Water and Sediments of an Acidic Hot Spring—Distinct Differentiation with Regard to the Microbial Community Composition and Functions
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of the Sampling Site
2.2. DNA Extraction and 16S rRNA Gene Sequencing
2.3. Sequencing and Analysis of the 4229s Metagenome
2.4. DNA Stable-Isotope Probing (DNA-SIP)
2.5. Radioisotopic Tracing Experiments
3. Results
3.1. The Microbial Communities of Sediments and Water in Hot Spring #4229
3.2. Metagenome Analysis of the Sediments Samples 4229s
3.3. DNA Stable-Isotope Probing of the Water Samples 4229w
3.4. Rates of Carbon Assimilation in the Water Samples 4229w
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kochetkova, T.V.; Podosokorskaya, O.A.; Elcheninov, A.G.; Kublanov, I.V. Diversity of thermophilic prokaryotes inhabiting Russian natural hot springs. Microbiology 2022, 91, 3–31. [Google Scholar] [CrossRef]
- Perevalova, A.A.; Kolganova, T.V.; Birkeland, N.K.; Schleper, C.; Bonch-Osmolovskaya, E.A.; Lebedinsky, A.V. Distribution of Crenarchaeota representatives in terrestrial hot springs of Russia and Iceland. Appl. Environ. Microbiol. 2008, 74, 7620–7628. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rozanov, A.S.; Bryanskaya, A.V.; Malup, T.K.; Meshcheryakova, I.A.; Lazareva, E.V.; Taran, O.P.; Ivanisenko, T.V.; Ivanisenko, V.A.; Zhmodik, S.M.; Kolchanov, N.A.; et al. Molecular analysis of the benthos microbial community in Zavarzin thermal spring (Uzon Caldera, Kamchatka, Russia). BMC Genom. 2014, 15, S12. [Google Scholar] [CrossRef] [PubMed]
- Dobretsov, N.L.; Lazareva, E.V.; Zhmodik, S.M.; Bryanskaya, A.V.; Morozova, V.V.; Tikunova, N.V.; Peltek, S.E.; Karpov, G.A.; Taran, O.P.; Ogorodnikova, O.L.; et al. Geological, hydrogeochemical, and microbiological characteristics of the Oil site of the Uzon caldera (Kamchatka). Russ. Geol. Geophys. 2015, 56, 39–63. [Google Scholar] [CrossRef]
- Bonch-Osmolovskaya, E.A. Studies of thermophilic microorganisms at the Institute of Microbiology, Russian Academy of Sciences. Microbiology 2004, 73, 644–658. [Google Scholar] [CrossRef]
- Brock, T.D. Thermophilic Microorganisms and Life at High Temperatures; Springer: Berlin, Germany, 1978; p. 233. [Google Scholar]
- Boyd, E.S.; Leavitt, W.D.; Geesey, G.G. CO2 uptake and fixation by a thermoacidophilic microbial community attached to precipitated sulfur in a geothermal spring. Appl. Environ. Microbiol. 2009, 75, 4289–4296. [Google Scholar] [CrossRef]
- Kees, E.D.; Murugapiran, S.K.; Bennett, A.C.; Hamilton, T.L. Distribution and Genomic Variation of Thermophilic Cyanobacteria in Diverse Microbial Mats at the Upper Temperature Limits of Photosynthesis. mSystems 2022, 26, e0031722. [Google Scholar] [CrossRef]
- Bonch-Osmolovskaya, E.A.; Gorlenko, V.M.; Karpov, G.A.; Starynin, D.A. Anaerobic destruction of the organic matter in microbial mats of the Thermofillnyi Spring (Uzon Caldera, Kamchatka). Microbiology 1987, 56, 812–818. [Google Scholar]
- Gorlenko, V.M.; Kachalkin, V.A.; Bonch-Osmolovskaya, E.A.; Starynin, D.A. Production processes in microbial cenoses of the Thermofilnyi hot spring. Microbiology 1987, 56, 692–697. [Google Scholar]
