Free-Living Protozoa and Legionella spp. Coexistence and Bacterial Diversity in Drinking Water Systems in Apartment Buildings and Hotels in Riga and Its Surroundings
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Detection of Legionella spp. and FLP
2.3. 18S and 16S rRNA Amplicon Sequencing and Sequence Analysis
2.4. Statistical Analysis of Data
3. Results and Discussion
3.1. Water Temperature in Apartment Houses and Hotels; Water Consumption
3.2. Presence of FLP and Legionella spp. and Their Co-Existence
3.3. 18S rRNA Amplicon Sequencing Results
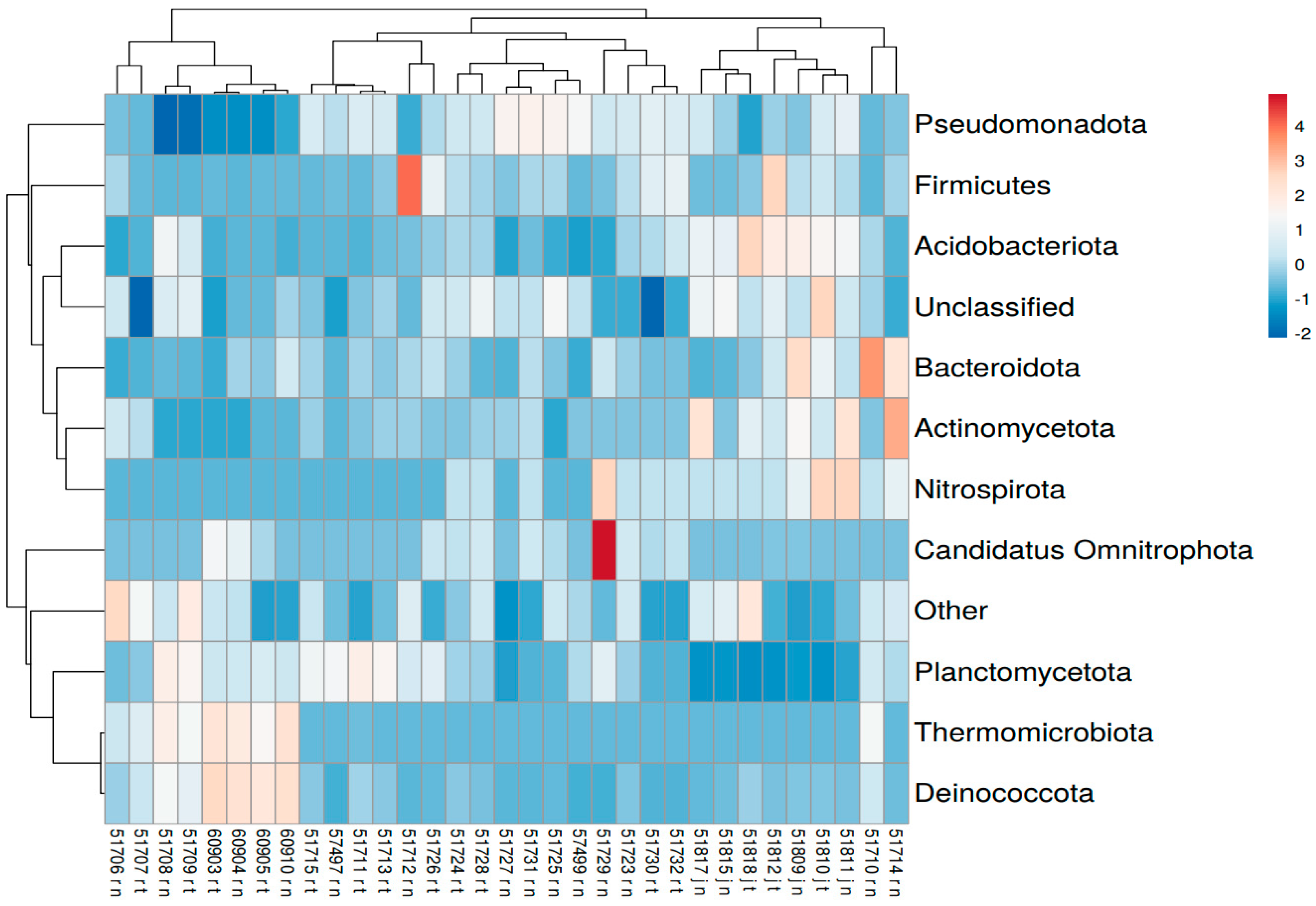
3.4. 16S rRNA Amplicon Sequencing Results
4. Conclusions
- Acanthamoeba, Vahlkampfia, and Hartmanella (Vermamoeba) free-living amoebas were identified in water samples, and 72.20% of cold water samples and 45.80% of hot water Legionella-positive samples were also FLP positive.
