Identification and Distribution of Antibiotic Resistance Genes and Antibiotic Resistance Bacteria in the Feces Treatment Process: A Case Study in a Dairy Farm, China
Abstract
1. Introduction
2. Sampling and Methods
2.1. Waste Treatment Process
2.2. Sampling and Sample Preparation
2.3. Chemical Properties of the Samples
2.4. Metagenomic Sequencing and Bioinformatics Analysis
2.4.1. Library Preparation and Sequencing
2.4.2. Analysis of Sequencing Information
2.5. Metagenomic Sequencing Data Statistical Analysis
3. Results
3.1. Chemical Properties of the Sampled Wastewater and Manure
3.2. Metagenome Assembly Results
3.3. Metagenomic Sequencing Results
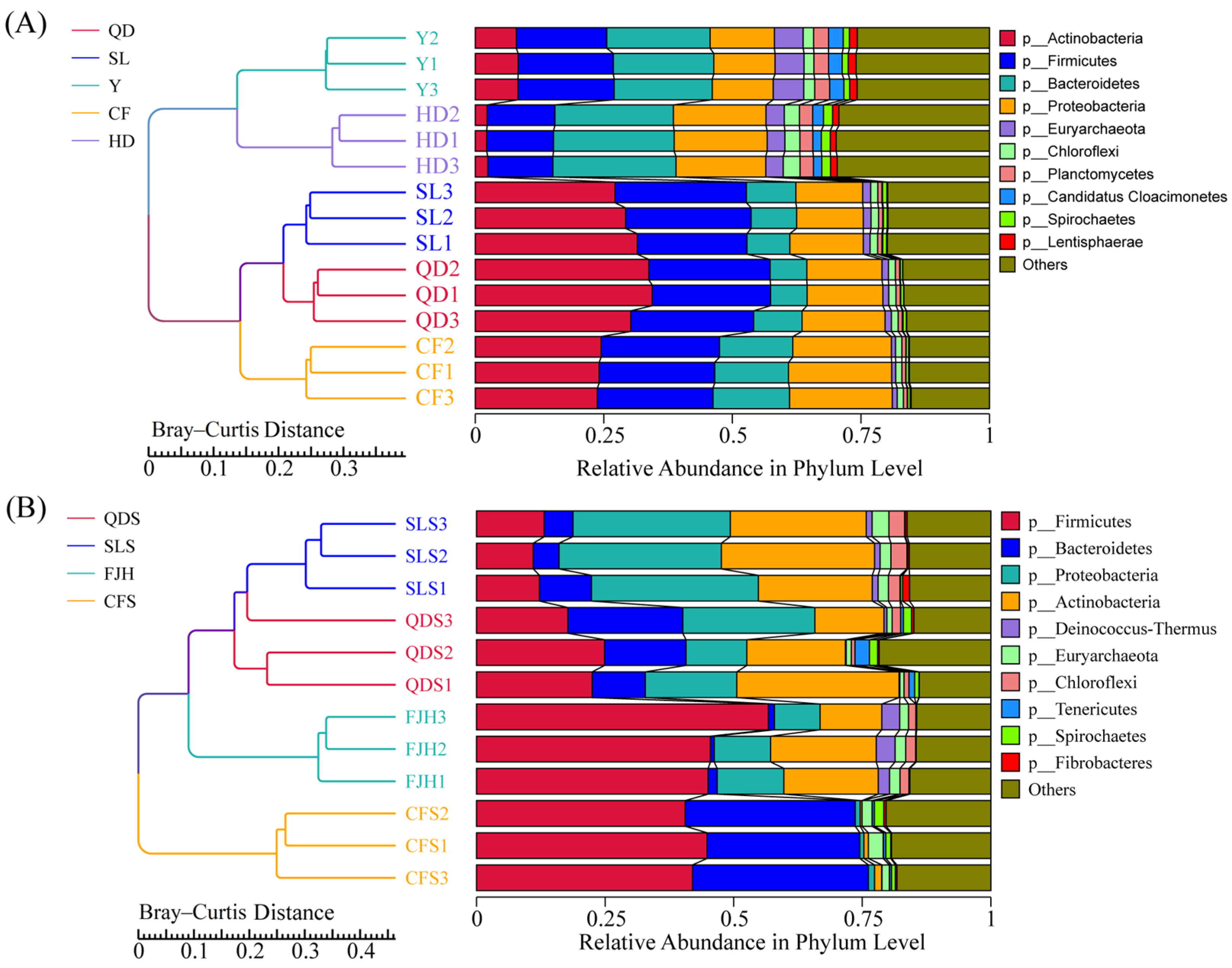
3.3.1. Diversity and Composition of Bacteria in the Wastewater and Manure Samples
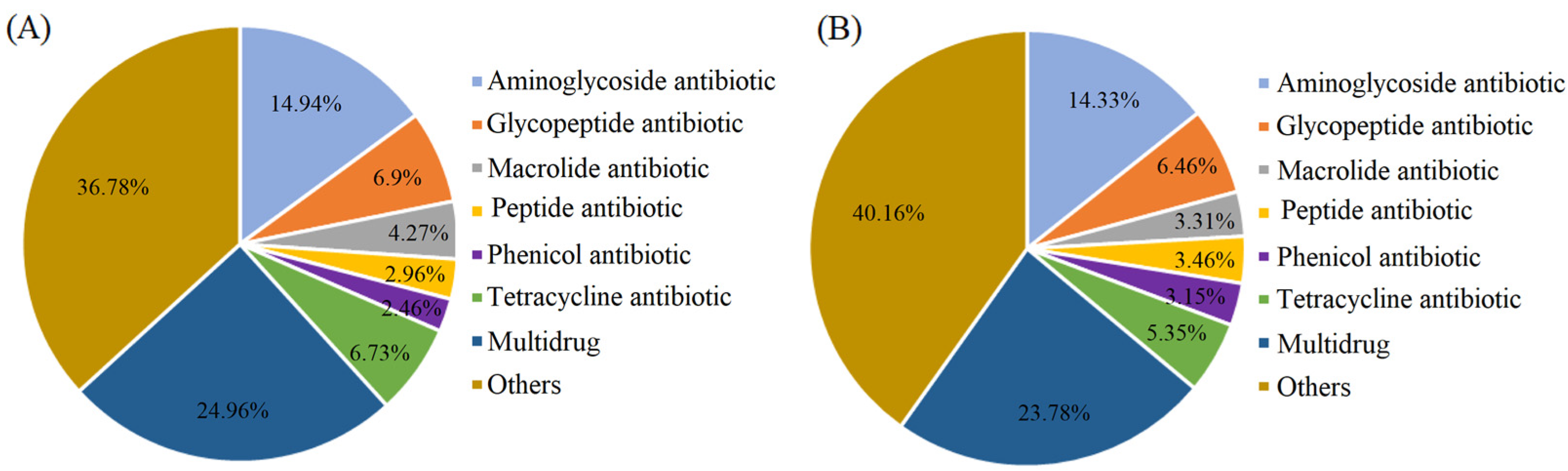
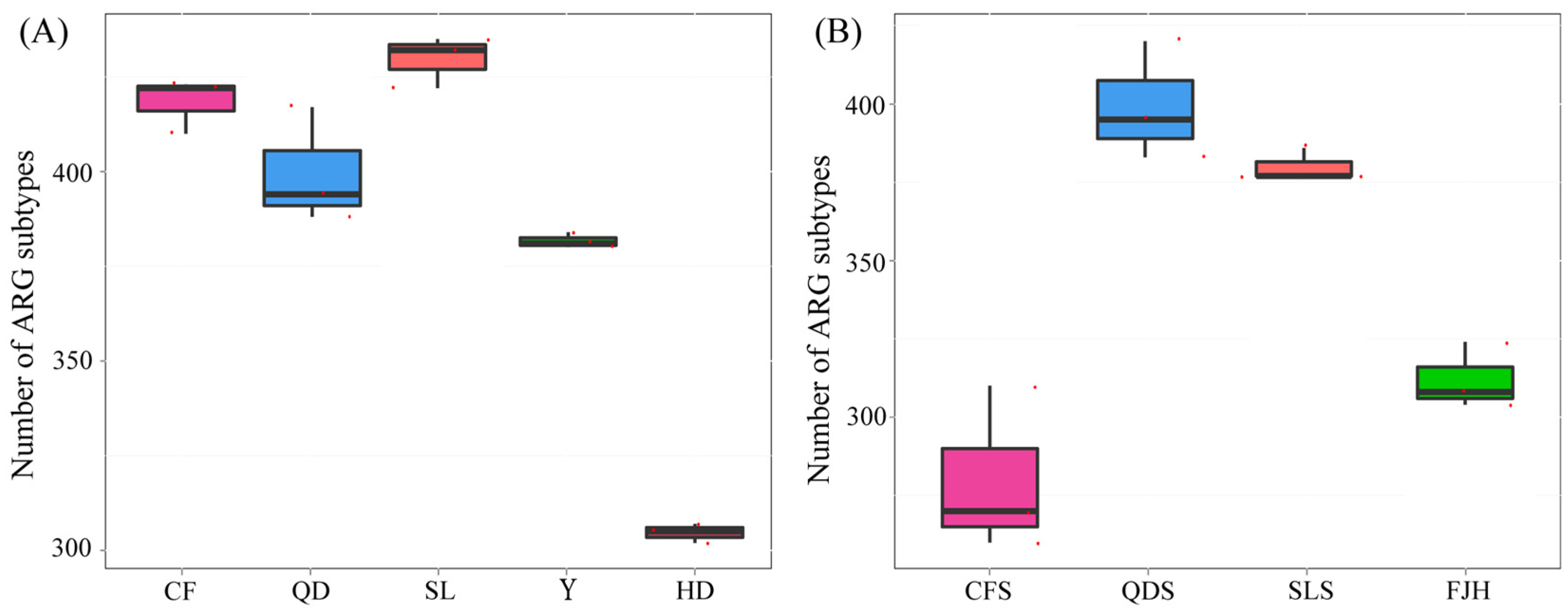
3.3.2. Abundance and Diversity of ARGs in the Wastewater and Manure Samples
3.3.3. Abundance and Diversity of the ARGs in the Surrounding Farmland Soil
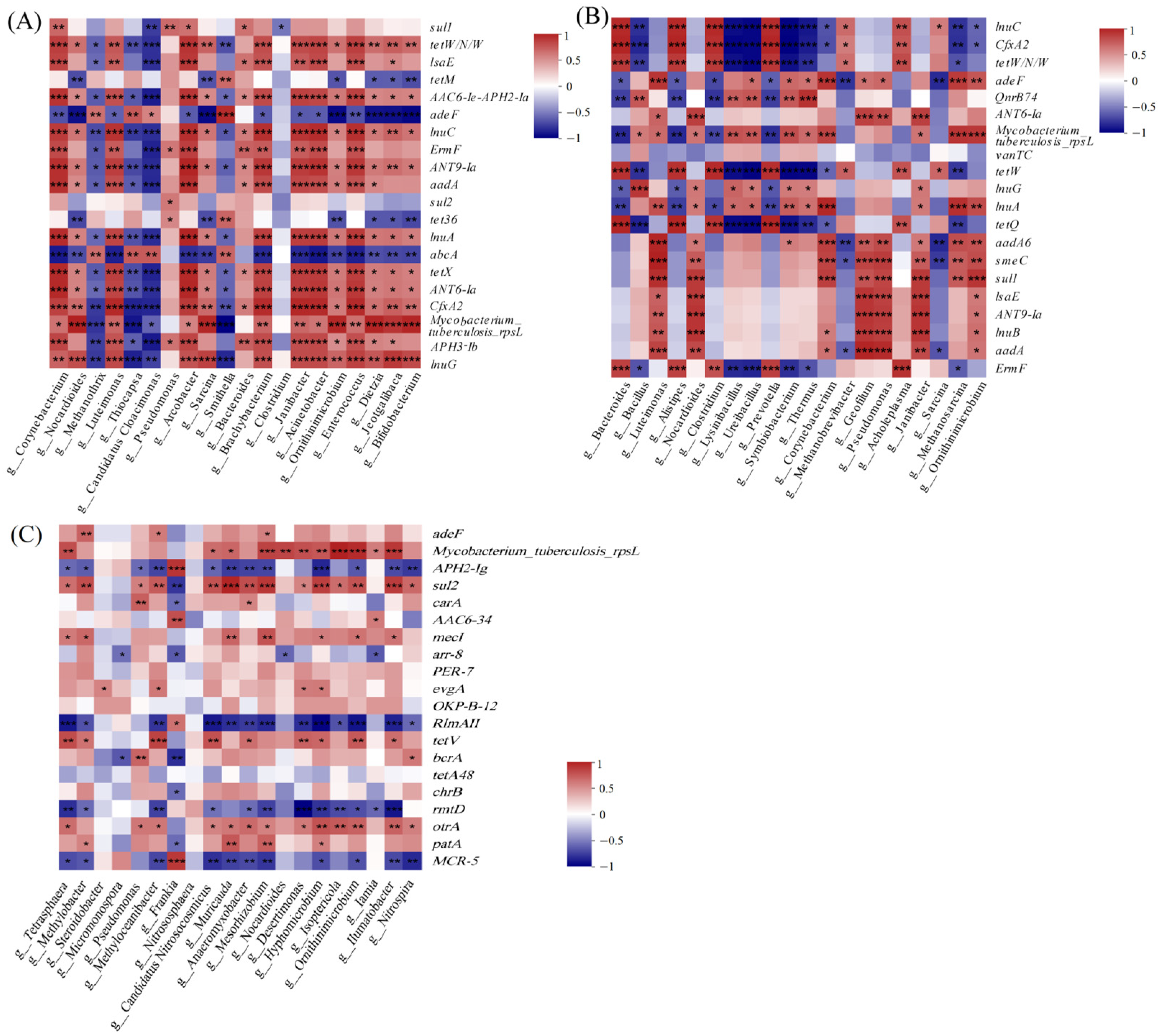
3.3.4. Correlation Analysis between the Bacterial Communities and the ARGs
4. Discussion
4.1. Occurrence and Distribution of the ARGs in Livestock Farms
4.2. ARGs Removal Efficiency of Current Livestock Wastewater and Manure Treatment Process
4.3. Relationship between ARB and ARGs in Livestock Wastewater, Manure, and Farmland Soil Samples
4.4. Resource Utilization and Risk of Livestock Wastewater and Manure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhao, H.; Wang, Z.; Liang, Y.; Wu, T.; Chen, Y.; Yan, J.; Zhu, Y.; Ding, D. Adsorptive decontamination of antibiotics from livestock wastewater by using alkaline-modified biochar. Environ. Res. 2023, 226, 115676. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Ying, G.G.; Pan, C.G.; Liu, Y.S.; Zhao, J.L. A comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modelling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Inda-Díaz, J.S.; Lund, D.; Parras-Moltó, M.; Johnning, A.; Bengtsson-Palme, J.; Kristiansson, E. Latent antibiotic resistance genes are abundant, diverse, and mobile in human, animal, and environmental microbiomes. Microbiome 2023, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai, China. J. Hazard. Mater. 2012, 235–236, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Zhang, X.X.; Miao, Y.; Zhao, Y.; Ye, L.; Li, B.; Zhang, T. Fate of antibiotic resistance genes and their associations with bacterial community in livestock breeding wastewater and its receiving river water. Water Res. 2017, 124, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Allen, H.; Donato, J.; Wang, H.; Cloud-Hansen, K.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef]
- Qin, K.; Wei, L.; Li, J.; Lai, B.; Zhu, F.; Yu, H.; Zhao, Q.; Wang, K. A review of ARGs in WWTPs: Sources, stressors and elimination. Chin. Chem. Lett. 2020, 31, 2603–2613. [Google Scholar] [CrossRef]
- Varma, V.S.; Parajuli, R.; Scott, E.; Canter, T.; Lim, T.T.; Popp, J.; Thoma, G. Dairy and swine manure management—Challenges and perspectives for sustainable treatment technology. Sci. Total Environ. 2021, 778, 146319. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Chambers, L.; Yang, Y.; Littier, H.; Ray, P.; Zhang, T.; Pruden, A.; Strickland, M.; Knowlton, K. Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic administration of third generation cephalosporin. PLoS ONE 2015, 10, e0133764. [Google Scholar] [CrossRef]
- Sun, J.; Liao, X.P.; D’Souza, A.W.; Boolchandani, M.; Li, S.H.; Cheng, K.; Martínez, J.L.; Li, L.; Feng, Y.J.; Fang, L.X.; et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 2020, 11, 1427. [Google Scholar] [CrossRef]