- Jackson, C.R.; Langner, H.W.; Donahoe-Christiansen, J.; Inskeep, W.P.; McDermott, T.R. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 2001, 3, 532–542. [Google Scholar] [CrossRef]
- Reigstad, L.J.; Jorgensen, S.L.; Schleper, C. Diversity and abundance of Korarchaeota in terrestrial hot springs of Iceland and Kamchatka. ISME J. 2010, 4, 346–356. [Google Scholar] [CrossRef]
- Hou, W.; Wang, S.; Dong, H.; Jiang, H.; Briggs, B.R.; Peacock, J.P.; Huang, Q.; Huang, L.; Wu, G.; Zhi, X.; et al. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS ONE 2013, 8, e53350. [Google Scholar] [CrossRef] [PubMed]
- Inskeep, W.P.; Jay, Z.J.; Tringe, S.G.; Herrgård, M.J.; Rusch, D.B. The YNP Metagenome project: Environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front. Microbiol. 2013, 6, 67. [Google Scholar] [CrossRef]
- Takacs-Vesbach, C.; Inskeep, W.P.; Jay, Z.J.; Herrgard, M.J.; Rusch, D.B.; Tringe, S.G.; Kozubal, M.A.; Hamamura, N.; Macur, R.E.; Fouke, B.W.; et al. Metagenome sequence analysis of filamentous microbial communities obtained from geochemically distinct geothermal channels reveals specialization of three aquificales lineages. Front. Microbiol. 2013, 29, 84. [Google Scholar] [CrossRef] [PubMed]
- Chernyh, N.A.; Mardanov, A.V.; Gumerov, V.M. Microbial life in Bourlyashchy, the hottest thermal pool of Uzon Caldera, Kamchatka. Extremophiles 2015, 19, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.H.; Liao, B.Y.; Chang, H.W.; Huang, S.W.; Chang, T.Y.; Yang, C.Y.; Wang, Y.B.; Lin, Y.T.; Wu, Y.W.; Tang, S.L.; et al. Metabolic characteristics of dominant microbes and key rare species from an acidic hot spring in Taiwan revealed by metagenomics. BMC Genom. 2015, 16, 1029. [Google Scholar] [CrossRef]
- Merkel, A.Y.; Pimenov, N.V.; Rusanov, I.I.; Slobodkin, A.I.; Slobodkina, G.B.; Tarnovetckii, I.Y.; Frolov, E.N.; Dubin, A.V.; Perevalova, A.A.; Bonch-Osmolovskaya, E.A. Microbial diversity and autotrophic activity in Kamchatka hot springs. Extremophiles 2017, 21, 307–317. [Google Scholar] [CrossRef]
- Cousins, C.R.; Fogel, M.; Bowden, R.; Crawford, I.; Boyce, A.; Cockell, C.; Gunn, M. Biogeochemical probing of microbial communities in a basalt-hosted hot spring at Kverkfjöll volcano, Iceland. Geobiology 2018, 16, 507–521. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Gumerov, V.M.; Beletsky, A.V.; Ravin, N.V. Microbial diversity in acidic thermal pools in the Uzon Caldera, Kamchatka. Antonie Van Leeuwenhoek 2018, 111, 35–43. [Google Scholar] [CrossRef]
- O’Neill, A.H.; Liu, Y.; Ferrera, I.; Beveridge, T.J.; Reysenbach, A.L. Sulfurihydrogenibium rodmanii sp. nov., a sulfur-oxidizing chemolithoautotroph from the Uzon Caldera, Kamchatka Peninsula, Russia, and emended description of the genus Sulfurihydrogenibium. Int. J. Syst. Evol. Microbiol. 2008, 58, 1147–1152. [Google Scholar] [CrossRef]
- Wemheuer, B.; Taube, R.; Akyol, P.; Wemheuer, F.; Daniel, R. Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka peninsula. Archaea 2013, 2013, 136714. [Google Scholar] [CrossRef]
