Abstract
Zooplankton, integral to aquatic ecosystems, face diverse environmental influences. To comprehend their dynamics, critical for ecological insights and fisheries management, traditional morphological analysis proves laborious. Recent advances include automated systems like ZooScan and DNA metabarcoding. This study examines two methods on the same samples to identify similarities and dependencies between them, potentially reducing the required workload and enhancing the quality of the results. Ten Lake Starnberg vertical tows in September 2021 provided zooplankton samples preserved in ethanol. Subsamples underwent ZooScan morphological identification and subsequent DNA metabarcoding. High concordance between ZooScan counts and DNA reads (86.8%) was observed, while biomass calculations from body length (major axis) and equivalent spherical diameter (ESD) showed slightly lower agreement (78.1% and 79.6%, respectively). Linear regression analysis revealed a correlation between counts and DNA reads (r2 = 0.59). This study underscores the complementary strengths and limitations of ZooScan and DNA metabarcoding for zooplankton analysis. ZooScan aids biomass estimation and morphological differentiation, whereas DNA metabarcoding offers superior taxonomic resolution and low-abundance taxon detection. Combining both methods on the same sample enhances understanding and facilitates future advanced analyses.
1. Introduction
Zooplankton, a pivotal component of aquatic ecosystems, is exposed to a multitude of environmental parameters that influence its abundance, diversity, and ecological role. These parameters include for example water temperature, nutrient availability, predator-prey interactions, and climatic conditions [1,2,3]. Comprehending the intricate dynamics of zooplankton populations is essential for several reasons. It is important for deciphering the ecological complexities within zooplankton communities, including growth rates, distribution patterns, and community structure [4]. On the other hand, knowing the dynamics of zooplankton communities also provides valuable insights which are relevant for the development of effective fisheries management strategies [5]. Zooplankton are characterized by being suspended in the water column and exposed to water currents. Most zooplankton species also exhibit a certain degree of motility which additionally influences the distribution of zooplankton communities within a water body. Hence, both abiotic factors such as water temperature and currents, but also biotic factors such as predator foraging and associated anti-predator behaviors (for example diel vertical migration or swarming), strongly affect the distribution patterns of zooplankton communities and zooplankton groups within the pelagic zone of lakes. In addition, lake geomorphology and associated mixing regimes, as well as the presence of individual basins within lakes, can also play an important role in shaping zooplankton distribution [6,7,8,9,10,11]. The potential uneven distribution of zooplankton and its consequences for fish foraging behavior and activities require intensive, high-resolution monitoring programs. However, such monitoring analyses often depend on the skills of the operators and their taxonomic expertise, as well as the associated costs and time constraints [12,13,14]. Various modern approaches are now available for the taxonomically standardized, time- and cost-efficient identification of zooplankton. On one hand, the digitization of zooplankton through different optical systems offers a means to obtain reliable data on the quality, quantity, and size (biomass) of zooplankton samples [15,16,17]. On the other hand, non-optical molecular techniques such as high-throughput sequencing (HTS) provide the opportunity for rapid and standardized analyses of zooplankton samples [18,19,20]. Such molecular methods can detect organisms below the usual thresholds of optical identification methods [4] but do not easily allow necessary zooplankton biomass estimations [18]. Recently, studies combined automated optical/digitization methods with molecular methods to achieve the highest level of resolution in analyzing zooplankton samples [18,21]. There has been ongoing discussion about the discrepancy between these two methods, and attempts are made to establish correlations between the abundance of individuals, their biomass, and their taxonomically specific sequence reads determined through molecular approaches [18,20,21,22,23,24,25,26]. With the increasing use of advanced analytical techniques, two prominent methodologies, namely ZooScan and DNA metabarcoding, have emerged as potent automatized tools for investigating zooplankton diversity and abundance in lakes [15,27,28]. A notable aspect of ZooScan is its ability to rapidly facilitate the semi-automated morphological identification of zooplankton without sacrificing the ability to individually validate images of each captured zooplankton specimen. This validation process is comparable to visual microscopy analysis and increases the accuracy and reliability of the data obtained. Of course, the quality of the analysis depends on the optical resolution. Several studies have shown that optical detection methods provide results comparable to traditional microscopic analysis [29,30]. Sample volumes that have traditionally been manually assessed in counting chambers under a binocular or microscope can now be digitized and captured for analysis simultaneously using modern technology. However, the subsequent taxonomic determination or accuracy still relies on the expertise of the user for manual image validation and specialized software developed for automated image analysis. Conversely, metabarcoding offers a swift means to ascertain the species composition of zooplankton samples based on genetic information, in theory without the need of visual determination such as ZooScan or microscopy. Such a genetic-based taxonomy relies however on available data bases where genetic information and taxonomic/morphological species identification have been correctly aligned. In this comparative study, we analyze samples from a pre-alpine lake using both ZooScan-based identifications and DNA metabarcoding taxonomic assignments. In contrast to most other studies, we use the two different analysis methods on the exact same sample to explore concordance and complementarities but also discrepancies between the two methods. Additionally, we apply both methods along a 10-point sampling transect within the lake. Our main question is whether both methods can be not only compared in their qualitative estimation of zooplankton communities (zooplankton identity) but also in their possible quantitative estimations (zooplankton abundance/biomass proxies).
2. Materials and Methods
2.1. Sampling
During September 2021, ten vertical tows were taken along the north–south axis of Lake Starnberg, a pre-alpine lake in Germany (Figure 1), within a time span of 1.5 h using a plankton net (mesh size: 250 µm, length: 60 cm, diameter: 40 cm, sampling depth: 17 m, representing the euphotic zone). Lake Starnberg is a deep lake with a maximum depth of 127 m, extending across a relatively uniform deep basin from north to south (Figure 1). The collected zooplankton was preserved in ethanol (>98%). Subsequently, subsamples (1/32) were extracted from each vertical sample in the laboratory using a zooplankton splitter [31].
Figure 1.
Map of the zooplankton sampling locations at Lake Starnberg in Germany.
