A Classification-Based Machine Learning Approach to the Prediction of Cyanobacterial Blooms in Chilgok Weir, South Korea
Abstract
:1. Introduction
2. Materials and Methods
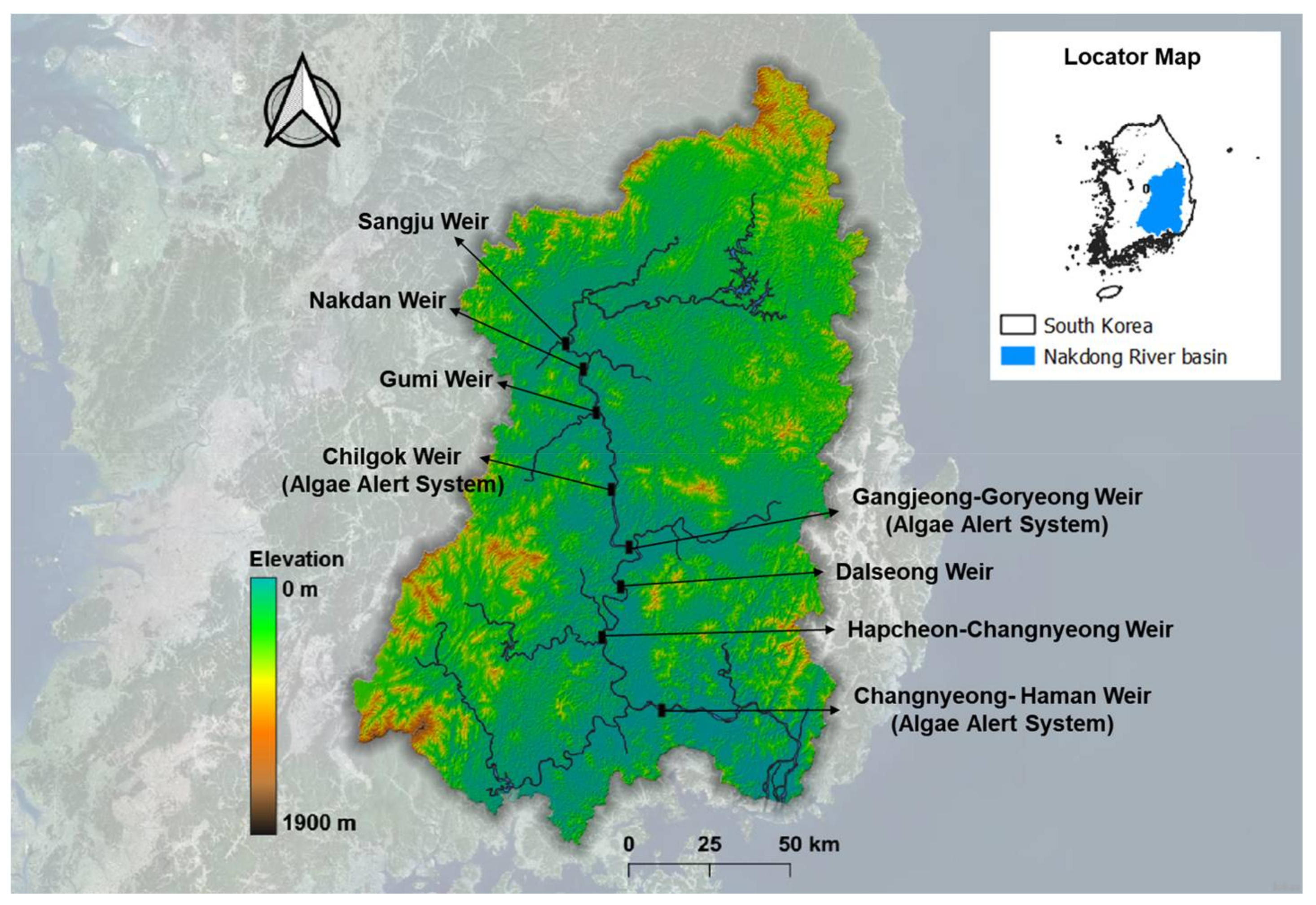
2.1. Study Area
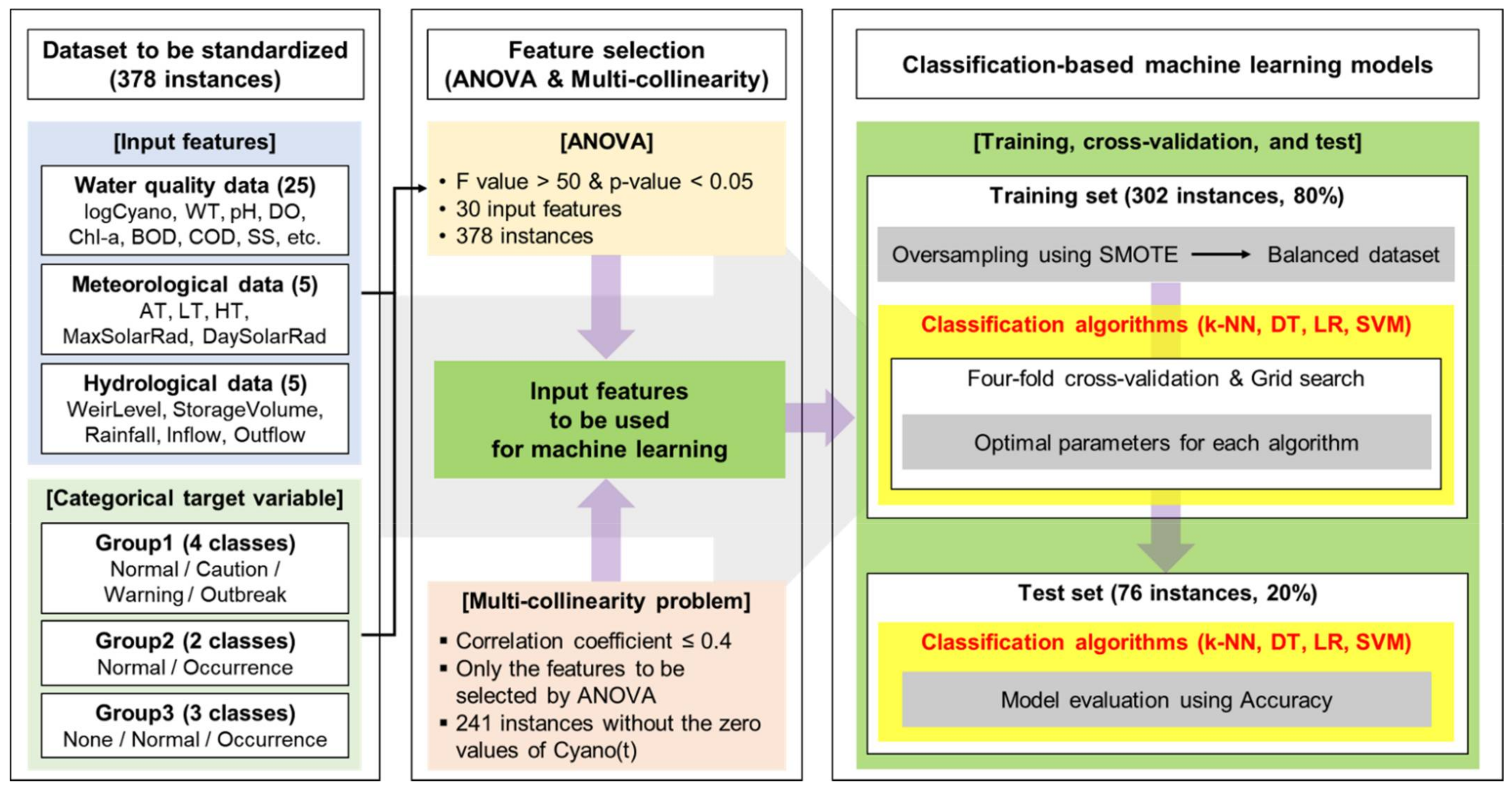
2.2. Dataset
2.2.1. Data Availability
2.2.2. Data Preprocessing
2.3. Feature Selection
2.3.1. Analysis of Variance (ANOVA)
2.3.2. Multi-Collinearity
2.4. Machine Learning
2.4.1. Classification Algorithms
2.4.2. Oversampling Using SMOTE (Synthetic Minority Oversampling Technique)
2.4.3. Training, Cross-Validation, and Test