- Wilkins, L.G.E.; Ettinger, C.L.; Jospin, G.; Eisen, J.A. Metagenome assembled genomes provide new insight into the microbial diversity of two thermal pools in Kamchatka, Russia. Sci. Rep. 2019, 9, 3059. [Google Scholar] [CrossRef] [PubMed]
- Zavarzin, G.A.; Svetlichny, V.A.; Bonch-Osmolovskaya, E.A. Extreme thermophiles from hydrotherm. In Recent Advances in Microbial Ecology; Hattori, T., Ed.; Japan Science Society Press: Tokyo, Japan, 1989; pp. 33–37. [Google Scholar]
- Kublanov, I.V.; Perevalova, A.A.; Slobodkina, G.B.; Lebedinsky, A.V.; Bidzhieva, S.K.; Kolganova, T.V.; Kaliberda, E.N.; Rumsh, L.D.; Haertlé, T.; Bonch-Osmolovskaya, E.A. Biodiversity of thermophilic prokaryotes with hydrolytic activities in hot springs of Uzon Caldera, Kamchatka (Russia). Appl. Environ. Microbiol. 2009, 75, 286–291. [Google Scholar] [CrossRef]
- Gumerov, V.M.; Mardanov, A.V.; Beletsky, A.V.; Bonch-Osmolovskaia, E.A.; Ravin, N.V. Molecular analysis of microbial diversity in the Zavarzin Spring, Uzon Caldera, Kamchatka. Microbiology 2011, 80, 244–251. [Google Scholar] [CrossRef]
- Burgess, E.A.; Unrine, J.M.; Mills, G.L.; Romanek, C.S.; Wiegel, J. Comparative geochemical and microbiological characterization of two thermal pools in the Uzon Caldera, Kamchatka, Russia. Microb. Ecol. 2012, 63, 471–489. [Google Scholar] [CrossRef]
- Menzel, P.; Gudbergsdóttir, S.R.; Rike, A.G.; Lin, L.; Zhang, Q.; Contursi, P.; Moracci, M.; Kristjansson, J.K.; Bolduc, B.; Gavrilov, S.; et al. Comparative metagenomics of eight geographically remote terrestrial hot springs. Microb. Ecol. 2015, 70, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Gohl, D.M.; MacLean, A.; Hauge, A.; Becker, A.; Walek, D.; Beckman, K.B. An optimized protocol for high-throughput amplicon-based microbiome profiling. Protoc. Exch. 2016. [Google Scholar] [CrossRef]
- Hugerth, L.W.; Wefer, H.A.; Lundin, S.; Jakobsson, H.E.; Lindberg, M.; Rodin, S.; Engstrand, L.; Andersson, A.F. DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl. Environ. Microbiol. 2014, 80, 5116–5123. [Google Scholar] [CrossRef]
- Merkel, A.Y.; Tarnovetskii, I.Y.; Podosokorskaya, O.A.; Toshchakov, S.V. Analysis of 16S rRNA primer systems for profiling of thermophilic microbial communities. Microbiology 2019, 88, 671–680. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP-a Flexible Pipeline for Genome-Resolved Metagenomic Data Analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An Automated Binning Algorithm to Recover Genomes from Multiple Metagenomic Datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 26, e7359. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory friendly classification with the Genome Taxonomy Database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Neufeld, J.D.; Vohra, J.; Dumont, M.G.; Lueders, T.; Manefield, M.; Friedrich, M.W.; Murrell, J.C. DNA stable-isotope probing. Nat. Protoc. 2007, 2, 860–866. [Google Scholar] [CrossRef]
- Dunford, E.A.; Neufeld, J.D. DNA stable-isotope probing (DNA-SIP). J. Vis. Exp. 2010, 2, 2027. [Google Scholar]