Morphological Taxonomic Identification
The subsamples were digitized and analyzed using the ZooScan system (HydroptiC, Version 2.4.0) [16]. Abundance, classification, and biomass determination of taxonomic groups followed the methodology outlined by Vogelmann et al. [15]. In short, the ZooScan system, coupled with the Zooprocess program (Version 7.22), digitizes and archives objects placed in a scanning cell. The program Zooprocess then segments the scanned zooplankton sample into individual images, creating vignettes for each object. The ethanol-fixed zooplankton samples were briefly stored in a freezer at −18 °C just before the scanning process. Subsequently, zooplankton were carefully separated from the ethanol using a gauze filter (250 µm) and placed on the scanning cell in cooled distilled water (7 °C) to avoid the potential buoyancy of zooplankton in the scanning cell. To prevent the overlap of individual objects in the scanning cell, a delicate separation of zooplankton was performed using scratch-free tools (cactus needles), with a maximum time allowance of 5 min per sample. Utilizing a re-established learning set (comprised of prior zooplankton analyses/scans from Lake Starnberg) with the Plankton Identifier (PkID, Version 1.2.6) program, in conjunction with ZooScan and Zooprocess enabled an automatic assignment of individual taxa [32]. The zooplankton sample was categorized into the following groups: Bosmina spp., Bythotrephes longimanus, Daphnia spp., calanoid copepods, cyclopoid copepods, and Leptodora kindtii. Certainly, it is important to note that despite automatic categorization into different classes using training sets, each sample additionally underwent subsequent manual validation by two zooplankton specialists. After digitization with the ZooScan system, the subsamples were again returned into >98% ethanol to enable subsequent molecular genetic analysis on the same samples.
2.2. Molecular Taxonomic Identification
For the sample preparation and DNA extraction, zooplankton samples were filtered to remove ethanol, air-dried, and then suspended in T1 lysis buffer (Macherey-Nagel, Düren, Germany). Samples were homogenized by adding sterile beads and by shaking on a homogenizer. Each sample was then spiked with Proteinase K and incubated overnight at 56 °C. Total DNA was extracted from the lysate using the NucleoSpin® Tissue Kit (Macherey-Nagel, Düren, Germany).
For the PCR amplification and HTS library preparation, DNA extracts were used as templates for the amplification of a 421 bp fragment of the mitochondrial cytochrome c oxidase gene (COI) using the primers BF2 and BR2 [33]. Each primer was modified to include 5´Illumina® overhang adapters (Illumina Inc., San Diego, CA, USA). PCR conditions were as follows: initial denaturation for 3 min at 94 °C, followed by 25 cycles of 20 s denaturation at 94 °C, annealing for 30 s at 48 °C, and extension for 40 s at 72 °C. The program ended with a 5 min extension step at 72 °C. The amplification of a correct amplicon was verified via electrophoresis on a 1.5% agarose gel, and the PCR products were then purified using the NucleoMag® NGS Clean-up kit (Macherey Nagel, Düren, Germany). Dual indexes and Illumina® sequencing adapters were attached to the amplicons with a second PCR using the Nextera® XT Index Kit (Illumina Inc., San Diego, CA, USA) following the manufacturer‘s protocol and then purified using the NucleoMag® NGS Clean-up (Macherey Nagel, Düren, Germany). Libraries were quantified using the Promega Quantifluor® ONE dsDNA system (Promega, Mannheim, Germany) and pooled equimolarly. Pooled libraries were sequenced on an Illumina® MiSeq system with the 2 × 300 bp reaction kit.
For the bioinformatic analysis, paired-end reads were assembled sample-wise using VSEARCH [34] setting a minimum overlap of 100 bases. Primer sequences were removed from the assemblies using cutadapt [35]. All contigs were then filtered for uncalled bases (“N”) and dereplicated with VSEARCH. Dereplicated sequences were clustered in operational taxonomic units (OTUs) using a 97% similarity threshold. Chimera sequences were detected using the UCHIME algorithm [36] as implemented in VSEARCH and removed. All non-chimeric OTUs were then taxonomically assigned using the blastn algorithm [37] on a local version of the BOLD reference database (status: December 2022).
2.3. Biomass Calculation
A proxy for zooplankton biomass was calculated through the body length (pixel, major axis of the best-fitting ellipse) or by the equivalent spherical diameter (ESD, pixel) as determined by the ZooScan method.
The determined abundance (counts of various taxa) was compared to the reads obtained through metabarcoding. As some objects on the ZooScan might overlap, potentially leading to distorted counts or Major/ESD values for specific taxa groups, connected objects were individually counted and corrected using the median Major/ESD value of the corresponding category. For further analyses, biomass data were transformed by log10 (x + 1).
2.4. Statistical Analysis
To compare the taxonomic distribution determined using ZooScan and metabarcoding, six groups were formed (Bosmina spp., B. longimanus, Daphnia spp., calanoid copepods, cyclopoid copepods, and L. kindtii). It should be noted that Diaphanosoma sp. was classified under the Daphnia category due to limitations in ZooScan’s morphological resolution. Detailed morphological identification at the species level is not feasible in most cases with ZooScan. Metabarcoding data were generally analyzed only to the genus level, as ZooScan’s capabilities are confined to the mentioned broader groups for analysis and comparison. A percentage similarity [38,39] was computed between the data obtained through ZooScan and metabarcoding. To infer potential relationships between DNA reads and the counts derived from ZooScan, a regression analysis was conducted.
3. Results
3.1. ZooScan
The ZooScan results for all 10 samples indicate an average of 506.5 individuals per sample (median = 525; SD = 109.5). For the calculated biomass proxies (pixel3) biomass length, the average is 4.01 × 109 pixels3 per sample (median = 3.89 × 109 pixels3; SD = 1.00 × 109 (pixels3), while for ESD biomass, the average is 9.03 × 108 pixels3 (median = 8.81 × 108 pixels3; SD = 2.20 × 108 (pixels3) (one pixel = 0.0053 mm). Detailed data can be found in Table A1 of Appendix A.
3.2. Metabarcoding
From sample sequencing, we obtained an average of 1.02 × 105 sequences per sample, which resulted in an average of 1.20 × 104 unique sequences per sample after the bioinformatic processing. Unique sequences were clustered in 421 OTUs, whose taxonomic assignments are reported in the Supplementary Materials (Table S1). For the present analyses, the taxonomic assignments were condensed to the genus level and the read counts of congeneric species were merged. Detailed data can be found in Table A1 of Appendix A.