- Legionella’s existence in water supply systems is still relevant in Riga City, and almost 66% of water samples were Legionella positive using at least one detection method.
- 72.20% of the cold water and 45.80% of the hot water samples, which were Legionella spp. positive, were also positive for FLP, which can possibly show that in cold water samples there were more FLP and Legionella spp. co-existence occurrences that can be discovered.
- Average hot water temperature in Riga is lower than 50 °C (48.11 °C), and sometimes it is lower even than 30 °C.
- There was a medium-strong positive correlation between building floor and Legionella quantity, which was stronger in cold water samples.
- More FLP- and Legionella-positive samples were collected from the kitchens than the showers.
- There are no big differences between water supply system samples from the left and right Daugava banks regarding Legionella and FLP. However, in the left bank samples, there is a much bigger quantity of Chaetonotida (hairybellies) (38.09%).
- Using 16S rRNA sequencing, a long list of bacteria was detected, the most common of which were Pseudomonadota, Planctomycetota, Bacillota, Acidobacteriota, and Candidatus Omnitrophota. Some were potentially pathogenic to humans (Coxiella, Leptospira, Listeria, Corynebacterium, Mycobacterium, and Pseudomonas).
- It was detected that each of the five hotel buildings in this study has its own bacterial diversity inside its water supply system.
- Several methods are needed to analyze water samples completely: microbiological methods, microscopy PCR, and sequencing. Then, we can see the whole picture of bacterial and FLP diversity.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muchesa, P.; Leifels, M.; Jurzik, L.; Barnard, T.G.; Bartie, C. Detection of amoeba-associated Legionella pneumophila in hospital water networks of Johannesburg. S. Afr. J. Infect. Dis. 2018, 33, 72–75. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Legionella. In Guidelines for Drinking Water. Addendum: Microbiological Agents in Drinking Water, 2nd ed.; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Valciņa, O.; Pūle, D.; Ķibilds, J.; Labecka, L.; Terentjeva, M.; Krūmiņa, A.; Bērziņš, A. Evaluation of genetic diversity and virulence potential of Legionella pneumophila isolated from water supply systems of residential buildings in Latvia. Pathogens 2023, 12, 884. [Google Scholar] [CrossRef]
- Shaheen, M.; Ashbolt, N. Free-Living Amoebae Supporting Intracellular Growth May Produce Vesicle-Bound Respirable Doses of Legionella Within Drinking Water Systems. Expo. Health 2017, 110, 201–209. [Google Scholar] [CrossRef]
- Armstrong, T.W.; Haas, C.N. A Quantitative Microbial Risk Assessment Model for Legionnaires’ Disease: Animal Model Selection and Dose-Response Modelling. Risk Anal. 2007, 27, 1581–1596. [Google Scholar] [CrossRef]
- Berendt, R.F.; Young, H.W.; Allen, R.G.; Knutsen, G.L. Dose-Response of Guinea Pigs Experimentally Infected with Aerosols of Legionella pneumophila. J. Infect. Dis. 1980, 141, 186–192. [Google Scholar] [CrossRef]
- Armstrong, T.W.; Haas, C.N. Legionnaires’ disease: Evaluation of a quantitative microbial risk assessment model. J. Water Health 2008, 6, 149–166. [Google Scholar] [CrossRef]