- Pimenov, N.V.; Bonch-Osmolovskaya, E.A. In situ activity studies in thermal environments. Methods Microbiol. 2006, 35, 29–53. [Google Scholar]
- Mardanov, A.V.; Gumerov, V.M.; Beletsky, A.V.; Perevalova, A.A.; Karpov, G.A.; Bonch-Osmolovskaya, E.A.; Ravin, N.V. Uncultured archaea dominate in the thermal groundwater of Uzon Caldera, Kamchatka. Extremophiles 2011, 15, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Hohn, M.J.; Hedlund, B.P.; Huber, H. Detection of 16S rDNA sequences representing the novel phylum “Nanoarchaeota”: Indication for a wide distribution in high temperature biotopes. Syst. Appl. Microbiol. 2002, 25, 551–554. [Google Scholar] [CrossRef][Green Version]
- Auchtung, T.A.; Shyndriayeva, G.; Cavanaugh, C.M. 16S rRNA phylogenetic analysis and quantification of Korarchaeota indigenous to the hot springs of Kamchatka, Russia. Extremophiles 2011, 15, 105–116. [Google Scholar] [CrossRef]
- Perevalova, A.A.; Kublanov, I.V.; Baslerov, R.V.; Zhang, G.; Bonch-Osmolovskaya, E.A. Brockia lithotrophica gen. nov., sp. nov., an anaerobic thermophilic bacterium from a terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 479–483. [Google Scholar] [CrossRef]
- Golyshina, O.V.; Bargiela, R.; Toshchakov, S.V.; Chernyh, N.A.; Ramayah, S.; Korzhenkov, A.A.; Kublanov, I.V.; Golyshin, P.N. Diversity of “Ca. Micrarchaeota” in two distinct types of acidic environments and their associations with Thermoplasmatales. Genes 2019, 10, 461. [Google Scholar] [CrossRef]
- Kozubal, M.A.; Romine, M.; Jennings, R.D.; Jay, Z.J.; Tringe, S.G.; Rusch, D.B.; Beam, J.P.; McCue, L.A.; Inskeep, W.P. Geoarchaeota: A new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J. 2012, 7, 622–634. [Google Scholar] [CrossRef]
- Kato, S.; Ohnishi, M.; Nagamori, M.; Yuki, M.; Takashina, T.; Ohkuma, M.; Itoh, T. Conexivisphaera calida gen. nov., sp. nov., a thermophilic sulfur- and iron-reducing archaeon, and proposal of Conexivisphaeraceae fam. nov., Conexivisphaerales ord. nov., and Conexivisphaeria class. nov. in the phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 2021, 71, 004595. [Google Scholar] [CrossRef]
- Shima, S.; Suzuki, K.I. Hydrogenobacter acidophilus sp. nov., a thermoacidophilic, aerobic, hydrogen-oxidizing bacterium requiring elemental sulfur for growth. Int. J. Syst. Bacteriol. 1993, 43, 703–708. [Google Scholar] [CrossRef]
- Eder, W.; Huber, R. New isolates and physiological properties of the Aquificales and description of Thermocrinis albus sp. nov. Extremophiles 2002, 6, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Donahoe-Christiansen, J.; D’Imperio, S.; Jackson, C.R.; Inskeep, W.P.; McDermott, T.R. Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 2004, 70, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Reysenbach, A.L.; Banta, A.; Civello, S.; Daly, J.; Mitchel, K.; Lalonde, S.; Konhauser, K.; Rodman, A.; Rustercholdz, K.; Takacs-Vesbach, C. Aquificales in Yellowstone National Park. In Geothermal Biology and Geochemistry in YNP; Inskeep, W.P., McDermott, T.R., Eds.; Montana State University Publications: Bozeman, MT, USA, 2005; pp. 129–142. [Google Scholar]