- Li, T.; Wang, Z.; Guo, J.; de la Fuente-Nunez, C.; Wang, J.; Han, B.; Tao, H.; Liu, J.; Wang, X. Bacterial resistance to antibacterial agents: Mechanisms, control strategies, and implications for global health. Sci. Total Environ. 2023, 860, 160461. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jia, S.; He, X.; Zhang, X.; Ye, L. Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 2017, 188, 455–464. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, C.; Zhao, Q.; Wang, Y.; Huo, M.; Wang, J.; Wang, S. Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J. Hazard. Mater. 2016, 320, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef] [PubMed]
- Zhi, S.; Shen, S.; Zhou, J.; Ding, G.; Zhang, K. Systematic analysis of occurrence, density and ecological risks of 45 veterinary antibiotics: Focused on family livestock farms in Erhai Lake basin, Yunnan, China. Environ. Pollut. 2020, 267, 115539. [Google Scholar] [CrossRef]
- Zhi, S.; Zhou, J.; Yang, F.; Tian, L.; Zhang, K. Systematic analysis of occurrence and variation tendency about 58 typical veterinary antibiotics during animal wastewater disposal processes in Tianjin, China. Ecotoxicol. Environ. Saf. 2018, 165, 376–385. [Google Scholar] [CrossRef]
- Pereira, A.R.; Paranhos, A.G.; de Aquino, S.F.; Silva, S.D. Distribution of genetic elements associated with antibiotic resistance in treated and untreated animal husbandry waste and wastewater. Environ. Sci. Pollut. Res. 2021, 28, 26380–26403. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Qian, X.; Gu, J.; Wang, X.J.; Duan, M.L. Mechanism and effect of temperature on variations in antibiotic resistance genes during anaerobic digestion of dairy manure. Sci. Rep. 2016, 6, 30237. [Google Scholar] [CrossRef]
- Kim, W.; Shin, S.G.; Cho, K.; Lee, C.; Hwang, S. Performance of methanogenic reactors in temperature phased two-stage anaerobic digestion of swine wastewater. J. Biosci. Bioeng. 2012, 114, 635–639. [Google Scholar] [CrossRef]
- Meegoda, J.; Li, B.; Patel, K.; Wang, L. A review of the processes, parameters, and optimization of anaerobic digestion. Int. J. Environ. Res. Public Health 2018, 15, 2224. [Google Scholar] [CrossRef]