3.3. Comparison ZooScan vs. DNA Metabarcoding
Table 1 displays the percentage similarity between ZooScan counts, biomass (bodylength), biomass (ESD), and read counts from the metabarcoding analyses. The results indicate the highest percentage similarity between ZooScan, counts, and metabarcoding (mean = 86.8%; median = 87.5%; standard deviation = 2.5%). In contrast, the calculated biomass using the length of individual objects (major axis) exhibits a somewhat lower percentage similarity (mean = 78.1%; median = 79.4%; standard deviation = 3.6%), as does the equivalent spherical diameter (ESD) (mean = 79.6%; median = 79.6%; standard deviation = 4.9%), when compared to metabarcoding.

Table 1.
The percentage similarity between the metabarcoding approach and ZooScan analyses for the same 10 zooplankton samples from Lake Starnberg was assessed. The comparison involved counts, biomass (calculated using the major axis and ESD value), and the read count from metabarcoding. All data have been transformed (log10 (x + 1)).
The determined count of distinct zooplankton taxa using ZooScan and the number of reads obtained through metabarcoding (Figure 2) exhibit a linear correlation with an r2 value of 0.59 (p-value < 0.0001). The regression of bodylength biomass and reads shows a linear regression with an r2 value of 0.21 (p-value = 0.0003). The r2 value of the linear regression of ESD biomass and reads is 0.32 (p-value < 0.0001).
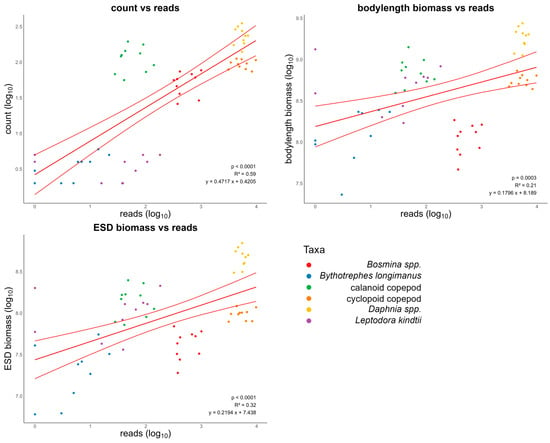
Figure 2.
Relationships between counts, bodylength biomass (major axis), and ESD biomass of zooplankton organisms by using ZooScan and the number of reads obtained through metabarcoding. The solid lines depict significant relationships, accompanied by 95% confidence intervals.
Along the sampling transect within the lake, a similar picture emerged (Figure 3). Both methods gave similar results for major zooplankton groups along the transect; within the ZooScan calculations, counts seem to be most similar to metabarcoding reads. Detailed data can be found in the Supplementary Materials (Table S2). Figure 4 shows the deviation between the two methods along the transect, usually being consistent in being higher or lower in estimating relative zooplankton group abundances from metabarcoding reads compared to ZooScan counts or biomass estimates. An exception was predatory cladocerans which were usually present in very low abundances and distributed unevenly along the transect. Detailed data can be found in Table A2 of Appendix B.
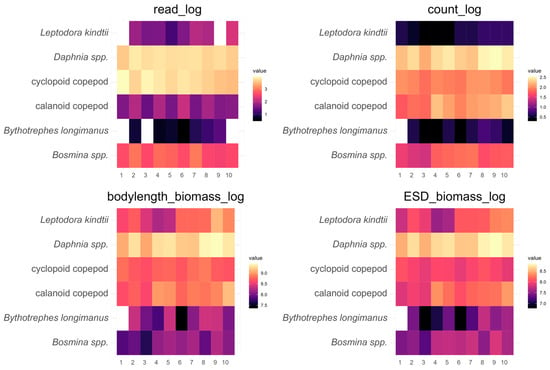
Figure 3.
Zooplankton distribution based on 10 samples (each collected in September 2021) along a north–south sampling transect in Lake Starnberg. The transformed zooplankton abundances from metabarcoding (read_log) and ZooScan (count_log) are presented, along with the calculated biomass estimations (bodylength_biomass_log, ESD_biomass_log).
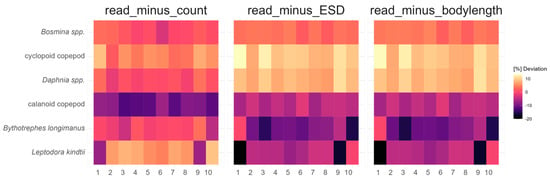
Figure 4.
Deviation (%) of relative zooplankton abundance between metabarcoding and morphological identification using ZooScan (count), along with the calculated biomass estimation (major and ESD), from 10 samples in Lake Starnberg (September 2021) along a north–south sampling transect. Detailed data can be found in Table A2 of Appendix B.
4. Discussion
Here, we compared two different methods of zooplankton quantification (the optical method ZooScan and the molecular method metabarcoding) on a series of zooplankton samples from a deep pre-alpine lake. Both methods were performed on the same sample for each location, allowing a rigid comparison of results. Several studies have demonstrated that sequence reads can depict relative differences in terms of quantity of individuals and biomass among various taxa [20,22,23,24]. However, studies [22] by Sun et al. [24] and Lindeque et al. [40] have also shown that HTS-based measurements correlate better with biomass than with the numerical abundance (counts) of individual organisms. Given that ZooScan provides the capacity to calculate biomass proxies through metrics such as the body length (major axis) or ESD value [15], we assumed that this could potentially offer better insights into the relationship between metabarcoding read count and the quantitative distribution of taxonomic groups via body length (major axis) and ESD values.
Our results present a slightly different perspective (Table 1). In our case, the ZooScan count vs. DNA reads exhibits the highest similarity at around 86%. Both biomass determinations, whether through the major axis or ESD, display an average similarity of 78% (bodylength biomass) and 79% (ESD biomass) when compared to DNA reads. The results (Figure 2) are consistent with previous findings by Sun et al. [24], indicating that the number of reads reflects species’ qualitative abundance reasonably well, but drawing conclusions about quantity is challenging. Similar to the results shown by Yang et al. [23], our results support the idea that the metabarcoding approach can provide indications of relative abundances in zooplankton communities. This can be performed in significantly shorter timeframes than traditional methods, such as manual counting or biomass calculation based on the dry weight of zooplankton individuals. Both the study by Yang et al. [23] and our study quantified freshwater zooplankton biomass by multiplying the abundance of a taxa by its body length raised to the power of three. In contrast to the study of Yang et al. [23], our results show the lowest similarity (Table 1) between the calculated biomass analyzed with the ZooScan and molecular read abundances. In addition, our results also show lower relationships between the calculated biomass of different zooplankton groups and molecular reads compared to the relationship between zooplankton abundances (counts) and molecular reads (Figure 2).