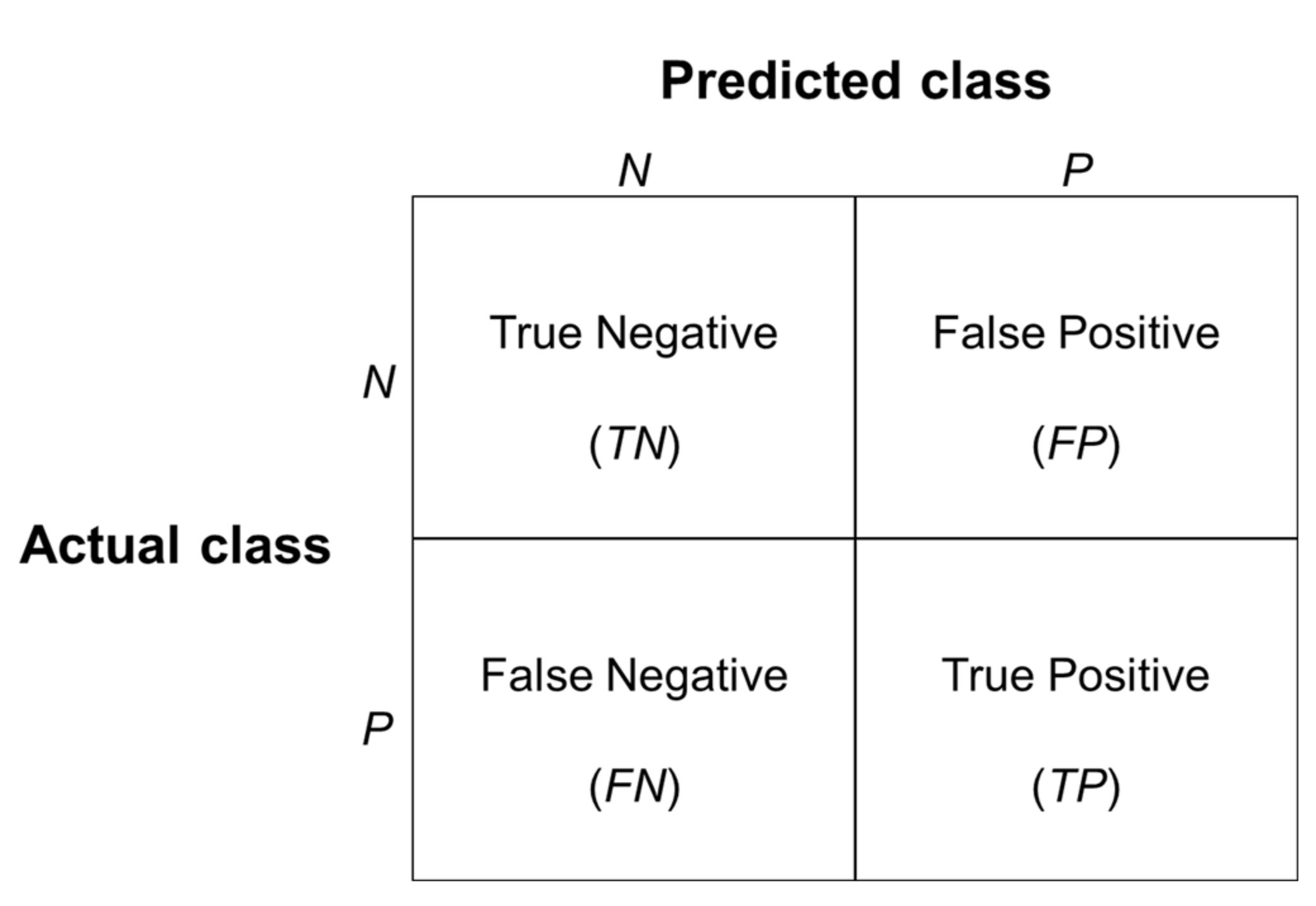
2.4.4. Model Evaluation
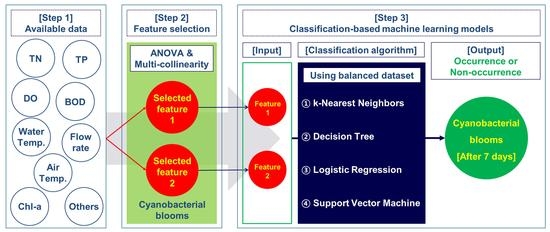
2.5. Summary of the Modeling Procedure
- One-way ANOVA was carried out using 30 features including logCyano of a standardized dataset with 378 instances for three groups (Group1, Group2, and Group3). For the purpose of selecting the features having a strong correlation with the target variable, F values of more than 50 and p-values of less than 0.05 [34,35] were applied. Here, the target variable was a class based on Cyano(t+1) for each group; Normal/Caution/Warning/Outbreak for Group1 (which was actually divided into three classes because the number of Outbreak elements was zero), Normal/Occurrence for Group2, and None/Normal/Occurrence for Group3.
- To address the multi-collinearity problem, the correlation analysis was performed among the features selected in the first step. As the final process for the feature selection, the paired features with low inter-correlation coefficients (0.4 or less [48,49]) were selected. Here, Pearson’s correlation analysis was performed with only 241 instances by excluding the zero values of Cyano(t) in 378 instances, as the zero values were able to distort the analysis result.