- Spear, J.R.; Walker, J.J.; McCollom, T.M.; Pace, N.R. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 2005, 15, 102–107. [Google Scholar] [CrossRef]
- Mathur, J.; Bizzoco, R.W.; Ellis, D.G.; Lipson, D.A.; Poole, A.W.; Levine, R.; Kelley, S.T. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl. Environ. Microbiol. 2007, 73, 2612–2623. [Google Scholar] [CrossRef]
- D’Imperio, S.; Lehr, C.R.; Oduro, H.; Druschel, G.; Kühl, M.; McDermott, T.R. Relative importance of H2 and H2S as energy sources for primary production in geothermal springs. Appl. Environ. Microbiol. 2008, 74, 5802–5808. [Google Scholar] [CrossRef]
- Hall, J.R.; Mitchell, K.R.; Jackson-Weaver, O.; Kooser, A.S.; Cron, B.R.; Crossey, L.J.; Takacs-Vesbach, C.D. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl. Environ. Microbiol. 2008, 74, 4910–4922. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, N.; Macur, R.E.; Korf, S.; Ackerman, G.; Taylor, W.P.; Kozubal, M.; Reysenbach, A.L.; Inskeep, W.P. Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments. Environ. Microbiol. 2009, 11, 421–431. [Google Scholar] [CrossRef]
- Inskeep, W.P.; Rusch, D.B.; Jay, Z.J.; Herrgard, M.J.; Kozubal, M.A.; Richardson, T.H.; Macur, R.E.; Hamamura, N.; Jennings, R.; Fouke, B.W.; et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE 2010, 5, e9773. [Google Scholar] [CrossRef]
- Jennings, R.M.; Moran, J.J.; Jay, Z.J.; Beam, J.P.; Whitmore, L.M.; Kozubal, M.A.; Kreuzer, H.W.; Inskeep, W.P. Integration of Metagenomic and Stable Carbon Isotope Evidence Reveals the Extent and Mechanisms of Carbon Dioxide Fixation in High-Temperature Microbial Communities. Front. Microbiol. 2017, 3, 88. [Google Scholar] [CrossRef]
- Valdes, J.; Quatrini, R.; Hallberg, K.; Dopson, M.; Valenzuela, P.D.; Holmes, D.S. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 2009, 191, 5877–5878. [Google Scholar] [CrossRef]
- Nunez, H.; Covarrubias, P.C.; Moya-Beltran, A.; Issotta, F.; Atavales, J.; Acuña, L.G.; Johnson, D.B.; Quatrini, R. Detection, identification and typing of Acidithiobacillus species and strains: A review. Res. Microbiol. 2016, 167, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Nunez, H.; Moya-Beltran, A.; Covarrubias, P.C.; Issotta, F.; Cárdenas, J.P.; González, M.; Atavales, J.; Acuña, L.G.; Johnson, D.B.; Quatrini, R. Molecular systematics of the genus Acidithiobacillus: Insights into the phylogenetic structure and diversification of the taxon. Front. Microbiol. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Fliermans, C.B.; Brock, T.D. Ecology of sulfur-oxidizing bacteria in hot acid soils. J. Bacteriol. 1972, 111, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Burton, N.P.; Norris, P.R. Microbiology of acidic, geothermal springs of Montserrat: Environmental rDNA analysis. Extremophiles 2000, 4, 315–320. [Google Scholar] [CrossRef]
- Hamamura, N.; Olson, S.H.; Ward, D.M.; Inskeep, W.P. Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl. Environ. Microbiol. 2005, 71, 5943–5950. [Google Scholar] [CrossRef]