- Zubair, M.; Li, Z.; Zhu, R.; Wang, J.; Liu, X.; Liu, X. The antibiotics degradation and its mechanisms during the livestock manure anaerobic digestion. Molecules 2023, 28, 4090. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.P.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Zhang, R.M.; Liu, X.; Wang, S.L.; Fang, L.X. Distribution patterns of antibiotic resistance genes and their bacterial hosts in pig farm wastewater treatment systems and soil fertilized with pig manure. Sci. Total Environ. 2021, 758, 143654. [Google Scholar] [CrossRef]
- Lu, X.M.; Lu, P.Z. Synergistic effects of key parameters on the fate of antibiotic resistance genes during swine manure composting. Environ. Pollut. 2019, 252, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, X.; Gu, J.; Zhang, S.; Yin, Y.; Li, Y. Effects of different swine manure to wheat straw ratios on antibiotic resistance genes and the microbial community structure during anaerobic digestion. Bioresour. Technol. 2017, 231, 1–8. [Google Scholar] [CrossRef]
- Pu, C.; Liu, H.; Ding, G.; Sun, Y.; Yu, X.; Chen, J.; Gong, X. Impact of direct application of biogas slurry and residue in fields: In situ analysis of antibiotic resistance genes from pig manure to fields. J. Hazard. Mater. 2018, 344, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Jiao, Q.; Cheng, L.; Song, L.; Xun, M.; Yang, H. Occurrence and prevalence of antibiotic resistance genes in apple orchard after continual application of anaerobic fermentation residues of pig manure. Environ. Sci. Pollut. Res. 2022, 30, 29229–29242. [Google Scholar] [CrossRef] [PubMed]
- Babić, S.; Periša, M.; Škorić, I. Photolytic degradation of norfloxacin, enrofloxacin and ciprofloxacin in various aqueous media. Chemosphere 2013, 91, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Li, Z.T.; Lin, Q.Y.; Wang, P.; Liu, W.; Yuan, J.; Hong, Z.S.; Chen, Y. Development and clinical