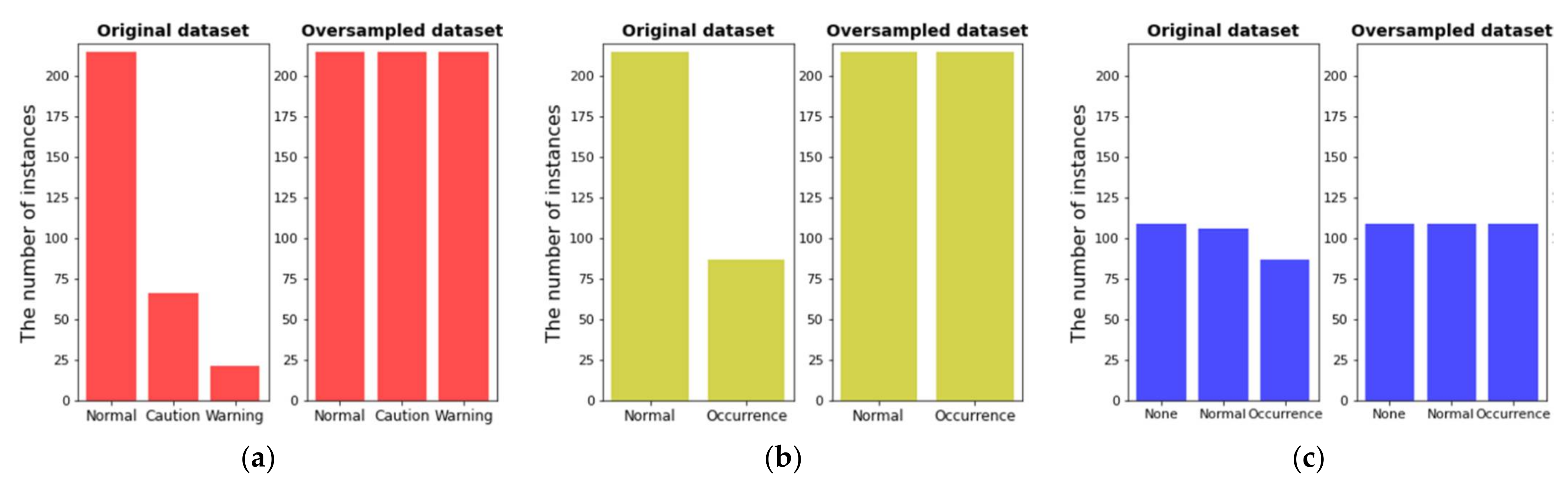
- The dataset consisting of the input features selected in the second step and the target variable was split into a training set and a test set by 80% and 20%. Therefore, 302 and 76 out of 378 instances were used as the training set and the test set, respectively. After that, oversampling for the training set was performed [31] by applying SMOTE. As a result of the oversampling, the number of instances by class became the same.
- Using the balanced datasets of three groups acquired in the third step, four classification-based machine learning algorithms including k-NN, DT, LR, and SVM, were applied. The models with optimal parameters for each machine learning method were constructed through four-fold cross-validation and grid search using the training set.
- The optimal combination of input features and machine learning algorithms for predicting the categorical target variable was presented by evaluating the performance (Accuracy) from the test set using the models developed in the fourth step.
3. Results
3.1. Determination of the Modeling Cases
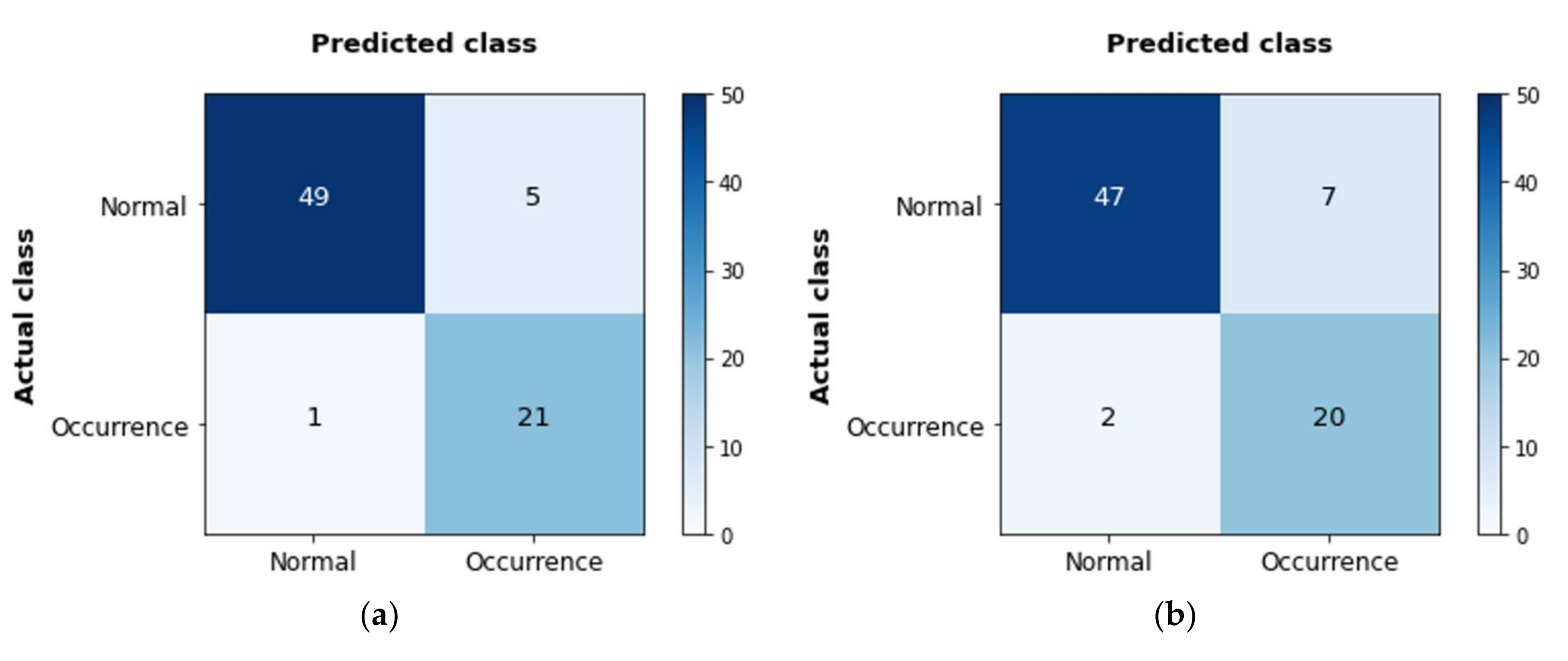
3.2. Accuracy of the Models
3.3. Summary of the Modeling Results
- We had nine input features including logCyano, WT, DO, TN, TDN, NO3-N, AT, LT, and HT from 30 input features by applying one-way ANOVA.
- Seven input features except for WT and DO were available finally for model construction due to the multi-collinearity problem.
- By using only two input features, we could build a model with a prediction accuracy of more than 80%.
- The models using Group2 with two classes surpassed the other models using Group1 and Group3 that were divided into three classes in terms of model performance.
- The optimal combination, developing the most accurate model was SVM-Group2-Case1, whose accuracy was the highest at 92%.
- All the models with the highest accuracy for each of the four machine learning algorithms (k-NN, DT, LR, and SVM) included logCyano as a feature.
- Among the models that did not use logCyano as a feature, the ones in combination with air temperature (AT, LT, or HT) and NO3-N enabled high predictive accuracy of more than 80%.
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, Y.D.; Xu, X.W.; Qi, M.; Sun, J.J.; Zhang, Y.Y.; Zhang, W.; Wang, M.Z.; Wang, X.J.; Zhang, Y. Lake warming intensifies the seasonal pattern of internal nutrient cycling in the eutrophic lake and potential impacts on algal blooms. Water Res. 2021, 188, 116570. [Google Scholar]
- Park, H.K.; Lee, H.J.; Heo, J.; Yun, J.H.; Kim, Y.J.; Kim, H.M.; Hong, D.G.; Lee, I.J. Deciphering the key factors determining spatio-temporal heterogeneity of cyanobacterial bloom dynamics in the Nakdong River with consecutive large weirs. Sci. Total Environ. 2021, 755, 143079. [Google Scholar] [CrossRef] [PubMed]
- Kosten, S.; Huszar, V.L.; Bécares, E.; Costa, L.S.; van Donk, E.; Hansson, L.A.; Jeppesen, E.; Kruk, C.; Lacerot, G.; Mazzeo, N. Warmer climates boost cyanobacterial dominance in shallow lakes. Glob. Chang. Biol. 2012, 18, 118–126. [Google Scholar] [CrossRef]