Additionally, our findings indicate that samples containing taxa with very low numbers of individuals can lead to a distorted outcome. Some taxonomic groups were morphologically detected in single-digit numbers within samples (Bythotrephes and Leptodora), which were not always confirmed through metabarcoding (Figure 2). On the other hand, predatory cladocerans were identified by metabarcoding in certain samples but could not be detected by ZooScan. It is plausible that during the splitting process using the plankton splitter [31], or during sample preparation [15], predatory cladocerans might have been excluded from the subsample, but parts of such large individuals such as spina, antenna, or eggs could have been separated through mechanical forces. These parts would not be identifiable by ZooScan but could still be detected through metabarcoding. If predatory cladocerans are excluded due to their low numbers, an even higher percentage similarity between DNA reads and ZooScan distribution is achieved. Detailed data can be found in Table A3 of Appendix C.
Given that ZooScan’s resolution extends only up to 200 µm [16] and does not allow species-level identification in all cases, grouping at the genus level is more than sufficient in our case. Another potential issue that may arise is the scanning of zooplankton organisms in an unsuitable position, hindering analysis by both the software and the specialist. Similar findings and potential solutions to avoid such problems were reported by Vogelmann et al. [15]. The taxonomic resolution on the species level is in most cases not necessary for fishery management. In the context of the ZooScan’s resolution constrained to 200 µm, it is worth noting that digital imaging techniques and optical tracking (such as the ZooScan method), are currently undergoing rapid advancements and continuous improvements. This technical progress supports the increasing precision and efficiency of optical methods, which are thereby becoming an increasingly valuable tool for the study of zooplankton and the development of comprehensive ecological and fisheries management strategies. Modern molecular methods such as metabarcoding can be of additional help in elucidating zooplankton composition [26,41], particularly benefiting from quantitative DNA analyses. On the other hand, numerous investigations highlight the potential for misinterpretation in sequence frequency [42]. A study by Thomas et al. [20] also underscores the substantial impact of primer selection on the quantitative composition of zooplankton.
Studies have highlighted complications in sequence frequency when species occur in very low numbers within a sample. In such cases, these species can be overrepresented compared to more abundant ones [20,43]. However, our results show that this must not always be the case. While individual species like B. longimanus and L. kindtii were morphologically classified, they could not be detected through metabarcoding. Similar findings are evident in a study by Harvey et al. [22] identifying different copepods and fish eggs. In some cases, specific taxa were identified through NGS but could not be morphologically confirmed, and vice versa. In their studies, the discrepancy might also be attributed to the nature of the sampling process, as two different samples were collected in close temporal proximity, unlike our approach where exactly the same sample was analyzed.
The high concordance in zooplankton composition (Table 1, Figure 4) when compared to visual morphological identification through ZooScan underlines that metabarcoding can offer a time-efficient complement for the ZooScan method. Additionally, it is important to highlight that Lake Starnberg boasts a considerable water volume. Our results (Figure 3) reveal a relatively uniform distribution of zooplankton within the sampling depth of 17 m (euphotic zone) across the north–south gradient, with the exception of predatory cladocerans. Such a result was anticipated due to the relatively uncomplicated geomorphology of Lake Starnberg, resulting in a single large deep water basin with a uniform mixing regime. Our investigation illustrates that zooplankton assessment by both methods, particularly regarding the distribution of different zooplankton groups with different nutritional value for fish, yields promising results. This suggests that both approaches offer extensive capabilities for elucidating zooplankton compositions, particularly regarding the ratio of copepods to cladocerans which can often be an important zooplankton community characteristic in terms of food quality of zooplankton for fish [44]. The high number of samples that can be processed in comparison to classical microscopic counting and identification allows a much higher temporal and spatial resolution of analyses of zooplankton dynamics in lakes [18,30]. Such a better temporal and spatial resolution can offer more detailed insights into pelagic ecosystem processes highly relevant for lake and fisheries management [5,45]. Our sampling transects including 10 sampling points showed a high persistency of results but also demonstrated sampling-point-specific community composition. For example, predatory cladocerans, which can be a very important source of food for fish and thereby can influence fish foraging patterns [46], were distributed unevenly along the sampling transect. Obviously, by comparing both methods one can observe some differences between detection probabilities along the transect. For more common zooplankton groups, the detection differences were however comparable along the transect. Depending on the research question (necessary taxonomic resolution of zooplankton, high-frequency spatial/temporal analyses, etc.), each method offers specific advantages. ZooScan shows some advantages in cases where a high taxonomic resolution is not required, but biomass or other image-analysis-derived proxies are necessary (Figure 3). For example, ZooScan can, in contrast to metabarcoding approaches, differentiate between egg-bearing and non-egg-bearing zooplankton and different juvenile stages (copepodites) [15]. Metabarcoding’s advantages compared to ZooScan include a higher taxonomic resolution and the ability to detect taxa that are below thresholds for detection by optical methods [22].
5. Conclusions
Our results show that automated optical and molecular methods can give comparable results on zooplankton abundances. ZooScan provides a detailed proxy for biomass and individual zooplankton counts. It proves to be less time-consuming than traditional manual zooplankton analyses. This method still requires a high amount of labor, which limits the number of possible samples. In contrast, metabarcoding provides the possibility to analyze multiple samples at once or composite samples, or a multitude of samples collected from waterbodies varying in size, in a short timeframe, thereby reducing labor and costs. The molecular data (reads) were clearly related to abundance and biomass estimates from automated imaging. Both analyses complement each other and using both automatized methods combined on the same sample can result in more precise calculations of biomass proxies and taxonomic resolution than either method on its own. The comparison of both methods can give insights which either method individually cannot deliver.