- Kilvington, S.; Gray, T.; Dart, J.; Morlet, M.; Beeching, J.R.; Frazer, D.G.; Matheson, M. Acanthamoeba Keratitis: The Role of Domestic Tap Water Contamination in the United Kingdom. Investig. Ophthalmol. Vis. Sci. 2004, 45, 165–169. [Google Scholar] [CrossRef]
- Ji, P.; Rhoads, W.J.; Edwards, M.A.; Pruden, A. Effect of heat shock on hot water plumbing microbiota and Legionella pneumophila control. Microbiome 2018, 6, 30–44. [Google Scholar] [CrossRef]
- Bertelli, C.; Courtois, S.; Rosikiewicz, M.; Piriou, P.; Aeby, S.; Robert, S.; Loret, J.-F.; Greub, G. Reduced Chlorine in drinking water distribution systems impacts bacterial biodiversity in biofilms. Front. Microbiol. 2018, 9, 2520. [Google Scholar] [CrossRef]
- Ling, F.; Whitaker, R.; Le Chevallier, M.W.; Liu, W.-T. Drinking water microbiome assembly induced by water stagnation. ISME 2018, 12, 1520–1531. [Google Scholar] [CrossRef]
- Zimoch, I.; Paciej, J. Microbiological contaminants of Legionella spp. in hot water in public buildings in Silesia province. In Water Supply and Wastewater Disposal; Lublin University of Technology: Lublin, Poland, 2018; pp. 344–357. [Google Scholar]
- Delafont, V.; Bouchon, D.; Héchard, Y.; Moulin, L. Environmental factors shaping cultured free-living amoebae and their associated bacterial community within drinking water network. Water Res. 2016, 100, 382–392. [Google Scholar] [CrossRef]
- Scaturro, M.; Buffoni, M.; Girolamo, A.; Cristino, S.; Girolamini, L.; Mazzotta, M.; Bucci Sabattini, M.A.; Zaccaro, C.M.; Chetti, L.; Laboratory, M.A.N.; et al. Performance of Legiolert test vs. ISO 11731 to confirm Legionella pneumophila contamination in potable water samples. Pathogens 2020, 9, 690. [Google Scholar] [CrossRef]
- Vaerewijck, M.J.M.; Sabbe, K.; Hende, J.; Bare, J.; Houf, K. Sampling strategy, occurence and diversity of free-living protozoa in domestic refrigerators. J. App. Microbiol. 2010, 109, 1556–1578. [Google Scholar]
- Schroeder, J.M.; Booton, G.C.; Hay, J. Use of Subgenic 18S Ribosomal DNA PCR and Sequencing for Genus and Genotype Identification of Acanthamoebae from Humans with Keratitis and from Sewage Sludge. J. Clin. Microbiol. 2001, 39, 1903–1911. [Google Scholar] [CrossRef]
- Calvez, T.; Trouilhe, M.C.; Humeau, O. Detection of free-living amoebae by using multiplex quantitative PCR. Mol. Cell. Probes. 2012, 26, 116–120. [Google Scholar] [CrossRef]
- Solgi, R.; Niyyati, M.; Haghihgi, A.; Mojarad, N.E. Occurence of thermotolerant Hartmanella vermiformis and Naegleria Spp. in Hot Springs of Ardebil Province, northwest Iran. Iran. J. Parasitol. 2012, 7, 47–52. [Google Scholar]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.; Breiner, H.W.; Richards, T.A. Multiple marker parallel tag environmental dna sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef]
- Bradley, I.M.; Pinto, A.J.; Guest, J.S. Design and evaluation of illumina miseq compatible, 18s rRNA gene-specific primers for improved characterization of mixed phototrophic communities. Appl. Environ. Microbiol. 2016, 82, 5878–5891. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M. Evaluation of general 16s ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Legionella and Prevention of Legionellosis; WHO: Geneva, Switzerland, 2007; p. 30. [Google Scholar]
- Li, H.; Li, S.; Tang, W.; Yang, Y.; Zhao, J.; Xia, S.; Zhang, W.; Wang, H. Influence of secondary water supply systems on microbial community structure and opportunistic pathogen gene markers. Water Res. 2018, 136, 160–168. [Google Scholar] [CrossRef]