- Paerl, H.W.; Huisman, J. Climate change: A catalyst for global expansion of harmful cyanobacterial blooms. Environ. Microbiol. Rep. 2009, 1, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Scott, J.T. Throwing fuel on the fire: Synergistic effects of excessive nitrogen inputs and global warming on harmful algal blooms. Environ. Sci. Technol. 2010, 44, 7756–7758. [Google Scholar] [CrossRef]
- Smith, G.J.; Daniels, V. Algal blooms of the 18th and 19th centuries. Toxicon 2018, 142, 42–44. [Google Scholar] [CrossRef] [PubMed]
- Plaas, H.E.; Paerl, H.W. Toxic Cyanobacteria: A Growing Threat to Water and Air Quality. Environ. Sci. Technol. 2021, 55, 44–64. [Google Scholar] [CrossRef]
- Ho, L.; Goethals, P. Research hotspots and current challenges of lakes and reservoirs: A bibliometric analysis. Scientometrics 2020, 124, 603–631. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kim, S.; Mehrotra, R.; Sharma, A. Predicting cyanobacteria occurrence using climatological and environmental controls. Water Res. 2020, 175, 115639. [Google Scholar] [CrossRef]
- Song, H.; Lynch, M.J. Restoration of Nature or Special Interests? A Political Economy Analysis of the Four Major Rivers Restoration Project in South Korea. Crit. Criminol. 2018, 26, 251–270. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.; Choi, B.; Kim, G.; Lee, J. Harmful algal blooms and liver diseases: Focusing on the areas near the four major rivers in South Korea. J. Environ. Sci. Health C 2019, 37, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W. Controlling cyanobacterial harmful blooms in freshwater ecosystems. Microb. Biotechnol. 2017, 10, 1106–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurtsbaugh, W.A.; Paerl, H.W.; Dodds, W.K. Nutrients, eutrophication and harmful algal blooms along the freshwater to marine continuum. Wiley Interdiscip. Rev. Water 2019, 6, e1373. [Google Scholar] [CrossRef]
- Ahn, J.M.; Kim, J.; Park, L.J.; Jeon, J.; Jong, J.; Min, J.H.; Kang, T. Predicting Cyanobacterial Harmful Algal Blooms (CyanoHABs) in a Regulated River Using a Revised EFDC Model. Water 2021, 13, 439. [Google Scholar] [CrossRef]
- Park, Y.; Pyo, J.; Kwon, Y.S.; Cha, Y.; Lee, H.; Kang, T.; Cho, K.H. Evaluating physico-chemical influences on cyanobacterial blooms using hyperspectral images in inland water, Korea. Water Res. 2017, 126, 319–328. [Google Scholar] [CrossRef]
- Rousso, B.Z.; Bertone, E.; Stewart, R.; Hamilton, D.P. A systematic literature review of forecasting and predictive models for cyanobacteria blooms in freshwater lakes. Water Res. 2020, 182, 115959. [Google Scholar] [CrossRef]
- Kim, J.; Lee, T.; Seo, D. Algal bloom prediction of the lower Han River, Korea using the EFDC hydrodynamic and water quality model. Ecol. Model. 2017, 366, 27–36. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, T.T.; Shi, Y.Z.; Wendroth, O.; Liu, B.Y. Comparing the Performance of An Autoregressive State-Space Approach to the Linear Regression and Artificial Neural Network for Streamflow Estimation. J. Environ. Inf. 2021, 37, 36–48. [Google Scholar] [CrossRef]
- Zeng, Q.H.; Liu, Y.; Zhao, H.T.; Sun, M.D.; Li, X.Y. Comparison of models for predicting the changes in phytoplankton community composition in the receiving water system of an inter basin water transfer project. Environ. Pollut. 2017, 223, 676–684. [Google Scholar] [CrossRef]