application of a rapid and visual loop-mediated isothermal amplification test for gene in strains cultured from feces. Int. J. Infect. Dis. 2022, 122, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Q.; Wang, X.M.; Li, H.; Shang, Y.H.; Pan, Y.S.; Wu, C.M.; Wang, Y.; Du, X.D.; Shen, J.Z. Novel lnu(G) gene conferring resistance to lincomycin by nucleotidylation, located on Tn6260 from Enterococcus faecalis E531. J. Antimicrob. Chemoth. 2017, 72, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, M.K.; Liu, T.; Chen, H.; Verma, S.; Duan, Y.; Awasthi, S.K. The behavior of antibiotic resistance genes and their associations with bacterial community during poultry manure composting. Bioresour. Technol. 2019, 280, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sun, R.; Zhang, C.; Ding, S.; Ying, M.; Shan, S. In situ analysis of antibiotic resistance genes in anaerobically digested dairy manure and its subsequent disposal facilities. Bioresour. Technol. 2021, 333, 124988. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2014, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Wang, J.; Liu, Y.; Wang, G.; Yang, Y.; Liu, Y.; Kong, Y.; Lin, J.; Li, Q.; Li, G.; et al. Dynamics of antibiotic resistance genes and bacterial community during pig manure, kitchen waste, and sewage sludge composting. J. Environ. Manag. 2023, 345, 118651. [Google Scholar] [CrossRef]
- Yang, Q.; Tian, T.; Niu, T.; Wang, P. Molecular characterization of antibiotic resistance in cultivable multidrug-resistant bacteria from livestock manure. Environ. Pollut. 2017, 229, 188–198. [Google Scholar] [CrossRef]
- Yuan, X.; Lv, Z.; Zhang, Z.; Han, Y.; Liu, Z.; Zhang, H. A review of antibiotics, antibiotic resistant bacteria, and resistance genes in aquaculture: Occurrence, contamination, and transmission. Toxics 2023, 11, 420. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Wang, Y.; Yu, D.; Sui, Q.; Wang, R.; Chen, M.; Tong, J.; Wei, Y. Profiles and drivers of antibiotic resistance genes distribution in one-stage and two-stage sludge anaerobic digestion based on microwave-H2O2 pretreatment. Bioresour. Technol. 2017, 241, 573–581. [Google Scholar] [CrossRef]