- Ni, Y.Q.; He, K.Y.; Bao, J.T.; Yang, Y.; Wan, D.S.; Li, H.Y. Genomic and phenotypic heterogeneity of Acidithiobacillus spp. strains isolated from diverse habitats in China. FEMS Microbiol. Ecol. 2008, 64, 248–259. [Google Scholar] [CrossRef][Green Version]
- Aditiawati, P.; Yohandini, H.; Madayanti, F. Microbial diversity of acidic hot spring (kawah hujan B) in geothermal field of kamojang area, west java-Indonesia. Open Microbiol. J. 2009, 3, 58–66. [Google Scholar] [CrossRef]
- Urbieta, M.S.; González, T.E.; Aguilera, A.; Giaveno, M.A.; Donati, E. First prokaryotic biodiversity assessment using molecular techniques of an acidic river in Neuquén, Argentina. Microb. Ecol. 2012, 64, 91–104. [Google Scholar] [CrossRef]
- Sahm, K.; John, P.; Nacke, H.; Wemheuer, B.; Grote, R.; Daniel, R.; Antranikian, G. High abundance of heterotrophic prokaryotes in hydrothermal springs of the Azores as revealed by a network of 16S rRNA gene-based methods. Extremophiles 2013, 17, 649–662. [Google Scholar] [CrossRef]
- Kelly, D.P.; Wood, A.P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2000, 2, 511–516. [Google Scholar] [CrossRef]
- Hallberg, K.B.; Lindstrom, E.B. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 1994, 140, 3451–3456. [Google Scholar] [CrossRef] [PubMed]
- Shooner, F.; Bousquet, J.; Tyagi, R.D. Isolation, phenotypic characterization, and phylogenetic position of a novel, facultatively autotrophic, moderately thermophilic bacterium, Thiobacillus thermosulfatus sp. nov. Int. J. Syst. Bacteriol. 1996, 46, 409–415. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kelly, D.P.; Uchino, Y.; Huber, H.; Amils, R.; Wood, A.P. Reassessment of the phylogenetic relationships of Thiomonas cuprina. Int. J. Syst. Evol. Microbiol. 2007, 57, 2720–2724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vesteinsdottir, H.; Reynisdottir, D.B.; Orlygsson, J. Thiomonas islandica sp. nov., a moderately thermophilic, hydrogen- and sulfur-oxidizing betaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2011, 61, 132–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- London, J.; Rittenberg, S.C. Thiobacillus perometabolis nov. sp., a non-autotrophic thiobacillus. Arch. Mikrobiol. 1967, 59, 218–225. [Google Scholar] [CrossRef]
- Moreira, D.; Amils, R. Phylogeny of Thiobacillus cuprinus and other mixotrophic thiobacilli: Proposal for Thiomonas gen. nov. Int. J. Syst. Bacteriol. 1997, 47, 522–528. [Google Scholar] [CrossRef]
- Panda, S.K.; Jyoti, V.; Bhadra, B.; Nayak, K.C.; Shivaji, S.; Rainey, F.A.; Das, S.K. Thiomonas bhubaneswarensis sp. nov., an obligately mixotrophic, moderately thermophilic, thiosulfate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 2009, 59, 2171–2175. [Google Scholar] [CrossRef]
- Slyemi, D.; Moinier, D.; Brochier-Armanet, C.; Bonnefoy, V.; Johnson, D. Characteristics of a phylogenetically ambiguous, arsenic-oxidizing Thiomonas sp., Thiomonas arsenitoxydans strain 3As(T) sp. nov. Arch. Microbiol. 2011, 193, 439–449. [Google Scholar] [CrossRef]