- Yajima, H.; Derot, J. Application of the Random Forest model for chlorophyll-a forecasts in fresh and brackish water bodies in Japan, using multivariate long-term databases. J. Hydroinform. 2018, 20, 206–220. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.S.; Park, S.; An, K.G.; Kwak, K.C. Algal Bloom Prediction Using Extreme Learning Machine Models at Artificial Weirs in the Nakdong River, Korea. Int. J. Environ. Res. Public Health 2018, 15, 2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellios, N.; Moe, S.J.; Laspidou, C. Machine Learning Approaches for Predicting Health Risk of Cyanobacterial Blooms in Northern European Lakes. Water 2020, 12, 1191. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Lee, H.K.; Shin, J.K.; Chon, K.; Kim, S.; Cho, K.H.; Kim, J.H.; Baek, S.S. A machine learning approach for early warning of cyanobacterial bloom outbreaks in a freshwater reservoir. J. Environ. Manag. 2021, 288, 112415. [Google Scholar] [CrossRef] [PubMed]
- Gnana, D.A.A.; Balamurugan, S.A.A.; Leavline, E.J. Literature review on feature selection methods for high-dimensional data. Int. J. Comput. Appl. 2016, 136, 9–17. [Google Scholar]
- Jiang, S.J.; Zheng, Y.; Solomatine, D. Improving AI System Awareness of Geoscience Knowledge: Symbiotic Integration of Physical Approaches and Deep Learning. Geophys. Res. Lett. 2020, 47, e2020GL088229. [Google Scholar] [CrossRef]
- Moreido, V.; Gartsman, B.; Solomatine, D.P.; Suchilina, Z. How Well Can Machine Learning Models Perform without Hydrologists? Application of Rational Feature Selection to Improve Hydrological Forecasting. Water 2021, 13, 1696. [Google Scholar] [CrossRef]
- Al-Abadi, A.M.; Handhal, A.M.; Al-Ginamy, M.A. Evaluating the Dibdibba Aquifer Productivity at the Karbala-Najaf Plateau (Central Iraq) Using GIS-Based Tree Machine Learning Algorithms. Nat. Resour. Res. 2020, 29, 1989–2009. [Google Scholar] [CrossRef]
- Yoo, C.; Cho, E. Effect of Multicollinearity on the Bivariate Frequency Analysis of Annual Maximum Rainfall Events. Water 2019, 11, 905. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Yoon, S.; Cha, Y. Prediction of cyanobacteria blooms in the lower Han River (South Korea) using ensemble learning algorithms. Desalin. Water Treat. 2017, 84, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Raschka, S.; Mirjalili, V. Python Machine Learning: Machine Learning and Deep Learning with Python, Scikit-Learn, and TensorFlow 2, Third Edition. Int. J. Knowl.-Based Organ. 2017, 10, 3175783. [Google Scholar]
- Choi, J.-H.; Kim, J.; Won, J.; Min, O. Modelling chlorophyll-a concentration using deep neural networks considering extreme data imbalance and skewness. In Proceedings of the 2019 21st International Conference on Advanced Communication Technology (ICACT), PyeongChang, Korea, 17–20 February 2019; pp. 631–634. [Google Scholar]
- Kim, S.; Chung, S.; Park, H.; Cho, Y.; Lee, H. Analysis of Environmental Factors Associated with Cyanobacterial Dominance after River Weir Installation. Water 2019, 11, 1163. [Google Scholar] [CrossRef] [Green Version]