- Zhang, R.; Gu, J.; Wang, X.J.; Li, Y.; Liu, J.Y.; Lu, C.; Qiu, L. Response of antibiotic resistance genes abundance by graphene oxide during the anaerobic digestion of swine manure with copper pollution. Sci. Total Environ. 2019, 654, 292–299. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Y.; Li, X.; Zhang, Y.; Ye, J.; Huang, H.; Zhu, C. Temporal effects of repeated application of biogas slurry on soil antibiotic resistance genes and their potential bacterial hosts. Environ. Pollut. 2020, 258, 113652. [Google Scholar] [CrossRef]
- You, Y.; Hilpert, M.; Ward, M.J. Detection of a common and persistent tet(L)—Carrying plasmid in chicken-waste-impacted farm soil. Appl. Environ. Microbiol. 2012, 78. [Google Scholar] [CrossRef]
- Zhu, N.; Jin, H.; Ye, X.; Liu, W.; Li, D.; Shah, G.M.; Zhu, Y. Fate and driving factors of antibiotic resistance genes in an integrated swine wastewater treatment system: From wastewater to soil. Sci. Total Environ. 2020, 721, 137654. [Google Scholar] [CrossRef]
- Beauchemin, J.; Fréchette, A.; Thériault, W.; Dufour, S.; Fravalo, P.; Thibodeau, A. Comparison of microbiota of recycled manure solids and straw bedding used in dairy farms in eastern Canada. Indian J. Dairy. Sci. 2022, 105, 389–408. [Google Scholar] [CrossRef]
- Rowbotham, R.F.; Ruegg, P.L. Bacterial counts on teat skin and in new sand, recycled sand, and recycled manure solids used as bedding in freestalls. J. Dairy Sci. 2016, 99, 6594–6608. [Google Scholar] [CrossRef]
- Zhang, T.; Boonyayatra, S.; Niu, G. Association of mastitis and farm management with contamination of antibiotics in bulk tank milk in southwest, China. Animals 2022, 12, 3392. [Google Scholar] [CrossRef]
