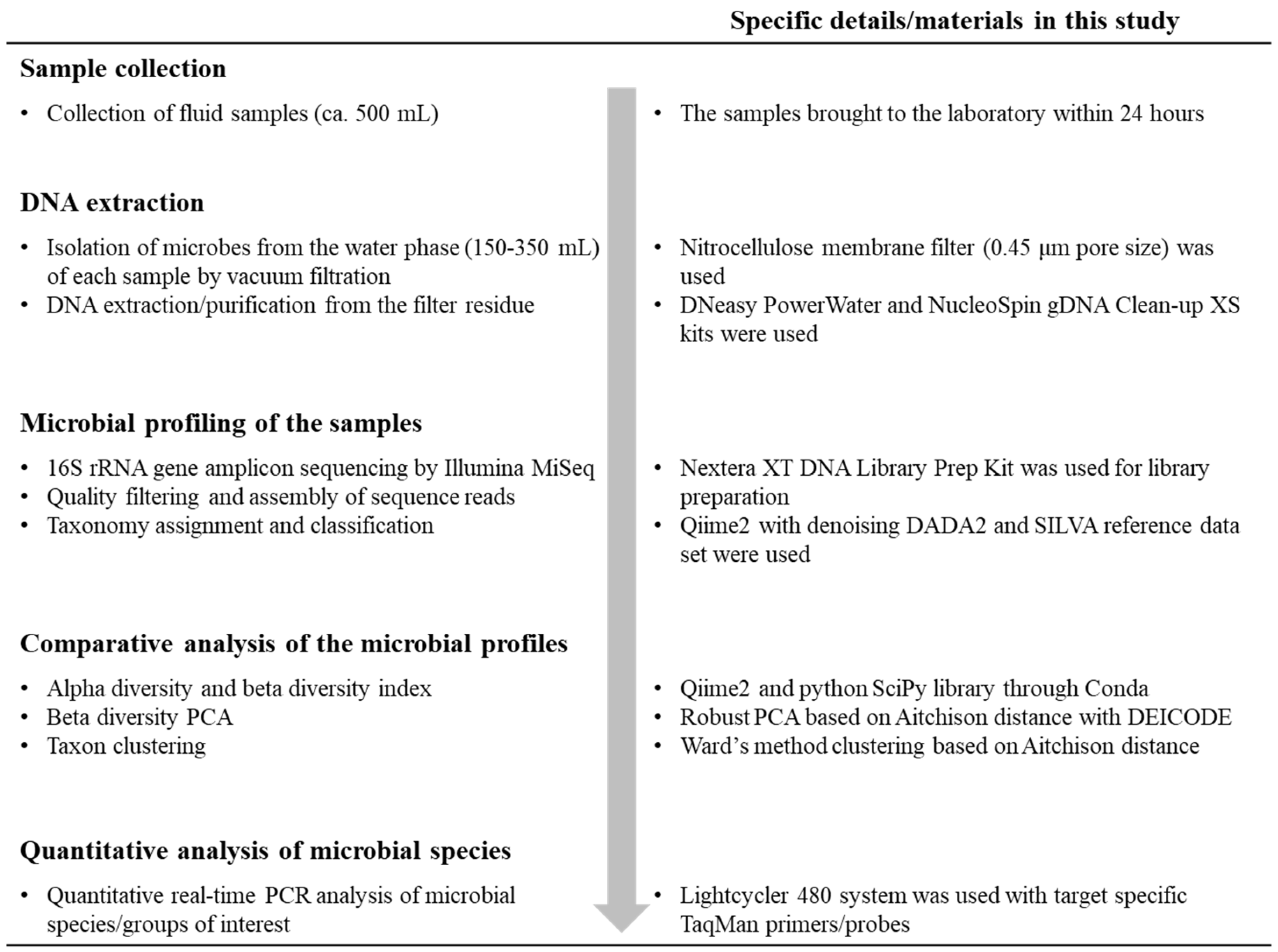
2.2. Sample Collection
2.3. DNA Extraction
2.4. Sequencing of 16S rRNA Gene Amplicons
2.5. Phylogenetic Characterization of the 16S rRNA Gene Amplicon Sequences
2.6. Statistical Analyses of the Microbial Profiles of Samples
2.7. Real-Time Quantitative Polymerase Chain Reaction
3. Results
3.1. Microbial Community Composition
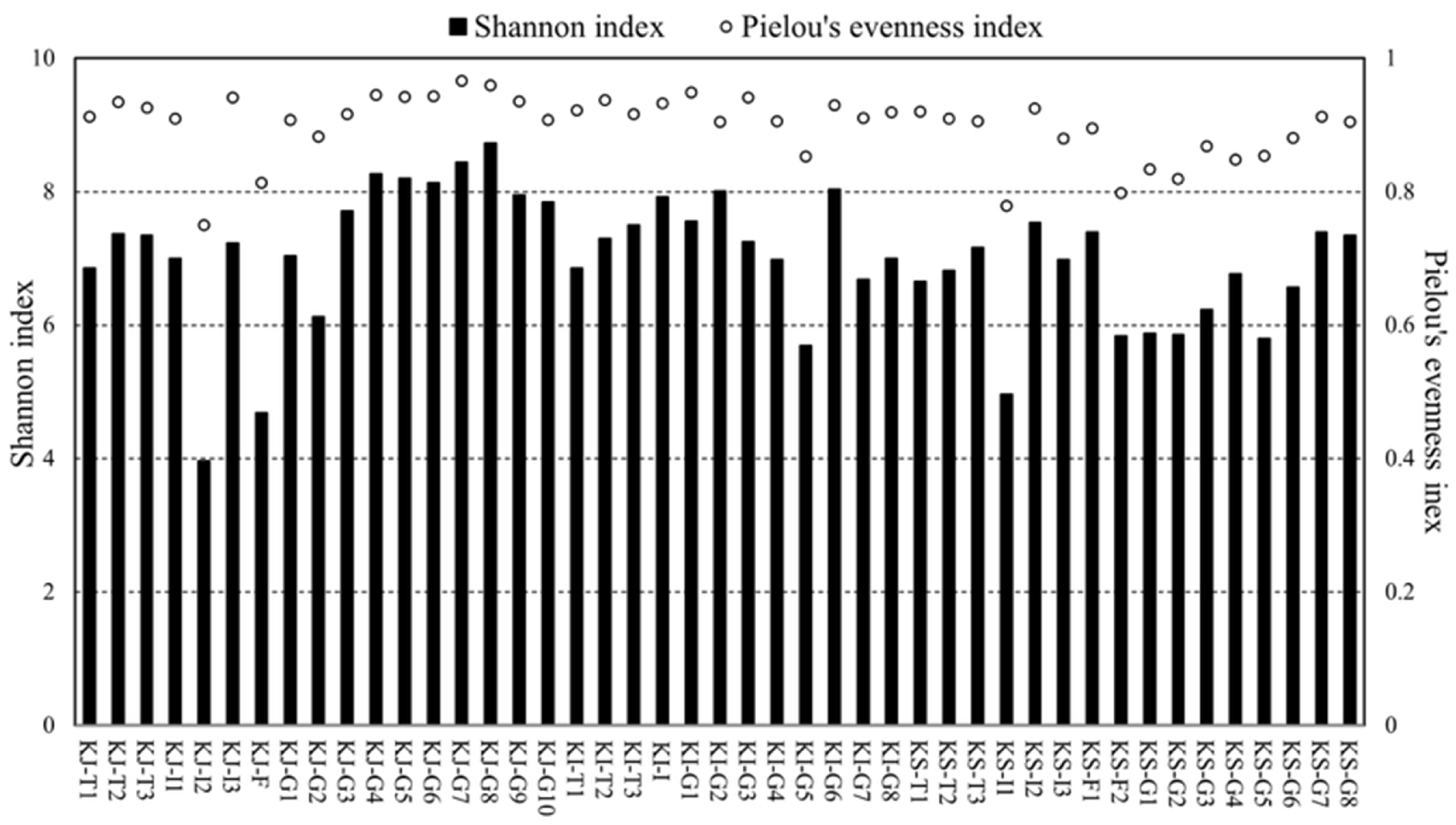
3.2. Data Trend in Alpha Diversity Index
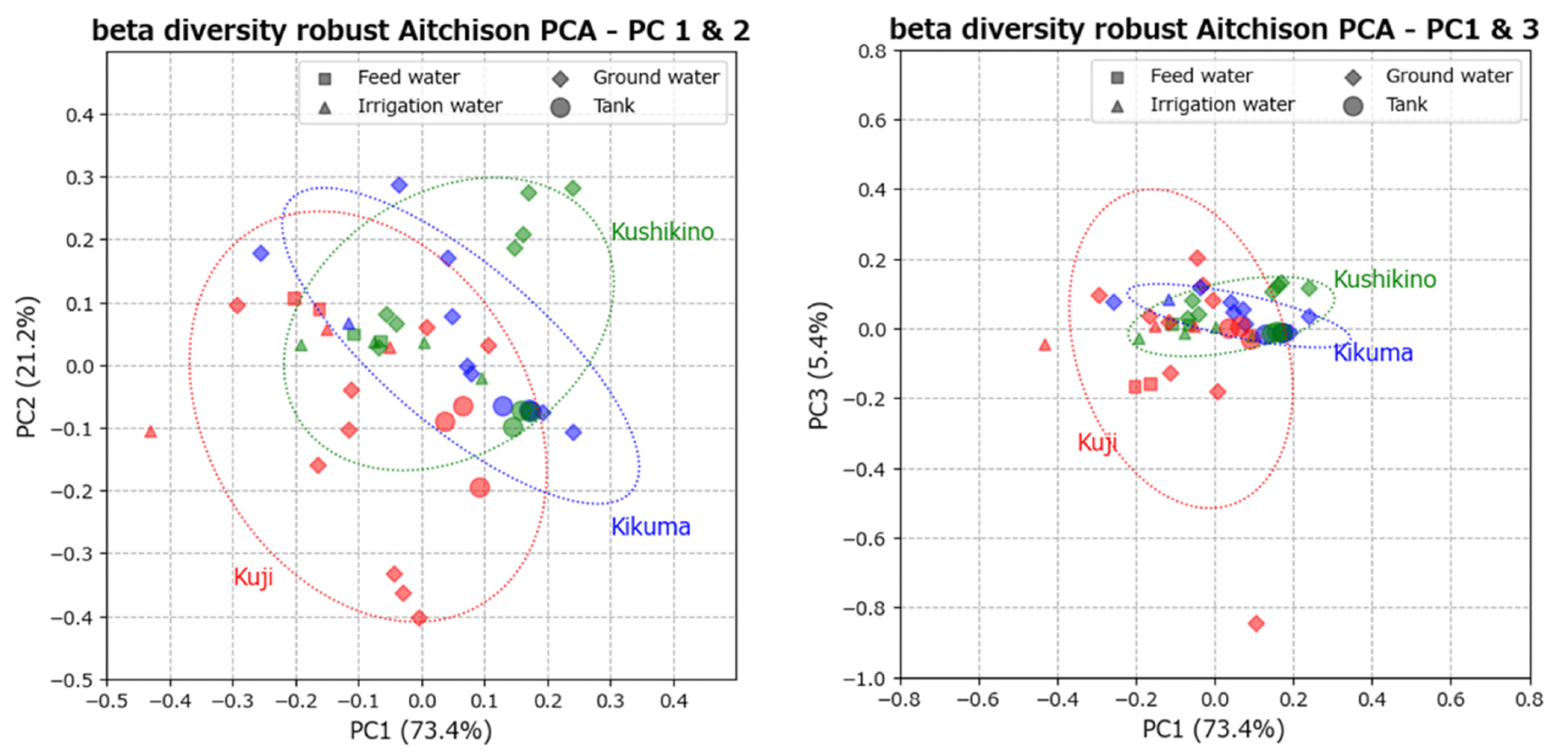
3.3. Data Visualization of Beta Diversity with PCA of Robust Aitchison Distances
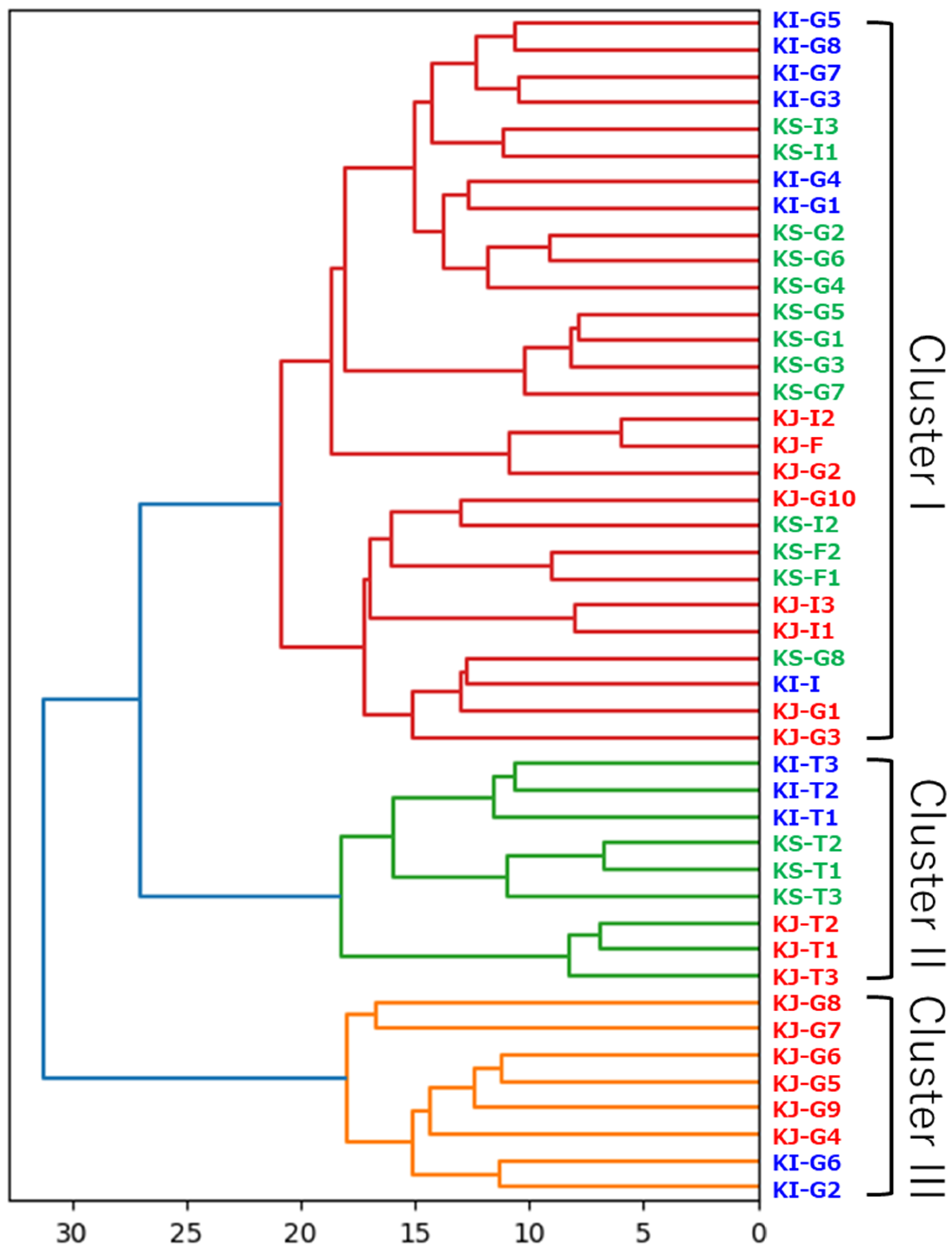
3.4. Data Visualization Using Aitchison Distances and Ward’s Linkage Method
3.5. Data Visualization Using Robust Z-Score Heatmap and qPCR Quantification of 16S rRNA Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| h | hours |
| min | minutes |
| m | meter |
| s | seconds |
| kL | kiloliter |
| mL | milliliter (10−3 dm3) |
| µL | milliliter (10−3 dm3) |
| µM | micro mol/dm3 |
| ppm | 10−6 g per kilogram water |
| ng | nanogram (10−9 g) |
| °C | degree Celsius |
| % | percent |
References
- International Energy Agency (IEA). Japan Oil Security Policy. Part of Oil Security Policy. 18 August 2022. Available online: https://www.iea.org/articles/japan-oil-security-policy (accessed on 1 November 2024).
- Japan, Ministry of Economy, Trade and Industry. Japan’s Energy White Paper 2023. 2023. Available online: https://www.meti.go.jp/english/press/2023/0606_003.html (accessed on 1 November 2024).
- Petroleum & LP Gas Stockpiling, Japan Organization for Metals and Energy Security (JOGMEC). 2018. Available online: https://www.jogmec.go.jp/content/300320107.pdf (accessed on 1 November 2023).
- Kiyoyama, S. The Present State of Underground Crude Oil Storage Technology in Japan. Tunn. Undergr. Space Technol. 1990, 5, 343–349. [Google Scholar] [CrossRef]