- Gerasimchuk, A.L.; Ivasenko, D.A.; Kasymova, A.A.; Frank, Y.A. Selective cultivation of bacterial strains with lipolytic and hydrocarbon-oxidizing activity from bottom sediments of the Ob River, Western Siberia. Vavilov J. Genet. Breed. 2022, 26, 449–457. [Google Scholar] [CrossRef]
- Bruneel, O.; Personné, J.C.; Casiot, C.; Leblanc, M.; Elbaz-Poulichet, F.; Mahler, B.J.; Le Flèche, A.; Grimont, P.A. Mediation of arsenic oxidation by Thiomonas sp. in acid-mine drainage (Carnoulès, France). J. Appl. Microbiol. 2003, 95, 492–499. [Google Scholar] [CrossRef]
- Duquesne, K.; Lieutaud, A.; Ratouchniak, J.; Muller, D.; Lett, M.C.; Bonnefoy, V. Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: From the strain isolation to the gene study. Environ. Microbiol. 2008, 10, 228–237. [Google Scholar] [CrossRef]
- Akob, D.M.; Hallenbeck, M.; Beulig, F.; Fabisch, M.; Küsel, K.; Keffer, J.L.; Woyke, T.; Shapiro, N.; Lapidus, A.; Klenk, H.P.; et al. Mixotrophic iron-oxidizing Thiomonas isolates from an acid mine drainage-affected creek. Appl. Environ. Microbiol. 2022, 86, e01424–20. [Google Scholar]
- Macur, R.E.; Langer, H.W.; Kocar, B.D.; Inskeep, W.P. Linking geochemical processes with microbial community analysis: Successional dynamics in an arsenic-rich, acid-sulphate-chloride geothermal spring. Geobiology 2004, 2, 163–177. [Google Scholar] [CrossRef]
- Inskeep, W.P.; McDermott, T.R. Geomicrobiology of acidsulfate-chloride springs in Yellowstone National Park. In Geothermal Biology and Geochemistry in YNP; Inskeep, W.P., McDermott, T.R., Eds.; Thermal Biology Institute: Bozeman, MT, USA, 2005; pp. 143–162. [Google Scholar]
- Wilson, M.S.; Siering, P.L.; White, C.L.; Hauser, M.E.; Bartles, A.N. Novel archaea and bacteria dominate stable microbial communities in North America’s Largest Hot Spring. Microb. Ecol. 2008, 56, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Bonch-Osmolovskaya, E.A.; Sokolova, T.G.; Kostrikina, N.A.; Zavarzin, G.A. Desulfurella acetivorans gen. nov., sp. nov., a new thermophilic sulfur-reducing bacterium. Arch. Microbiol. 1990, 153, 151–155. [Google Scholar] [CrossRef]
- Miroshnichenko, M.L.; Rainey, F.A.; Hippe, H.; Chernyh, N.A.; Kostrikina, N.A.; Bonch-Osmolovskaya, E.A. Desulfurella kamchatkensis sp. nov. and Desulfurella propionica sp. nov., new sulfur-respiring thermophilic bacteria from Kamchatka thermal environments. Int. J. Syst. Bacteriol. 1998, 2, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Florentino, A.P.; Brienza, C.; Stams, A.J.M.; Sánchez-Andrea, I. Desulfurella amilsii sp. nov., a novel acidotolerant sulfur-respiring bacterium isolated from acidic river sediments. Int. J. Syst. Evol. Microbiol. 2016, 66, 1249–1253. [Google Scholar] [CrossRef]
- Bonch-Osmolovskaya, E.A.; Myroshichenko, M.L.; Slobodkin, A.I.; Sokolova, T.G.; Kostrikina, N.A.; Prokofeva, M.I.; Rusanov, I.I.; Pimenov, N.V.; Karpov, G.A.; Zavarzina, D.G. Biodiversity of anaerobic lithotrophic prokaryotes in terrestrial hot springs of Kamchatka. Microbiology 1999, 68, 398–406. [Google Scholar]