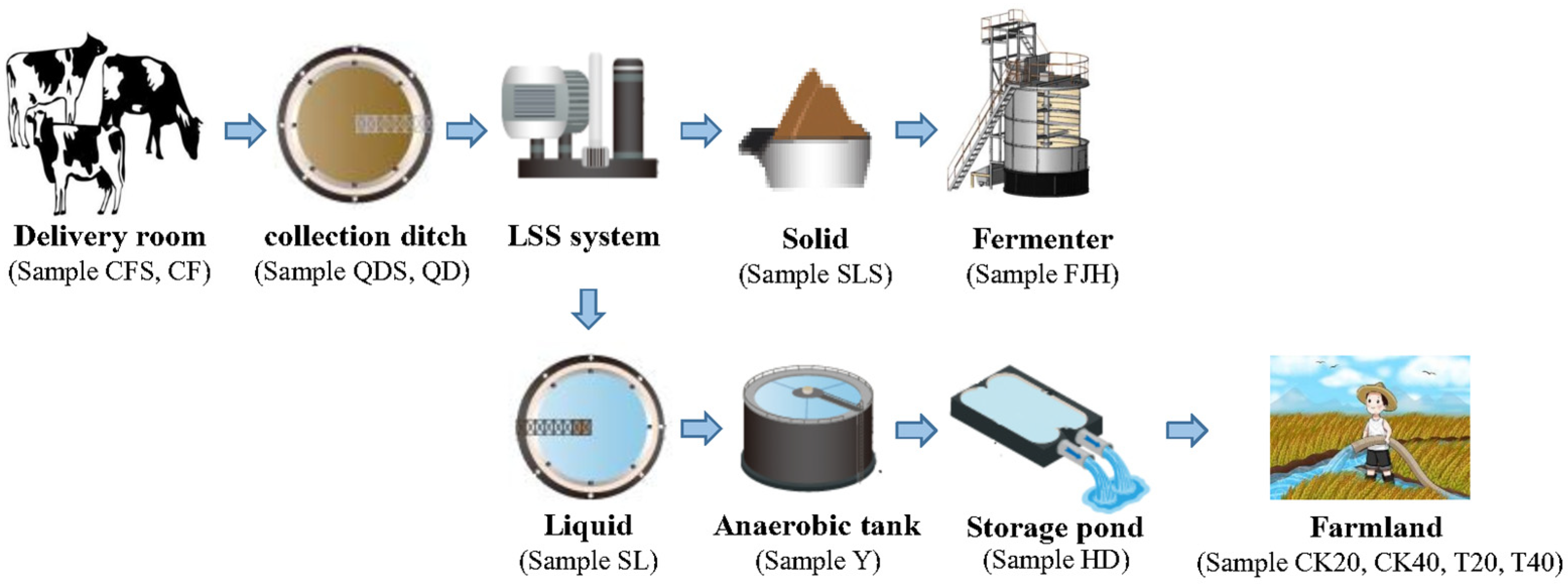

| Sample Type | Sample Name | TOC | TN | TP |
|---|---|---|---|---|
| Livestock wastewater (mg/L) | CF | 2869.8 ± 85.4 | 1283.3 ± 7.9 | 147.7 ± 5.6 |
| QD | 2428.2 ± 130.9 | 1040.7 ± 32.4 | 148.7 ± 4.7 | |
| SL | 3403.8 ± 188.7 | 1058.0 ± 25.7 | 235.3 ± 43.7 | |
| Y | 2983.2 ± 156.0 | 1173.0 ± 20.0 | 157.4 ± 34.0 | |
| HD | 2295.0 ± 79.0 | 552.0 ± 29.2 | 119.1 ± 24.6 | |
| Manure (mg/kg) | CFS | 54,550.0 ± 4450.0 | 829.3 ± 8.8 | 525.5 ± 78.5 |
| QDS | 38,100.0 ± 2400.0 | 722.0 ± 162.0 | 535.5 ± 11.5 | |
| SLS | 78,800.0 ± 5800.0 | 671.7 ± 31.5 | 498.5 ± 85.5 | |
| FJH | 57,300.0 ± 4300.0 | 676.0 ± 45.0 | 513.7 ± 80.4 |
| Sample | tetW/N/W | sul1 | lsaE | tetM | AAC6-Ie-APH2-Ia | adeF | lnuC | ErmF | ANT9-Ia | aadA | Others | Total ARGs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CF | 63.3 ± 2.3 | 101.5 ± 4.4 | 59.3 ± 1.8 | 53.2 ± 2.6 | 51.2 ± 1.4 | 15.3 ± 0.5 | 45.5 ± 2.3 | 39.5 ± 1.4 | 36.6 ± 1.8 | 32.2 ± 2.5 | 947.7 ± 12.4 | 1445.3 ± 10.1 |
| QD | 59.1 ± 0.6 | 22.5 ± 0.2 | 24.6 ± 1.3 | 9.7 ± 0.6 | 3.9 ± 0.5 | 12.0 ± 1.0 | 27.1 ± 3.0 | 8.4 ± 1.1 | 15.4 ± 0.8 | 7.6 ± 0.1 | 422.2 ± 8.1 | 612.6 ± 12.5 |
| SL | 62.6 ± 4.0 | 28.8 ± 2.1 | 34.1 ± 6.8 | 13.5 ± 1.8 | 6.6 ± 1.5 | 11.6 ± 0.7 | 26.1 ± 2.4 | 9.0 ± 1.0 | 19.8 ± 3.8 | 8.9 ± 0.9 | 465.1 ± 11.9 | 686.3 ± 30.3 |
| Y | 37.7 ± 3.1 | 18.3 ± 0.5 | 22.7 ± 0.7 | 23.1 ± 1.6 | 2.5 ± 0.2 | 19.3 ± 0.4 | 8.9 ± 0.3 | 6.6 ± 1.0 | 9.5 ± 0.9 | 6.7 ± 0.5 | 300.1 ± 6.9 | 455.3 ± 10.1 |
| HD | 18.7 ± 0.5 | 23.6 ± 0.6 | 22.8 ± 1.3 | 24.1 ± 1.5 | 2.4 ± 0.1 | 45.9 ± 3.1 | 5.2 ± 0.4 | 7.5 ± 0.3 | 7.3 ± 0.2 | 6.7 ± 0.6 | 326.9 ± 6.1 | 491.1 ± 8.0 |
| Sample | lnuC | CfxA2 | tetW/N/W | adeF | QnrB74 | ANT6-Ia | Mycobacterium tuberculosis rpsL | vanTC | tetW | lnuG | Others | Total ARGs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFS | 386.2 ± 53.2 | 87.1 ± 5.3 | 41.2 ± 7.2 | 0.7 ± 0.6 | 0.0 ± 0.0 | 1.3 ± 0.6 | 0.3 ± 0.0 | 0.6 ± 0.6 | 22.3 ± 5.2 | 0.6 ± 0.4 | 301.6 ± 24.5 | 841.9 ± 44.4 |