Additionally, raw data generated by both methods are open for later reanalyses in case improved methods (such as, for example, AI-based image analyses) become available. Therefore, long-term data series on zooplankton including imaging as well as molecular data are accessible for optimal reanalyses at later stages.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/w16030411/s1, Figure S1. Zooplankton distribution based on 10 samples (each collected in September 2021) along a north–south sampling transect in Lake Starnberg. The transformed zooplankton abundances from metabarcoding (read_log) and ZooScan (count_log) are presented, along with the calculated biomass estimations (maj_biomass_log, ESD_biomass_log). Table S1: List of OTUs and the correlating genus created from the metabarcoding raw data for every sampling location. Table S2: Transformed (log10 (x + 1) data from metabarcoding reads, ZooScan biomass (ESD, length major axis), and ZooScan count per taxa at every sampling location.
Author Contributions
Conceptualization, C.V. and H.S.; methodology, C.V., A.B., J.-M.K. and H.S.; formal analysis, C.V., A.B. and J.-M.K.; investigation, C.V. and A.B.; resources, H.S.; data curation, C.V., A.B. and J.-M.K.; writing—original draft, C.V., A.B. and H.S.; writing—review and editing, C.V., A.B., J.-M.K. and H.S.; visualization, C.V. and A.B.; supervision, H.S.; project administration, H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Bavarian State Ministry of the Environment and Consumer Protection (StMUV) and the Bavarian State Ministry of Food, Agriculture, Forestry, and Tourism (StMELF), A0730.0-2008/30-85.
Data Availability Statement
All raw data and supplementary data can be requested from the authors.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A

Table A1.
ZooScan analysis (ZS) of counts, biomass (bodylength/major axis, ESD), and the number of reads detected through metabarcoding.
Table A1.
ZooScan analysis (ZS) of counts, biomass (bodylength/major axis, ESD), and the number of reads detected through metabarcoding.
| Number of Sample Location | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Mean | ||
| Bosmina spp. | ZS count | 35 | 28 | 25 | 60 | 67 | 73 | 76 | 45 | 55 | 57 | 52.1 |
| ZS ESD [pixel] | 27,566,182.12 | 32,495,496.12 | 19,029,160.31 | 52,665,226.17 | 55,490,778.83 | 51,028,271.22 | 60,290,752.10 | 32,217,069.42 | 69,362,878.87 | 43,161,058.88 | 44,330,687.40 | |
| ZS length [pixel] | 70,587,289.32 | 85,142,052.44 | 46,622,046.35 | 132,396,936.54 | 157,732,885.05 | 133,450,427.75 | 162,635,117.60 | 81,453,143.99 | 184,883,536.82 | 118,796,394.16 | 117,369,983.00 | |
| reads | 410 | 916 | 377 | 788 | 680 | 415 | 1007 | 364 | 322 | 370 | 564.9 | |
| Bythotrephes longimanus | ZS count | 0 | 3 | 1 | 1 | 3 | 1 | 3 | 5 | 4 | 2 | 2.3 |
| ZS ESD [pixel] | 0 | 26,066,701.65 | 6,043,315.86 | 10,914,324.13 | 24,208,511.41 | 6,209,493.52 | 18,531,729.95 | 55,292,988.65 | 32,163,017.49 | 40,780,173.55 | 22,021,025.62 | |
| ZS length [pixel] | 0 | 223,473,643.05 | 94,010,182.89 | 64,481,201.00 | 232,910,637.39 | 23,124,764.56 | 118,665,461.00 | 245,621,937.54 | 232,317,861.20 | 104,543,339.72 | 133,914,902.84 | |
| reads | 0 | 6 | 0 | 4 | 5 | 2 | 9 | 13 | 21 | 0 | 6 | |
| calanoid copepod | ZS count | 55 | 72 | 67 | 176 | 125 | 90 | 120 | 132 | 143 | 194 | 117.4 |
| ZS ESD [pixel] | 72,098,081.70 | 90,008,701.24 | 78,486,813.29 | 229,454,031.17 | 165,574,001.42 | 112,693,383.91 | 147,953,500.99 | 163,016,513.95 | 168,342,631.00 | 248,853,829.59 | 147,648,148.83 | |
| ZS length [pixel] | 424,386,413.57 | 543,696,952.05 | 394,100,694.89 | 988,346,542.37 | 919,737,681.68 | 577,768,692.34 | 738,921,135.01 | 677,658,890.31 | 812,868,715.53 | 1,411,054,299.64 | 748,854,001.74 | |
| reads | 40 | 101 | 27 | 90 | 36 | 140 | 35 | 79 | 42 | 47 | 63.7 | |
| cyclopoid copepod | ZS count | 106 | 93 | 73 | 95 | 86 | 79 | 109 | 111 | 98 | 78 | 92.8 |
| ZS ESD [pixel] | 117,447,700.49 | 96,835,928.62 | 79,929,652.05 | 103,016,793.44 | 100,803,418.60 | 80,244,816.57 | 101,225,792.52 | 121,677,697.05 | 97,790,488.10 | 78,786,373.58 | 97,775,866.10 | |
| ZS length [pixel] | 640,100,884.52 | 507,669,510.88 | 439,185,225.17 | 563,845,827.32 | 597,904,269.80 | 491,577,917.21 | 646,761,576.30 | 732,360,853.05 | 519,144,921.68 | 471,955,804.21 | 561,050,679.01 | |
| reads | 9778 | 4666 | 8300 | 6681 | 4942 | 6426 | 5415 | 4370 | 3445 | 3128 | 5715.1 | |
| Daphnia spp. | ZS count | 173 | 235 | 139 | 233 | 274 | 202 | 183 | 316 | 350 | 288 | 239.3 |
| ZS ESD [pixel] | 306,893,805.18 | 499,874,607.97 | 314,201,460.96 | 472,928,350.61 | 519,948,737.81 | 392,833,635.37 | 401,175,718.37 | 622,046,569.80 | 695,818,379.94 | 556,503,158.23 | 478,222,442.42 | |
| ZS length [pixel] | 1,180,043,451.50 | 2,022,277,561.51 | 1,123,595,795.78 | 1,961,352,933.25 | 2,075,519,518.70 | 1,589,159,574.14 | 1,540,446,053.76 | 2,530,286,146.69 | 2,745,435,377.57 | 2,164,620,995.36 | 1,893,273,740.83 | |
| reads | 3890 | 7042 | 5512 | 6311 | 5522 | 5839 | 6006 | 4636 | 5471 | 4181 | 5441.0 | |
| Leptodora kindtii | ZS count | 3 | 2 | 1 | 1 | 1 | 3 | 3 | 4 | 4 | 4 | 3 |
| ZS ESD [pixel] | 59,423,738.91 | 110,460,445.39 | 83,242,428.14 | 36,287,042.20 | 42,822,250.27 | 128,466,750.81 | 128,466,750.81 | 133,006,482.04 | 200,730,084.96 | 213,002,171.28 | 113,590,814.48 | |
| ZS length [pixel] | 389,754,324.62 | 524,410,928.38 | 273,738,689.72 | 170,400,051.70 | 200,612,600.96 | 601,837,802.89 | 601,837,802.89 | 618,792,587.69 | 1,330,508,641.16 | 828,707,218.43 | 554,060,064.84 | |
| reads | 0 | 65 | 37 | 38 | 15 | 41 | 106 | 89 | 0 | 181 | 57.2 | |
Appendix B

Table A2.