- Jing, H.; Sahle-Demessie, E.; Sorial, A.G. Inhibition of biofilm growth on polymer -MWCNTs composites and metal surfaces. Sci. Total Environ. 2018, 633, 167–178. [Google Scholar] [CrossRef]
- Srinivasan, A.; Bova, G.; Ross, T.; Mackie, K.; Paquette, N.; Merz, W.; Perl, T.M. A 17-month evaluation of a chlorine dioxide water treatment system to control Legionella species in a hospital water supply. Infect. Control. Hosp. Epidemiol. 2003, 24, 575–579. [Google Scholar] [CrossRef]
- Valciņa, O.; Pūle, D.; Mališevs, A.; Trofimova, J.; Makarova, S.; Konvisers, G.; Bērziņš, A.; Krūmiņa, A. Co-Occurrence of Free-Living Amoeba and Legionella in Drinking Water Supply Systems. Medicina 2019, 55, 492. [Google Scholar] [CrossRef]
- Quandt, C.A.; Beaudet, D.; Corsaro, D.; Walochnik, J.; Michel, R.; Corradi, N.; James, T.Y. The genome of an intranuclear parasite, Paramicrosporidium saccamoebae, reveals alternative adaptations to obligate intracellular parasitism. eLife 2017, 6, e29594. [Google Scholar] [CrossRef]
- Sales-Ortells, H.; Medema, G. Screening-Level Risk Assessment of Coxiella burnetii (Q Fever) Transmission via Aeration of Drinking Water. Environ. Sci. Technol. 2012, 46, 4125–4133. [Google Scholar] [CrossRef]
- Wynwood, S.J.; Graham, G.C.; Weier, S.L.; Collet, T.D.; McKay, D.B.; Craig, S.B. Leptospirosis from water sources. Pathog. Glob. Health 2014, 108, 334–338. [Google Scholar] [CrossRef]
- Rusin, P.A.; Rose, J.B.; Gerba, C.P. Health significance of pigmented bacteria in drinking water. War. Sci. Tech. 1997, 11–12, 21–27. [Google Scholar] [CrossRef]
- Vaerewijck, M.J.M.; Huys, G.; Palomino, J.C.; Swings, J.; Portaels, F. Mycobacteria in drinking water distribution systems: Ecology and significance for human health. FEMS Microbiol. Rev. 2005, 29, 911–934. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O., III; Norton, C.D.; Le Chevallier, M.W. Factors Influencing Numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in Drinking Water Distribution Systems. Appl. Environ. Microbiol. 2000, 3, 1225–1231. [Google Scholar] [CrossRef]
- Walker, J.; Moore, G. Pseudomonas aeruginosa in hospital water systems: Biofilms, guidelines, and practicalities. J. Hosp. Infect. 2014, 89, 324–327. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, K.; Struewing, I.; Buse, H.; Domingo, J.S.; Lytle, D.; Lu, J. The bacterial community diversity of bathroom hot tap water was significantly lower than that of cold tap and shower water. Front. Microbiol. 2021, 12, 625324. [Google Scholar] [CrossRef]
- Kori, J.A.; Mahar, R.B.; Vistro, M.R.; Tariq, H.; Khan, I.A.; Goel, R. Metagenomic analysis of drinking water samples collected from treatment plants of Hyderabad City and Mehran University Employees Cooperative Housing Society. Environ. Sci. Pollut. Res. 2019, 26, 29052–29064. [Google Scholar] [CrossRef]
- Mehari, Y.T.; Hayes, J.; Redding, K.S.; Mariappan, P.V.G.; Gunderson, J.H.; Farone, A.L.; Farone, M.B. Description of ‘Candidatus Berkiella aquae’ and ‘Candidatus Berkiella cookevillensis’, two intranuclear bacteria of freshwater amoebae. Int. J. Syst. Evol. Microbiol. 2016, 66, 536–541. [Google Scholar] [CrossRef]
- Mrva, M. Mayorella vespertilioides Page, 1983 (Amoebozoa)—New host for the ectoparasitic fungus Amoebophilus simplex (Zygomycota). Biologia 2011, 66, 645–647. [Google Scholar] [CrossRef]