- Watanabe, K.; Jodai, K.; Miyashita, K.; Oono, K.; Yamamoto, N.; Hirooka, S.; Kataoka, S. Influence of Earthquakes on the Rock Caverns of Underground Petroleum Stockpiling Bases. J. Groundw. Hydrol. 2022, 64, 21–34. (In Japanese) [Google Scholar] [CrossRef]
- Yoshida, H.; Ohto, Y.; Furui, D. Regarding the tsunami damage and the crisis measures by the earthquake on March 11, 2011 in the national oil stockpiling base in Kuji, Japan. Explosion 2011, 21, 111–116. (In Japanese) [Google Scholar] [CrossRef]
- Jodai, K.; Uede, K.; Honda, M.; Hasegawa, M.; Kojima, K.; Ogawa, T. Simplified Criteria for Daily Inspection of Water-Sealing Function Stability of Underground Oil Cavern. J. Jpn. Soc. Civ. Eng. Collect. Pap. C 2007, 63, 624–634. (In Japanese) [Google Scholar] [CrossRef]
- Yamashita, M.; Amadomari, T.; Hirai, T.; Bouno, H.; Suenaga, H. An Application of Fiber-Optic Temperature Measurement for Groundwater Behavior Monitoring at Underground Petroleum Stockpiling Base. J. Groundw. Hydrol. 2022, 64, 35–48. (In Japanese) [Google Scholar] [CrossRef]
- Morfeldf, C. Storage of Oil and Gas in Unlined Caverns. In Proceedings of the SPE European Spring Meeting, Amsterdam, The Netherlands, 29–30 May 1974. [Google Scholar] [CrossRef]
- Lindblom, U.E. History and Present Status of Hydrocarbon Storage in Excavated Rock Caverns. In Proceedings of the Rock Mechanics in Petroleum Engineering, Delft, The Netherlands, 29–31 August 1994. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, B.; Shi, L.; Ye, Y. Dynamic Variation Characteristics of Seawater Intrusion in Underground Water-Sealed Oil Storage Cavern under Island Tidal Environment. Water 2019, 11, 130. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, B.; Li, Y.; Wang, L.; Li, Y.; Shi, L.; Wang, H. Probabilistic analysis of water-sealed performance in underground oil storage considering spatial variability of hydraulic conductivity. Sci. Rep. 2022, 12, 13782. [Google Scholar] [CrossRef]