Percentage sample deviation between DNA read minus ZooScan biomass via ESD, DNA read minus ZooScan count and DNA read minus ZooScan biomass via major axis (bodylength) for each taxa.
Table A2.
Percentage sample deviation between DNA read minus ZooScan biomass via ESD, DNA read minus ZooScan count and DNA read minus ZooScan biomass via major axis (bodylength) for each taxa.
| Taxa | Comparison | Sample Deviation [%] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Bosmina spp. | read_minus_ESD | 3.4 | 3.8 | 3.7 | 3.3 | 4.3 | 1.8 | 3.7 | 1.7 | 3.6 | 3.1 |
| read_minus_count | 3.1 | 2.8 | 1.4 | −0.3 | 0.6 | −2.8 | 0.0 | 0.3 | 1.9 | 0.5 | |
| read_minus_maj | 3.9 | 4.2 | 4.2 | 3.8 | 4.7 | 2.2 | 4.1 | 2.1 | 4.0 | 3.4 | |
| calanoid copepod | read_minus_ESD | −6.2 | −3.5 | −6.2 | −4.4 | −5.9 | −2.2 | −6.7 | −4.1 | −4.1 | −4.9 |
| read_minus_count | −7.7 | −8.0 | −12.4 | −11.8 | −11.4 | −7.0 | −11.7 | −8.8 | −9.1 | −11.5 | |
| read_minus_maj | −6.4 | −3.6 | −6.2 | −4.3 | −5.9 | −2.3 | −6.8 | −4.1 | −4.1 | −5.2 | |
| cyclopoid copepod | read_minus_ESD | 13.4 | 7.4 | 12.5 | 9.0 | 10.0 | 9.6 | 8.0 | 7.8 | 11.3 | 9.4 |
| read_minus_count | 9.0 | 1.7 | 5.8 | 3.8 | 5.7 | 5.1 | 3.2 | 3.6 | 7.5 | 5.9 | |
| read_minus_maj | 13.3 | 7.4 | 12.4 | 8.8 | 9.8 | 9.3 | 7.8 | 7.5 | 11.2 | 9.0 | |
| Daphnia spp. | read_minus_ESD | 9.0 | 7.1 | 9.8 | 7.4 | 8.8 | 7.9 | 7.1 | 6.5 | 11.1 | 8.6 |
| read_minus_count | 3.0 | −1.7 | 0.9 | −0.6 | 0.6 | 0.3 | 1.1 | −0.8 | 3.4 | 0.9 | |
| read_minus_maj | 9.3 | 7.4 | 10.2 | 7.6 | 9.1 | 8.0 | 7.3 | 6.6 | 11.4 | 8.7 | |
| Bythotrephes longimanus | read_minus_ESD | 0.0 | −10.0 | −14.7 | −10.1 | −9.8 | −11.1 | −8.5 | −8.3 | −4.9 | −15.7 |
| read_minus_count | 0.0 | −1.3 | −3.8 | 1.4 | −0.9 | −0.1 | 0.3 | −0.2 | 3.4 | −5.0 | |
| read_minus_maj | 0.0 | −10.6 | −15.8 | −10.5 | −10.5 | −11.2 | −8.9 | −8.4 | −5.3 | −15.4 | |
| Leptodora kindtii | read_minus_ESD | −19.6 | −4.9 | −5.2 | −5.2 | −7.3 | −6.0 | −3.5 | −3.6 | −17.0 | −0.5 |
| read_minus_count | −7.4 | 6.6 | 8.1 | 7.4 | 5.4 | 4.5 | 7.1 | 6.0 | −7.1 | 9.3 | |
| read_minus_maj | −20.0 | −4.8 | −4.8 | −5.3 | −7.3 | −6.0 | −3.5 | −3.7 | −17.3 | −0.5 | |
Appendix C

Table A3.
Percentage similarity without predatory cladocerans between ZooScan count and DNA reads, ZooScan biomass via bodylenght and DNA reads and ZooScan biomass via ESD and DNA reads for each sampling location at Starnberg Lake.
Table A3.
Percentage similarity without predatory cladocerans between ZooScan count and DNA reads, ZooScan biomass via bodylenght and DNA reads and ZooScan biomass via ESD and DNA reads for each sampling location at Starnberg Lake.