- Deepika, D.S.; Sowmya, K.L. Bioindices of Bacterial Communities. Int. J. Curr. Microbiol. Appl. Sci. 2016, 12, 219–233. [Google Scholar] [CrossRef]
- Henne, K.; Kahlisch, L.; Brettar, I.; Höfle, M.G. Analysis of Structure and Composition of Bacterial Core Communities in Mature Drinking Water Biofilms and Bulk Water of a Citywide Network in Germany. Appl. Environ. Microbiol. 2012, 78, 3530–3538. [Google Scholar] [CrossRef]
- Revetta, R.P.; Pemberton, A.; Lamendella, R.; Iker, B.; Domingo, J.W.S. Identification of bacterial populations in drinking water using 16S rRNA-based sequence analyses. Water Res. 2010, 44, 1353–1360. [Google Scholar] [CrossRef]
- Gavarāne, I.; Trofimova, J.; Mališevs, A.; Valciņa, O.; Kirjušina, M.; Rubeniņa, I.; Bērziņš, A. DNA extraction from amoebal isolates and genotype determination of Acanthamoeba from tap water in Latvia. Parasitol. Res. 2018, 117, 3299–3303. [Google Scholar] [CrossRef]
- Vilne, B.; Grantiņa-Ieviņa, L.; Ķibilds, J.; Mališevs, A.; Konvisers, G.; Makarova, S.; Pūle, D.; Valciņa, O. Investigating Bacterial and Free-Living Protozoa Diversity in Biofilms of Hot Water Pipes of Apartment Buildings in the City of Riga (Latvia). Front. Water 2021, 3, 799840. [Google Scholar] [CrossRef]
- Valciņa, O.; Pūle, D.; Ķibilds, J.; Lazdāne, A.; Trofimova, J.; Makarova, S.; Konvisers, G.; Ķimse, L.; Krūmiņa, A.; Bērziņš, A. Prevalence and Genetic Diversity of Legionella spp. in Hotel Water-Supply Systems in Latvia. Microorganisms 2023, 11, 596. [Google Scholar] [CrossRef]
- Lasjerdi, Z.; Niyyati, M.; Haghighi, A.; Shahabi, S.; Biderouni, F.T.; Taghipour, N.; Eftekhar, M.; Mojarad, E.N. Potentially pathogenic free-living amoebae isolated from hospital wards with immunodeficient patients in Tehran, Iran. Parasitol. Res. 2011, 109, 575–580. [Google Scholar] [CrossRef]
- Springe, G.; Juhna, T. Water supply and sanitation in Riga: Development, present, and future. In Environmental History of Water: Global Views on Community Water Supply and Sanitation; Juuti, P.S., Katko, T.S., Vuorinen, H.S., Eds.; Wiley: New York, NY, USA, 2007; pp. 401–410. [Google Scholar]





| Water Samples | Riga | Salaspils | Jurmala |
|---|---|---|---|
| Hot water | 37 | 2 | 1 |
| Cold water | 38 | 2 | 1 |
| Shower | 50 | 4 | 2 |
| Kitchen | 26 | 0 | 0 |
| Apartments | 38 | 2 | 1 |
| Primer | Primer Sequence | Reference |
|---|---|---|
| JDP1-F | “5′-GGCCCAGATCGTTTACCGTGAA-3′” | [16] |
| JDP2-R | “5′-TCTCACAAGCTGCTAGGGAGTCA-3′” | |
| Vahl_560_F | “5′-AGGTAGTGACAAGMYRTAGYGACT-3′” | [17] |
| Vahl_730_R | “5′-GGGCGTTTTAACTACARCAGTATTA-3′” | |
| Amo_1400_F | “5′-ATGCCGACCARSGATYMGGAG-3′” | |
| Amo_1540_R | “5′-CAAGSTGCYMGGGGAGTCAT-3′” | |
| Hartm Solgi F | “5′-GCT CCA ATA GCG TAT ATT AA-3′” | [18] |
| Hartm Solgi R | “5′-AGA AAG AGC TAT CAA TCT GT-3′” |
| Results | Cold Water Samples (n = 41) | Hot Water Samples (n = 40) | Total (n = 81) |
|---|---|---|---|
| Legionella spp. positive by ISO 11731: 2017 method | 18 (43.90%) | 23 (57.50%) | 41 (50.60%) |
| Legionella spp. positive by real-time PCR | 14 (34.15%) | 25 (62.5%) | 39 (48.10%) |
| Legionella spp. positive by at least one method | 22 (53.66%) | 31 (77.5%) | 53 (65.43%) |
| Legionella spp. positive by both methods | 10 (24.39%) | 17 (42.5%) | 27 (33.33%) |
| FLP positive by microscopy | 24 (58.54%) | 21 (52.50%) | 45 (55.60%) |
| FLP positive by PCR | 17 (41.46%) | 15 (37.5%) | 32 (39.50%) |