- Lascelles, P.; Wan, J.; Robinson, L.; Allmon, R.; Evans, G.; Ursell, L.; Scott, N.M.; Chase, J.; Jablanovic, J.; Karimi, M.; et al. Applying Subsurface DNA Sequencing in Wolfcamp Shales, Midland Basin. In Proceedings of the SPE Hydraulic Fracturing Technology Conference and Exhibition, Woodlands, TX, USA, 24–26 January 2017. [Google Scholar] [CrossRef]
- Vigneron, A.; Alsop, E.B.; Lomans, B.P.; Kyrpides, N.C.; Head, I.M.; Tsesmetzis, N. Succession in the Petroleum Reservoir Microbiome through an Oil Field Production Lifecycle. ISME J. 2017, 11, 2141–2154. [Google Scholar] [CrossRef]
- Silva, J.; Ursell, L.; Percak-Dennett, E. Applying Subsurface DNA Diagnostics and Data Science in the Delaware Basin. In Proceedings of the SPE Hydraulic Fracturing Technology Conference and Exhibition, Woodlands, TX, USA, 23–25 January 2018. [Google Scholar] [CrossRef]
- Percak-Dennett, E.; Liu, J.; Shojaei, H.; Luke, U.; Thomas, I. High Resolution Dynamic Drainage Height Estimations Using Subsurface DNA Diagnostics. In Proceedings of the SPE Western Regional Meeting, San Jose, CA, USA, 23–26 April 2019. [Google Scholar] [CrossRef]
- Ursell, L.; Hale, M.; Menendez, E.; Zimmerman, J.; Dombroski, B.; Hoover, K.; Everman, Z.; Liu, J.K.; Shojaei, H.; Percak-Dennett, E.M.; et al. High Resolution Fluid Tracking from Verticals and Laterals Using Subsurface DNA Diagnostics in the Permian Basin. In Proceedings of the SPE/AAPG/SEG Unconventional Resources Technology Conference, Denver, CO, USA, 22–24 July 2019. [Google Scholar] [CrossRef]