- Vien, B.S.; Wong, L.; Kuen, T.; Rose, L.F.; Chiu, W.K. A Machine Learning Approach for Anaerobic Reactor Performance Prediction Using Long Short-Term Memory Recurrent Neural Network. Struct. Health Monit. 8apwshm 2021, 18, 61. [Google Scholar]
- Gradilla-Hernandez, M.S.; de Anda, J.; Garcia-Gonzalez, A.; Meza-Rodriguez, D.; Montes, C.Y.; Perfecto-Avalos, Y. Multivariate water quality analysis of Lake Cajititlan, Mexico. Environ. Monit. Assess. 2020, 192, 5. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.P.; Khaled, U.; Al-Rashed, A.A.A.A.; Meer, R.; Goodarzi, M.; Sarafraz, M.M. Potential application of Response Surface Methodology (RSM) for the prediction and optimization of thermal conductivity of aqueous CuO (II) nanofluid: A statistical approach and experimental validation. Physica A 2020, 554, 124353. [Google Scholar] [CrossRef]
- Wu, S.S.; Hu, X.L.; Zheng, W.B.; He, C.C.; Zhang, G.C.; Zhang, H.; Wang, X. Effects of reservoir water level fluctuations and rainfall on a landslide by two-way ANOVA and K-means clustering. B Eng. Geol. Environ. 2021, 80, 5405–5421. [Google Scholar] [CrossRef]
- Xu, X.D.; Lin, H.; Liu, Z.H.; Ye, Z.L.; Li, X.Y.; Long, J.P. A Combined Strategy of Improved Variable Selection and Ensemble Algorithm to Map the Growing Stem Volume of Planted Coniferous Forest. Remote Sens. 2021, 13, 4631. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, X.D.; Yu, L.F.; Ren, L.L.; Luo, Y.Q. Combining WV-2 images and tree physiological factors to detect damage stages of Populus gansuensis by Asian longhorned beetle (Anoplophora glabripennis) at the tree level. Ecosyst 2021, 8, 35. [Google Scholar] [CrossRef]
- Nagawa, K.; Suzuki, M.; Yamamoto, Y.; Inoue, K.; Kozawa, E.; Mimura, T.; Nakamura, K.; Nagata, M.; Niitsu, M. Texture analysis of muscle MRI: Machine learning-based classifications in idiopathic inflammatory myopathies. Sci. Rep. 2021, 11, 9821. [Google Scholar] [CrossRef]
- Tousi, E.G.; Duan, J.N.G.; Gundy, P.M.; Bright, K.R.; Gerba, C.P. Evaluation of E. coli in sediment for assessing irrigation water quality using machine learning. Sci. Total Environ. 2021, 799, 149286. [Google Scholar] [CrossRef]
- Kim, Y.; Oh, S. Machine-learning insights into nitrate-reducing communities in a full-scale municipal wastewater treatment plant. J. Environ. Manag. 2021, 300, 113795. [Google Scholar] [CrossRef]
- Uma, K.V.; Balamurugan, S.A.A. C5.0 Decision Tree Model Using Tsallis Entropy and Association Function for General and Medical Dataset. Intell. Autom. Soft Comput. 2020, 26, 61–70. [Google Scholar] [CrossRef]
- Bourel, M.; Segura, A.M. Multiclass classification methods in ecology. Ecol. Indic. 2018, 85, 1012–1021. [Google Scholar] [CrossRef]
- Ahmed, M.; Mumtaz, R.; Mohammad, S. Analysis of water quality indices and machine learning techniques for rating water pollution: A case study of Rawal Dam, Pakistan. Water Supply 2021, 21, 3225–3250. [Google Scholar] [CrossRef]
- Fernandez, A.; Garcia, S.; Herrera, F.; Chawla, N.V. SMOTE for Learning from Imbalanced Data: Progress and Challenges, Marking the 15-year Anniversary. J. Artif. Intell. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- Arabgol, R.; Sartaj, M.; Asghari, K. Predicting Nitrate Concentration and Its Spatial Distribution in Groundwater Resources Using Support Vector Machines (SVMs) Model. Environ. Model. Assess. 2016, 21, 71–82. [Google Scholar] [CrossRef]