| FLP positive by at least one method | 24 (58.54%) | 21 (52.5%) | 45 (55.56%) |
| FLP positive by both methods | 17 (41.46%) | 15 (37.5%) | 32 (39.51%) |
| Parameter | Cold-Water Consumption Amount, m3 per Month | Hot Water Consumption Amount, m3 per Month |
|---|---|---|
| Legionella spp. positive by ISO 11731: 2017 method | 3.38 (1.00–6.00) | 2.71 (1.00–6.00) |
| Legionella spp. negative by ISO 11731: 2017 method | 4.11 (0.80–12.00) | 2.69 (2.00–3.20) |
| FLP positive by microscopy | 3.39 (0.80–12.00) | 2.75 (1.20–6.10) |
| FLP negative by microscopy | 4.37 (1.00–7.00) | 2.66 (1.00–4.10) |
| χ2 | p-Value | |
|---|---|---|
| Legionella | 0.01 | 0.94 |
| FLP | 0.4 | 0.53 |
| Water Sample | Legionella spp. Positive % | FLP Positive % |
|---|---|---|
| Shower | 46.4 | 48.2 |
| Kitchen | 57.7 | 73.1 |
| Protozoa | Left Bank % | Right Bank % | Hotels % |
|---|---|---|---|
| Vermamoeba | 7.56 | 10.25 | 13.64 |
| Uncultured eukaryote | 3.17 | 3.60 | 3.33 |
| Chromulinales | 2.88 | 4.04 | 11.39 |
| Spumella | 0.62 | 1.14 | 0.77 |
| Oligohymenophorea | 0.28 | 8.91 | 4.02 |
| Phyllopharyngea | 1.38 | 5.96 | 1.92 |
| Euamoebida | 3.69 | 4.09 | 0.76 |
| Chytridiomycetes | 1.75 | 1.14 | 2.56 |
| Haplotaxida | 2.68 | 8.26 | 6.09 |
| Chaetonotida (hairybellies) | 38.09 | 1.60 | 9.37 |
| Shannon’s Diversity Index | Simpson’s Diversity | Simpson’s Reciprocal Index | Berger–Parker’s Diversity | Fisher’s Index | |
|---|---|---|---|---|---|
| Average, cold | 3.19 | 0.922 | 15.263 | 0.186 | 89.419 |
| Average, hot | 2.72 | 0.861 | 11.187 | 0.264 | 75.433 |
| Average, hotel | 2.32 | 0.801 | 6.181 | 0.394 | 3.706 |
| Minimal, cold | 2.25 | 0.717 | 3.539 | 0.06 | 2.399 |
| Minimal, hot | 0.39 | 0.146 | 1.171 | 0.083 | 0.819 |
| Minimal, hotel | 1.34 | 0.562 | 2.282 | 0.133 | 1.499 |
| Maximal, cold | 3.90 | 0.971 | 35.018 | 0.521 | 535.168 |
| Maximal, hot | 3.81 | 0.967 | 30.514 | 0.923 | 443.079 |
| Maximal, hotel | 3.014 | 0.931 | 14.544 | 0.929 | 6.693 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mališevs, A.; Ķibilds, J.; Konvisers, G.; Pūle, D.; Valciņa, O.; Bērziņš, A.; Grantiņa-Ieviņa, L. Free-Living Protozoa and Legionella spp. Coexistence and Bacterial Diversity in Drinking Water Systems in Apartment Buildings and Hotels in Riga and Its Surroundings. Water 2025, 17, 1485. https://doi.org/10.3390/w17101485
Mališevs A, Ķibilds J, Konvisers G, Pūle D, Valciņa O, Bērziņš A, Grantiņa-Ieviņa L. Free-Living Protozoa and Legionella spp. Coexistence and Bacterial Diversity in Drinking Water Systems in Apartment Buildings and Hotels in Riga and Its Surroundings. Water. 2025; 17(10):1485. https://doi.org/10.3390/w17101485
Chicago/Turabian StyleMališevs, Artjoms, Juris Ķibilds, Genadijs Konvisers, Daina Pūle, Olga Valciņa, Aivars Bērziņš, and Lelde Grantiņa-Ieviņa. 2025. "Free-Living Protozoa and Legionella spp. Coexistence and Bacterial Diversity in Drinking Water Systems in Apartment Buildings and Hotels in Riga and Its Surroundings" Water 17, no. 10: 1485. https://doi.org/10.3390/w17101485
APA StyleMališevs, A., Ķibilds, J., Konvisers, G., Pūle, D., Valciņa, O., Bērziņš, A., & Grantiņa-Ieviņa, L. (2025). Free-Living Protozoa and Legionella spp. Coexistence and Bacterial Diversity in Drinking Water Systems in Apartment Buildings and Hotels in Riga and Its Surroundings. Water, 17(10), 1485. https://doi.org/10.3390/w17101485