- Schlecht, M.; Sawadogo, J.; Sadeghi, S.; Reeve, N.; Haggerty, M.; Liu, J.; Ursell, L. Improved Stacked Permian Development by Integrating DNA Diagnostics with Traditional Reservoir Analysis. In Proceedings of the SPE Annual Technical Conference and Exhibition, Virtual, 26–29 October 2020. [Google Scholar] [CrossRef]
- Sawadogo, J.; Haggerty, M.; Mallory, C.L.; Huchton, J.; DeAngelis, W.; Price, C. Impact of Completion Design and Interwell Communication on Well Performance in Full Section Development: A STACK Case Study Using DNA Based Diagnostics. In Proceedings of the SPE Annual Technical Conference and Exhibition, Virtual, 26–29 October 2020. [Google Scholar] [CrossRef]
- Sawadogo, J.; Ursell, L.; Reeve, N.; Schlecht, M. Mature Fields—Optimizing Waterflood Management through DNA Based Diagnostics. In Proceedings of the Abu Dhabi International Petroleum Exhibition & Conference, Abu Dhabi, United Arab Emirates, 9–12 November 2020. [Google Scholar] [CrossRef]
- Li, J.; Pang, Z.; Tian, L.; Zhao, H.; Bai, G. Variations of Stable Isotopes in Daily Precipitation in a Monsoon Region. Water 2022, 14, 2891. [Google Scholar] [CrossRef]
- Paruch, L.; Paruch, A.M. An Overview of Microbial Source Tracking Using Host-Specific Genetic Markers to Identify Origins of Fecal Contamination in Different Water Environments. Water 2022, 14, 1809. [Google Scholar] [CrossRef]
- Zhang, Y.; Dekas, A.E.; Hawkins, A.J.; Primo, J.C.; Gorbatenko, O.; Horne, R.N. Comparison of Microbial Profiling and Tracer Testing for the Characterization of Injector-Producer Interwell Connectivities. Water 2022, 14, 2921. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, T. DNA-Based Tracers for the Characterization of Hydrogeological Systems—Recent Advances and New Frontiers. Water 2022, 14, 3545. [Google Scholar] [CrossRef]