| Sampling Location | Similarity Count vs. Reads [%] | Similarity Bodylenght Biomass vs. Reads [%] | Similarity ESD Biomass vs. Reads [%] |
|---|---|---|---|
| 1 | 90.6 | 87.8 | 87.8 |
| 2 | 91.7 | 90.9 | 91.3 |
| 3 | 87.2 | 86.5 | 86.4 |
| 4 | 88.9 | 90.1 | 89.8 |
| 5 | 88.0 | 87.8 | 88.1 |
| 6 | 91.0 | 90.1 | 89.8 |
| 7 | 87.7 | 87.6 | 87.8 |
| 8 | 90.7 | 90.1 | 89.7 |
| 9 | 88.8 | 87.7 | 87.4 |
| 10 | 87.6 | 88.5 | 88.3 |
| Mean | 89.2 | 88.7 | 88.6 |
| Median | 88.8 | 88.2 | 88.2 |
| SD | 1.56 | 1.38 | 1.40 |
References
- Beisner, B.E.; Peres-Neto, P.R.; Lindström, E.S.; Barnett, A.; Longhi, M.L. The Role of Environmental and Spatial Processes in Structuring Lake Communities from Bacteria to Fish. Ecology 2006, 87, 2985–2991. [Google Scholar] [CrossRef]
- Hairston, N.G.; Smith, F.E.; Slobodkin, L.B. Community Structure, Population Control, and Competition. Am. Nat. 1960, 94, 421–425. [Google Scholar] [CrossRef]
- Sommer, U.; Adrian, R.; De Senerpont Domis, L.; Elser, J.J.; Gaedke, U.; Ibelings, B.; Jeppesen, E.; Lürling, M.; Molinero, J.C.; Mooij, W.M. Beyond the Plankton Ecology Group (PEG) Model: Mechanisms Driving Plankton Succession. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 429–448. [Google Scholar] [CrossRef]
- Lombard, F.; Boss, E.; Waite, A.M.; Vogt, M.; Uitz, J.; Stemmann, L.; Sosik, H.M.; Schulz, J.; Romagnan, J.-B.; Picheral, M.; et al. Globally Consistent Quantitative Observations of Planktonic Ecosystems. Front. Mar. Sci. 2019, 6, 196. [Google Scholar] [CrossRef]
- Anneville, O.; Lainé, L.; Benker, S.; Ponticelli, A.; Gerdeaux, D. Food Habits and Ontogenetic Changes in the Diet of Whitefish Larvae in Lake Annecy. Bull. Fr. Pêche Piscic. 2007, 387, 21–33. [Google Scholar] [CrossRef]
- Thomas, K.; Hansen, T.; Brophy, D.; Maoiléidigh, N.Ó.; Fjelldal, P.G. Experimental Investigation of the Effects of Temperature and Feeding Regime on Scale Growth in Atlantic Salmon Salmo salar Post-smolts. J. Fish Biol. 2019, 94, jfb.13971. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.; Rösch, R.; Eckmann, R. Seasonal and Long-Term Changes in Fishing Depth of Lake Constance Whitefish. Fish. Manag. Ecol. 2010, 17, 386–393. [Google Scholar] [CrossRef]
- O’Brien, W.J. The Predator-Prey Interaction of Planktivorous Fish and Zooplankton: Recent Research with Planktivorous Fish and Their Zooplankton Prey Shows the Evolutionary Thrust and Parry of the Predator-Prey Relationship. Am. Sci. 1979, 67, 572–581. [Google Scholar]
- Nisson, N. Seasonal Fluctuations in the Food Segregation of Trout, Char and Whitefish in 14 North-Swedish Lakes. Rep. Inst. Freshw. Res. Drottningholm 1960, 41, 185–205. [Google Scholar]
- Wagler, E. Der Blaufelchen Des Bodensees. (Coregonus wartmanni Bloch.) Versuch Einer Monographie. Int. Rev. Gesamten Hydrobiol. Hydrogr. 1927, 18, 129–230. [Google Scholar] [CrossRef]
- Peterson, W.T.; Fisher, J.L.; Peterson, J.O.; Morgan, C.A.; Burke, B.J.; Fresh, K.L. Applied Fisheries Oceanography: Ecosystem Indicators of Ocean Conditions Inform Fisheries Management in the California Current. Oceanography 2014, 27, 80–89. [Google Scholar] [CrossRef]
- Benfield, M.C.; Grosjean, P.; Culverhouse, P.F.; Irigoien, X.; Sieracki, M.E.; Lopez-Urrutia, A.; Dam, H.G.; Hu, Q.; Davis, C.S.; Hansen, A.; et al. RAPID: Research on Automated Plankton Identification. Oceanography 2007, 20, 172–187. [Google Scholar] [CrossRef]
- Davis, C.S.; Hu, Q.; Gallager, S.M.; Tang, X.; Ashjian, C.J. Real-Time Observation of Taxa-Specific Plankton Distributions: An Optical Sampling Method. Mar. Ecol. Prog. Ser. 2004, 284, 77–96. [Google Scholar] [CrossRef]
- Remsen, A.; Hopkins, T.L.; Samson, S. What You See Is Not What You Catch: A Comparison of Concurrently Collected Net, Optical Plankton Counter, and Shadowed Image Particle Profiling Evaluation Recorder Data from the Northeast Gulf of Mexico. Deep Sea Res. Part I Oceanogr. Res. Pap. 2004, 51, 129–151. [Google Scholar] [CrossRef]
- Vogelmann, C.; Teichert, M.; Schubert, M.; Martens, A.; Schultes, S.; Stibor, H. The Usage of a Zooplankton Digitization Software to Study Plankton Dynamics in Freshwater Fisheries. Fish. Res. 2022, 251, 106326. [Google Scholar] [CrossRef]
- Gorsky, G.; Ohman, M.D.; Picheral, M.; Gasparini, S.; Stemmann, L.; Romagnan, J.-B.; Cawood, A.; Pesant, S.; Garcia-Comas, C.; Prejger, F. Digital Zooplankton Image Analysis Using the ZooScan Integrated System. J. Plankton Res. 2010, 32, 285–303. [Google Scholar] [CrossRef]
- Grosjean, P.; Picheral, M.; Warembourg, C.; Gorsky, G. Enumeration, Measurement, and Identification of Net Zooplankton Samples Using the ZOOSCAN Digital Imaging System. ICES J. Mar. Sci. 2004, 61, 518–525. [Google Scholar] [CrossRef]
- Pierella Karlusich, J.J.; Lombard, F.; Irisson, J.-O.; Bowler, C.; Foster, R.A. Coupling Imaging and Omics in Plankton Surveys: State-of-the-Art, Challenges, and Future Directions. Front. Mar. Sci. 2022, 9, 878803. [Google Scholar] [CrossRef]
- Lamb, P.D.; Hunter, E.; Pinnegar, J.K.; Creer, S.; Davies, R.G.; Taylor, M.I. How Quantitative Is Metabarcoding: A Meta-analytical Approach. Mol. Ecol. 2019, 28, 420–430. [Google Scholar] [CrossRef]