- Pimenov, N.V. Radioisotope studies of microbial activity in the hot springs of the Uzon Caldera (Kamchatka). Proc. Winogradsky Inst. Microbiol. 2011, 16, 144–159. [Google Scholar]
- Frolov, E.N.; Kublanov, I.V.; Toshchakov, S.; Samarov, N.I.; Novikov, A.A.; Lebedinsky, A.V.; Bonch-Osmolovskaya, E.A.; Chernyh, N.A. Thermodesulfobium acidiphilum sp. nov., a new thermoacidophilic sulfate-reducing chemoautotrophic bacterium from a Kamchatkan thermal site. Int. J. Syst. Evol. Microbiol. 2016, 67, 1482–1485. [Google Scholar] [CrossRef]
- Urschel, M.R.; Kubo, M.D.; Hoehler, T.M.; Peters, J.W.; Boyd, E.S. Carbon source preference in chemosynthetic hot spring communities. Appl. Environ. Microbiol. 2015, 81, 3834–3847. [Google Scholar] [CrossRef] [PubMed][Green Version]


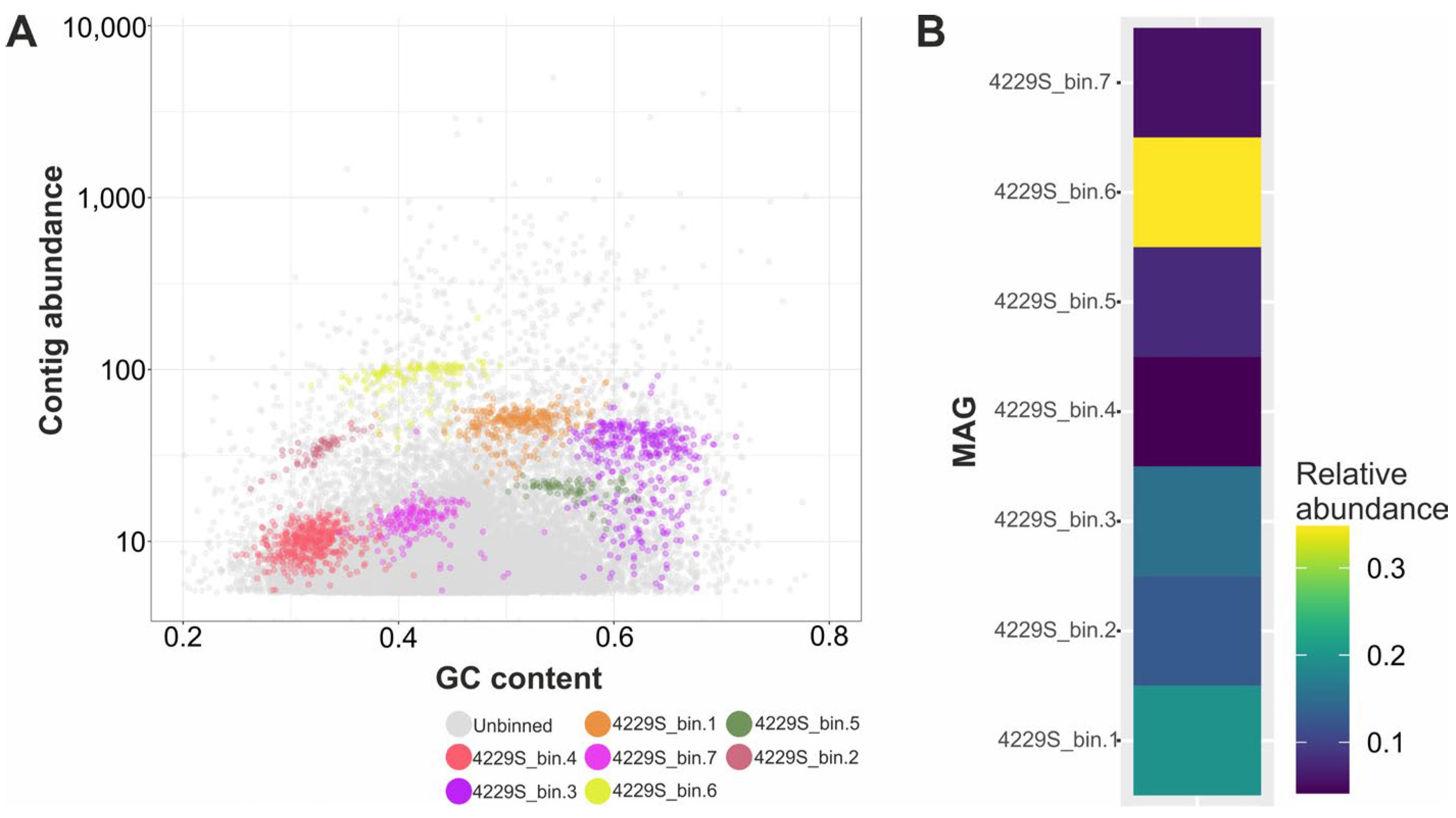
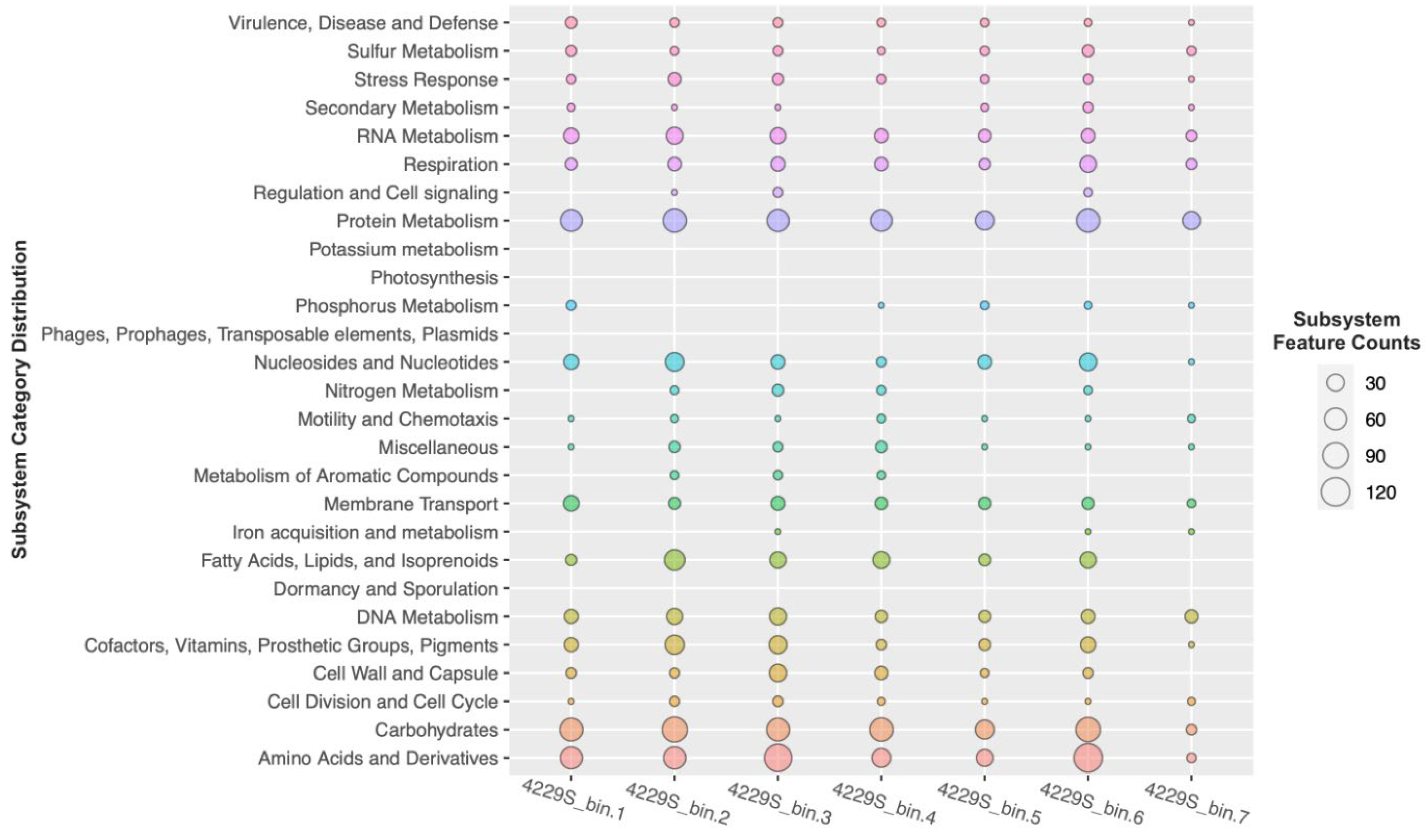
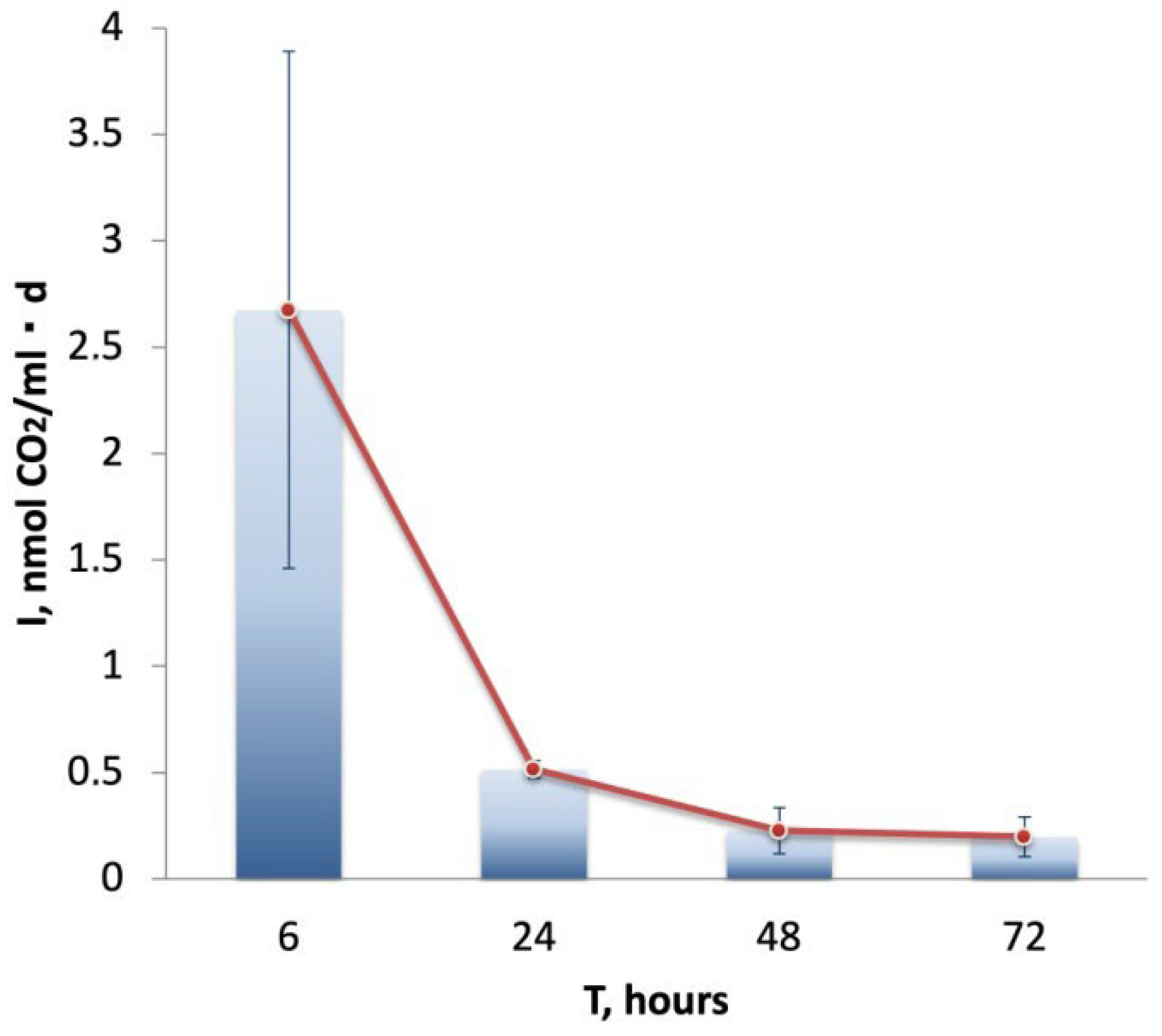
| MAG | Size, Mbp | Completeness, % | Contamination, % | Taxonomical Assignment (According to the GTDB Taxonomy) |
|---|---|---|---|---|
| 4229S_bin.1 | 1.36 | 87.62 | 0.93 | Thermoproteota/Thermoproteia/’Ca. Marsarchaeales’/NA |
| 4229S_bin.2 | 1.23 | 100 | 0.93 | Thermoplasmatota/Thermoplasmata/ARK-15/NA |
| 4229S_bin.3 | 1.54 | 91.59 | 0 | Thermoproteota/Nitrososphaeria/Conexivisphaerales/Conexivisphaeraceae/Conexivisphaera |
| 4229S_bin.4 | 1.14 | 80.61 | 0 | Thermoproteota/Thermoproteia/Sulfolobales/Acidilobaceae/Caldisphaera |
| 4229S_bin.5 | 0.99 | 86.99 | 0 | Thermoproteota/Thermoproteia/’Ca. Marsarchaeales’/NA |
| 4229S_bin.6 | 1.26 | 90.81 | 0 | Thermoproteota/Nitrososphaeria/Conexivisphaerales/NA |
| 4229S_bin.7 | 0.48 | 64.89 | 1.25 | Nanoarchaeota/Nanoarchaeia/Parvarchaeales/Parvarchaeacea/NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maltseva, A.I.; Klyukina, A.A.; Elcheninov, A.G.; Pimenov, N.V.; Rusanov, I.I.; Kublanov, I.V.; Kochetkova, T.V.; Frolov, E.N. Water and Sediments of an Acidic Hot Spring—Distinct Differentiation with Regard to the Microbial Community Composition and Functions. Water 2023, 15, 3415. https://doi.org/10.3390/w15193415
Maltseva AI, Klyukina AA, Elcheninov AG, Pimenov NV, Rusanov II, Kublanov IV, Kochetkova TV, Frolov EN. Water and Sediments of an Acidic Hot Spring—Distinct Differentiation with Regard to the Microbial Community Composition and Functions. Water. 2023; 15(19):3415. https://doi.org/10.3390/w15193415
Chicago/Turabian StyleMaltseva, Anastasia I., Alexandra A. Klyukina, Alexander G. Elcheninov, Nikolay V. Pimenov, Igor I. Rusanov, Ilya V. Kublanov, Tatiana V. Kochetkova, and Evgeny N. Frolov. 2023. "Water and Sediments of an Acidic Hot Spring—Distinct Differentiation with Regard to the Microbial Community Composition and Functions" Water 15, no. 19: 3415. https://doi.org/10.3390/w15193415
APA StyleMaltseva, A. I., Klyukina, A. A., Elcheninov, A. G., Pimenov, N. V., Rusanov, I. I., Kublanov, I. V., Kochetkova, T. V., & Frolov, E. N. (2023). Water and Sediments of an Acidic Hot Spring—Distinct Differentiation with Regard to the Microbial Community Composition and Functions. Water, 15(19), 3415. https://doi.org/10.3390/w15193415