- Zhang, Y.; Horne, R.N.; Hawkins, A.J.; Primo, J.C.; Gorbatenko, O.; Dekas, A.E. Geological activity shapes the microbiome in deep-subsurface aquifers by advection, Proc. Natl. Acad. Sci. USA 2022, 119, e2113985119. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Goto, A.; Feng, X.; Uruma, K.; Momoi, Y.; Watanabe, S.; Sato, K.; Zhang, Y.; Horne, R.N.; Shibuya, T.; et al. Long-Term Microbial DNA-Based Monitoring of the Mature Sarukawa Oil Field in Japan. SPE Reserv. Eval. Eng. 2023, 26, 1110–1119. [Google Scholar] [CrossRef]
- Tang, P.; Zhang, W.; Wang, H.; Zhou, J.; Dang, Y.; Chao, Z. Analysis of the Hydromechanical Properties of Compact Sandstone and Engineering Application. Water 2023, 15, 2011. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003, 55, 541–555. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, W.L.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 Years of Serving the Community with Ribosomal RNA Gene Reference Databases and Tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible Sequence Taxonomy Reference Database Management. PLOS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Champaign, IL, USA, 1949. [Google Scholar]
- Pielou, E.C. The Measurement of Diversity in Different Types of Biological Collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME 2 enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef]
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.; Grout, J.; Corlay, S.; et al. Jupyter Notebooks—A Publishing Format for Reproducible Computational Workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas; IOS Press: Amsterdam, The Netherlands, 2016; pp. 87–90. [Google Scholar] [CrossRef]
- Spruyt, V. How to Draw an Error Ellipse Representing the Covariance Matrix. Computer Vision for Dummies. 3 April 2014. Available online: https://www.visiondummy.com/2014/04/draw-error-ellipse-representing-covariance-matrix (accessed on 1 November 2023).