- Thomas, A.C.; Deagle, B.E.; Eveson, J.P.; Harsch, C.H.; Trites, A.W. Quantitative DNA Metabarcoding: Improved Estimates of Species Proportional Biomass Using Correction Factors Derived from Control Material. Mol. Ecol. Resour. 2016, 16, 714–726. [Google Scholar] [CrossRef]
- Ibarbalz, F.M.; Henry, N.; Brandão, M.C.; Martini, S.; Busseni, G.; Byrne, H.; Coelho, L.P.; Endo, H.; Gasol, J.M.; Gregory, A.C.; et al. Global Trends in Marine Plankton Diversity across Kingdoms of Life. Cell 2019, 179, 1084–1097.e21. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.B.J.; Johnson, S.B.; Fisher, J.L.; Peterson, W.T.; Vrijenhoek, R.C. Comparison of Morphological and next Generation DNA Sequencing Methods for Assessing Zooplankton Assemblages. J. Exp. Mar. Biol. Ecol. 2017, 487, 113–126. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, X.; Xie, Y.; Song, C.; Zhang, Y.; Yu, H.; Burton, G.A. Zooplankton Community Profiling in a Eutrophic Freshwater Ecosystem-Lake Tai Basin by DNA Metabarcoding. Sci. Rep. 2017, 7, 1773. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhao, Y.; Li, H.; Dong, Y.; MacIsaac, H.; Zhan, A. Unreliable Quantitation of Species Abundance Based on High-Throughput Sequencing Data of Zooplankton Communities. Aquat. Biol. 2015, 24, 9–15. [Google Scholar] [CrossRef]
- Elbrecht, V.; Leese, F. Can DNA-Based Ecosystem Assessments Quantify Species Abundance? Testing Primer Bias and Biomass—Sequence Relationships with an Innovative Metabarcoding Protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef] [PubMed]
- Riedel, A.; Sagata, K.; Suhardjono, Y.R.; Tänzler, R.; Balke, M. Integrative Taxonomy on the Fast Track—Towards More Sustainability in Biodiversity Research. Front. Zool. 2013, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation Sequencing Technologies for Environmental DNA Research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Pawlowski, J.; Christen, R.; Lecroq, B.; Bachar, D.; Shahbazkia, H.R.; Amaral-Zettler, L.; Guillou, L. Eukaryotic Richness in the Abyss: Insights from Pyrotag Sequencing. PLoS ONE 2011, 6, e18169. [Google Scholar] [CrossRef]
- Naito, A.; Abe, Y.; Matsuno, K.; Nishizawa, B.; Kanna, N.; Sugiyama, S.; Yamaguchi, A. Surface Zooplankton Size and Taxonomic Composition in Bowdoin Fjord, North-Western Greenland: A Comparison of ZooScan, OPC and Microscopic Analyses. Polar Sci. 2019, 19, 120–129. [Google Scholar] [CrossRef]
- Cornils, A.; Thomisch, K.; Hase, J.; Hildebrandt, N.; Auel, H.; Niehoff, B. Testing the Usefulness of Optical Data for Zooplankton Long-term Monitoring: Taxonomic Composition, Abundance, Biomass, and Size Spectra from ZooScan Image Analysis. Limnol. Ocean Methods 2022, 20, 428–450. [Google Scholar] [CrossRef]
- Motoda, S. Devices of Simple Plankton Apparatus. Mem. Fac. Fish. Hokkaido Univ. 1959, 7, 73–94. [Google Scholar]
- Gasparini, S.; Antajan, E. PLANKTON IDENTIFIER: A Software for Automatic Recognition of Planktonic Organisms. User Guide 2007–2013. Available online: http://www.obs-vlfr.fr/~gaspari/Plankton_Identifier/userguide (accessed on 5 December 2023).
- Elbrecht, V.; Leese, F. Validation and Development of COI Metabarcoding Primers for Freshwater Macroinvertebrate Bioassessment. Front. Environ. Sci. 2017, 5, 11. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-Parametric Multivariate Analyses of Changes in Community Structure. Austral Ecol 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Platell, M.E.; Potter, I.C.; Clarke, K.R. Resource Partitioning by Four Species of Elasmobranchs (Batoidea: Urolophidae) in Coastal Waters of Temperate Australia. Mar. Biol. 1998, 131, 719–734. [Google Scholar] [CrossRef]
- Lindeque, P.K.; Parry, H.E.; Harmer, R.A.; Somerfield, P.J.; Atkinson, A. Next Generation Sequencing Reveals the Hidden Diversity of Zooplankton Assemblages. PLoS ONE 2013, 8, e81327. [Google Scholar] [CrossRef]
- Hablützel, P.; Rombouts, I.; Dillen, N.; Lagaisse, R.; Mortelmans, J.; Ollevier, A.; Perneel, M.; Deneudt, K. Exploring New Technologies for Plankton Observations and Monitoring of Ocean Health. Oceanog 2021, 34, 20–25. [Google Scholar] [CrossRef]
- Deagle, B.E.; Thomas, A.C.; Shaffer, A.K.; Trites, A.W.; Jarman, S.N. Quantifying Sequence Proportions in a DNA-based Diet Study Using Ion Torrent Amplicon Sequencing: Which Counts Count? Mol. Ecol. Resour. 2013, 13, 620–633. [Google Scholar] [CrossRef]
- Kembel, S.W.; Wu, M.; Eisen, J.A.; Green, J.L. Incorporating 16S Gene Copy Number Information Improves Estimates of Microbial Diversity and Abundance. PLoS Comput. Biol. 2012, 8, e1002743. [Google Scholar] [CrossRef] [PubMed]
- Taipale, S.J.; Kahilainen, K.K.; Holtgrieve, G.W.; Peltomaa, E.T. Simulated eutrophication and browning alters zooplankton nutritional quality and determines juvenile fish growth and survival. Ecol. Evol. 2018, 8, 2671–2687. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.; Breitenstein, M.; Bia, M.M.; Rellstab, C.; Kirchhofer, A. Bottom-up Control of Whitefish Populations in Ultra-Oligotrophic Lake Brienz. Aquat. Sci. 2007, 69, 271–288. [Google Scholar] [CrossRef]
- Palmer, A.; Stich, H.-B.; Maier, G. Distribution Patterns and Predation Risk of the Coexisting Cladocerans Bythotrephes Longimanus and Leptodora Kindtii in a Large Lake—Lake Constance. Hydrobiologia 2001, 442, 301–307. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).