| QDS | 38.6 ± 7.2 | 15.4 ± 3.6 | 31.7 ± 6.9 | 12.0 ± 5.4 | 0.1 ± 0.1 | 27.0 ± 7.6 | 16.4 ± 5.0 | 0.1 ± 0.1 | 7.8 ± 1.6 | 17.2 ± 6.5 | 395.7 ± 32.3 | 562.0 ± 37.8 |
| SLS | 7.8 ± 2.0 | 0.8 ± 0.7 | 20 ± 4.5 | 42.6 ± 1.1 | 0.1 ± 0.1 | 18.0 ± 2.9 | 27.4 ± 4.7 | 0.1 ± 0.1 | 4.1 ± 0.4 | 9.8 ± 1.5 | 320.6 ± 15.4 | 451.2 ± 19.4 |
| FJH | 2.6 ± 0.3 | 0.1 ± 0.0 | 8.1 ± 0.4 | 19.7 ± 3.3 | 28.4 ± 9.8 | 10.2 ± 2.9 | 22.1 ± 5.1 | 12.9 ± 11.9 | 2.9 ± 0.5 | 19.6 ± 1.3 | 192.4 ± 20.9 | 319.1 ± 26.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Gao, Y.; Zheng, L.; Ji, L.; Kong, X.; Du, J.; Wang, H.; Duan, L.; Niu, T.; Liu, J.; et al. Identification and Distribution of Antibiotic Resistance Genes and Antibiotic Resistance Bacteria in the Feces Treatment Process: A Case Study in a Dairy Farm, China. Water 2024, 16, 1575. https://doi.org/10.3390/w16111575
Wang H, Gao Y, Zheng L, Ji L, Kong X, Du J, Wang H, Duan L, Niu T, Liu J, et al. Identification and Distribution of Antibiotic Resistance Genes and Antibiotic Resistance Bacteria in the Feces Treatment Process: A Case Study in a Dairy Farm, China. Water. 2024; 16(11):1575. https://doi.org/10.3390/w16111575
Chicago/Turabian StyleWang, Hailun, Yongchao Gao, Liwen Zheng, Lei Ji, Xue Kong, Jianhua Du, Hui Wang, Luchun Duan, Tian Niu, Jianhui Liu, and et al. 2024. "Identification and Distribution of Antibiotic Resistance Genes and Antibiotic Resistance Bacteria in the Feces Treatment Process: A Case Study in a Dairy Farm, China" Water 16, no. 11: 1575. https://doi.org/10.3390/w16111575
APA StyleWang, H., Gao, Y., Zheng, L., Ji, L., Kong, X., Du, J., Wang, H., Duan, L., Niu, T., Liu, J., & Shang, M. (2024). Identification and Distribution of Antibiotic Resistance Genes and Antibiotic Resistance Bacteria in the Feces Treatment Process: A Case Study in a Dairy Farm, China. Water, 16(11), 1575. https://doi.org/10.3390/w16111575