- CarstenSchelp.github.io. Bits, Pieces, Nuts and Bolts on Data Analysis and Machine Learning. Schelp, C. An Alternative Way to Plot the Covariance Ellipse. Available online: https://carstenschelp.github.io/2018/09/14/Plot_Confidence_Ellipse_001.html (accessed on 1 November 2023).
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Quinn, T.P.; Richardson, M.F.; Lovell, D.; Crowley, T.M. Propr: An R-Package for Identifying Proportionally Abundant Features Using Compositional Data Analysis. Sci. Rep. 2017, 7, 16252. [Google Scholar] [CrossRef]
- Jin, S.; Notredame, C.; Erb, I. Compositional Covariance Shrinkage and Regularised Partial Correlations. SORT 2023, 47, 245–268. [Google Scholar] [CrossRef]
- Rideout, J.R.; Caporaso, G.; Bolyen, E.; McDonald, D.; Baeza, Y.V.; Alastuey, J.C.; Pitman, A.; Morton, J.; Navas, J.; Gorlick, K.; et al. Scikit-Bio/Scikit-Bio: Scikit-Bio 0.6.0. Zenodo. 2024. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Everitt, B.S.; Landau, S.; Leese, M.; Stahl, D. Cluster Analysis, 5th ed.; Wiley Series in Probability and Statistics; King’s College London: London, UK, 2011. [Google Scholar]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-Specific Primer and Probe Sets to Detect Methanogenic Communities Using Quantitative Real-Time Polymerase Chain Reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Gaines, W.L.; Richy, H.J.; Lehmkuhl, J.F. Monitoring Biodiversity: Quantification and Interpretation; PNW-GTR-443. General Technical Report; U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station: Washington, DC, USA, 1999; p. 27. [Google Scholar] [CrossRef]
- Francé, J.; Varkitzi, I.; Stanca, E.; Cozzoli, F.; Skejić, S.; Ungaro, N.; Vascotto, I.; Mozetič, P.; Gladan, Ž.N.; Assimakopoulou, G.; et al. Large-Scale Testing of Phytoplankton Diversity Indices for Environmental Assessment in Mediterranean Sub-regions (Adriatic, Ionian and Aegean Seas). Ecol. Indic. 2021, 126, 107630. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Aitchison, J.; Barceló-Vidal, C.; Martín-Fernández, J.A.; Pawlowsky-Glahn, V. Logratio Analysis and Compositional Distance. Math. Geosci. 2000, 32, 271–275. [Google Scholar] [CrossRef]
- Martino, C.; Morton, J.T.; Marotz, C.A.; Thompson, L.R.; Tripathi, A.; Knight, R.; Zengler, K. A Novel Sparse Compositional Technique Reveals Microbial Perturbations. mSystems. 2019, 4, e00016-19. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Malo, N.; Hanley, J.A.; Cerquozzi, S.; Pelletier, J.; Nadon, R. Statistical Practice in High-Throughput Screening Data Analysis. Nat. Biotechnol. 2006, 24, 167–175. [Google Scholar] [CrossRef]
- Gibson, G.R. Physiology and Ecology of the Sulphate-Reducing Bacteria. J. Appl. Bacteriol. 1990, 69, 769–797. [Google Scholar] [CrossRef]
- Smith, G.J.; Angle, J.C.; Solden, L.M.; Borton, M.A.; Morin, T.H.; Daly, R.A.; Johnston, M.D.; Stefanik, K.C.; Wolfe, R.; Gil, B.; et al. Members of the Genus Methylobacter Are Inferred To Account for the Majority of Aerobic Methane Oxidation in Oxic Soils from a Freshwater Wetland. mBio 2018, 9, e00815-18. [Google Scholar] [CrossRef]
- Kodama, Y.; Watanabe, K. Isolation and Characterization of a Sulfur-Oxidizing Chemolithotroph Growing on Crude Oil under Anaerobic Conditions. Appl. Environ. Microbiol. 2003, 69, 107–112. [Google Scholar] [CrossRef]
- Kodama, Y.; Watanabe, K. Sulfuricurvum kujiense gen. nov., sp. nov., a Facultatively Anaerobic, Chemolithoautotrophic, Sulfur-Oxidizing Bacterium Isolated from an Underground Crude-Oil Storage Cavity. Int. J. Syst. Evol. Microbiol. 2004, 54, 2297–2300. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M.; Kobayashi, H.; Davies, B.W.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 2007, 5, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Ardissone, S.; Kobayashi, H.; Kambara, K.; Rummel, C.; Noel, K.D.; Walker, G.C.; Broughton, W.J.; Deakin, W.J. Role of BacA in Lipopolysaccharide Synthesis, Peptide Transport and Nodulation by Rhizobium sp. NGR234. J. Bacteriol. 2011, 193, 2218–2228. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-L.; Sekiguchi, Y.; Hanada, S.; Imachi, H.; Tseng, I.; Cheng, S.; Ohashi, A.; Harada, H.; Kamagata, Y. Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: Two Anaerobic Bacteria That Degrade Phthalate Isomers in Syntrophic Association with Hydrogenotrophic Methanogens. Arch. Microbiol. 2006, 185, 172–182. [Google Scholar] [CrossRef]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. The causes of Pseudomonas diversity. Microbiology 2000, 146, 2345–2350. [Google Scholar] [CrossRef]
- Silby, W.M.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]
- Watanabe, K.; Kodama, Y.; Syutsubo, K.; Harayama, S. Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude oil storage cavities. Appl. Environ. Microbiol. 2000, 66, 4803–4809. [Google Scholar] [CrossRef]
- Watanabe, K.; Kodama, Y.; Kaku, N. Diversity and abundance of bacteria in an underground oil-storage cavity. BMC Microbiol. 2002, 2, 23. [Google Scholar] [CrossRef]
- Nakamura, H.; Sumi, K. Geothermal Investigations of Matsukawa Hot Spring area. Bull. Geol. Surv. Jpn. 1958, 12, 73–84. (In Japanese) [Google Scholar]
- Imai, A.; Shikazono, N.; Shimizu, M.; Shimazaki, H. Sulfur Isotope Study on Hg and Sb Deposits in Japan. Resour Geol. 2006, 56, 37–48. [Google Scholar] [CrossRef]
- Aljubran, M.J.; Horne, R.N. Thermal Earth model for the conterminous United States using an interpolative physics-informed graph neural network. Geotherm. Energy 2024, 12, 25. [Google Scholar] [CrossRef]
- Yang, H.; Yu, C.; Wang, S.; Scheirer, A.H.; Kong, X.; Zhao, H.; Yang, X.; Wang, S.; Jiang, L. DNA-sequencing method maps subsurface fluid flow paths for enhanced monitoring. Commun. Earth Environ. 2025, 6, 306. [Google Scholar] [CrossRef]
- Sun, S.; Firoozabadi, A.; Wheeler, M. Introduction of COMPES: Finding directions in energy and computational science. Comput. Energy Sci. 2024, 1, 1–2. [Google Scholar] [CrossRef]

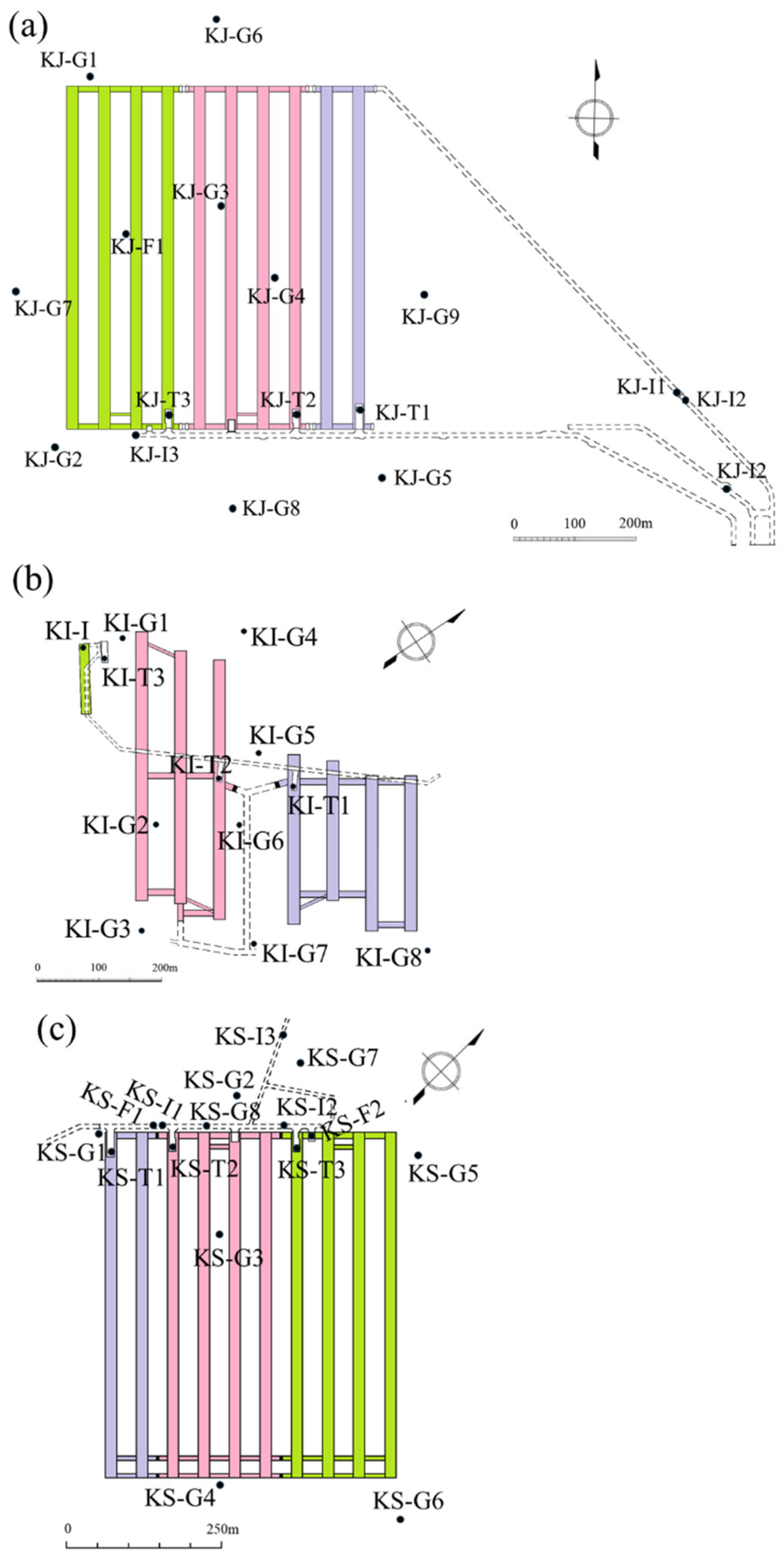



| Kuji | Kikuma | Kushikino | |
|---|---|---|---|
| Location (prefecture) | Iwate | Ehime | Kagoshima |
| Rock type | Granite | Granite | Andesite |
| Year of construction | 1989–1992 | 1989–1992 | 1988–1991 |
| Year of oil in | 1993 | 1994 | 1993 |
| Tank capacity (kL) | 1.75 million | 1.5 million | 1.75 million |
| Cavern dimension, W × H × L (m) | 18 × 22 × 540 | 20.5 × 30 × 230 − 448 | 18 × 22 × 555 |
| Mean sea level (m) | −20 to −42 | −35 to −65 | −20 to −42 |
| Subsurface depth (m) | Under −100 | −65 to −100 | Under −100 |
| Total water inflow (m3/day) * | 4600 | 200 | 320 |
| Feedwater supply (m3/day) * | 2000 | 40 to 50 | 20 to 40 |
| Kuji | Kikuma | Kushikino | |
|---|---|---|---|
| Tank (T) | KJ-T1/T2/T3 | KI-T1/T2/T3 | KS-T1/T2/T3 |
| Irrigation water (I) | KJ-I1/I2/I3 | KI-I | KS-I1/I2/I3 |
| Feedwater (F) | KJ-F | - | KS-F1/F2 |
| Groundwater (G) | KJ-G1 to G10 | KI-G1 to G8 | KS-G1 to G8 |
| Sample number | 17 | 12 | 16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goto, A.; Watanabe, S.; Uruma, K.; Momoi, Y.; Oomukai, T.; Kobayashi, H. Microbial DNA-Based Monitoring of Underground Crude Oil Storage Bases Using Water-Sealed Rock-Cavern Tanks. Water 2025, 17, 2197. https://doi.org/10.3390/w17152197
Goto A, Watanabe S, Uruma K, Momoi Y, Oomukai T, Kobayashi H. Microbial DNA-Based Monitoring of Underground Crude Oil Storage Bases Using Water-Sealed Rock-Cavern Tanks. Water. 2025; 17(15):2197. https://doi.org/10.3390/w17152197
Chicago/Turabian StyleGoto, Ayae, Shunichi Watanabe, Katsumasa Uruma, Yuki Momoi, Takuji Oomukai, and Hajime Kobayashi. 2025. "Microbial DNA-Based Monitoring of Underground Crude Oil Storage Bases Using Water-Sealed Rock-Cavern Tanks" Water 17, no. 15: 2197. https://doi.org/10.3390/w17152197
APA StyleGoto, A., Watanabe, S., Uruma, K., Momoi, Y., Oomukai, T., & Kobayashi, H. (2025). Microbial DNA-Based Monitoring of Underground Crude Oil Storage Bases Using Water-Sealed Rock-Cavern Tanks. Water, 17(15), 2197. https://doi.org/10.3390/w17152197