- Mulyani, E.; Hidayah, I.; Fauziati, S. Dropout Prediction Optimization through SMOTE and Ensemble Learning. In Proceedings of the 2019 International Seminar on Research of Information Technology and Intelligent Systems (ISRITI), Yogyakarta, Indonesia, 5–6 December 2019; pp. 516–521. [Google Scholar]
- Patil, V.B.; Pinto, S.M.; Govindaraju, T.; Hebbalu, V.S.; Bhat, V.; Kannanur, L.N. Multivariate statistics and water quality index (WQI) approach for geochemical assessment of groundwater quality—A case study of Kanavi Halla Sub-Basin, Belagavi, India. Environ. Geochem. Health 2020, 42, 2667–2684. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Yao, X.Y.; Wu, Q.; Huang, Y.B.; Zhou, Z.X.; Yang, J.; Liu, X.W. Turbidity prediction of lake-type raw water using random forest model based on meteorological data: A case study of Tai lake, China. J. Environ. Manag. 2021, 290, 112657. [Google Scholar] [CrossRef]
- Chou, J.S.; Pham, T.T.P.; Ho, C.C. Metaheuristic Optimized Multi-Level Classification Learning System for Engineering Management. Appl. Sci. 2021, 11, 5533. [Google Scholar] [CrossRef]
- Zhao, W.X.; Li, Y.Y.; Jiao, Y.J.; Zhou, B.; Vogt, R.D.; Liu, H.L.; Ji, M.; Ma, Z.; Li, A.D.; Zhou, B.H.; et al. Spatial and Temporal Variations in Environmental Variables in Relation to Phytoplankton Community Structure in a Eutrophic River-Type Reservoir. Water 2017, 9, 754. [Google Scholar] [CrossRef] [Green Version]




| Stage | Cyanobacterial Cell Density (cells mL−1) |
|---|---|
| Caution | ≥1000 |
| Warning | ≥10,000 |
| Outbreak | ≥1,000,000 |
| Category | Feature | Description | Unit | Frequency | Source |
|---|---|---|---|---|---|
| Water quality data | Cyano | Cyanobacterial cell density | cells mL−1 | Weekly | NIER |
| WT | Water temperature | °C | |||
| pH | Hydrogen ion concentration | - | |||
| DO | Dissolved oxygen | mg L−1 | |||
| Chl-a | Chlorophyll a | mg m−3 | |||
| BOD | Biochemical oxygen demand | mg L−1 | |||
| COD | Chemical oxygen demand | mg L−1 | |||
| SS | Suspended solids | mg L−1 | |||
| TN | Total nitrogen | mg L−1 | |||
| TP | Total phosphorus | mg L−1 | |||
| N/P | TN/TP ratio | - | |||
| TOC | Total organic carbon | mg L−1 | |||
| EC | Electrical conductivity | μS cm−1 | |||
| TotalColiform | Total coliforms | 100 mL−1 | |||
| TDN | Total dissolved nitrogen | mg L−1 | |||
| NH3-N | Ammonium nitrogen | mg L−1 | |||
| NO3-N | Nitrate nitrogen | mg L−1 | |||
| TDP | Total dissolved phosphorus | mg L−1 | |||
| PO4-P | Phosphate phosphorus | mg L−1 | |||
| FecalColiform | Fecal coliforms | - | |||
| Meteorological data | AT | Average air temperature | °C | Daily | KMA |
| LT | Lowest air temperature | °C | |||
| HT | Highest air temperature | °C | |||
| MaxSolarRad | Maximum amount of solar radiation for one hour | MJ m−2 | |||
| DaySolarRad | Total amount of solar radiation | MJ m−2 | |||
| Hydrological data | WeirLevel | Water level of weir | EL.m | Daily | K-water |
| StorageVolume | Storage volume of weir | million m3 | |||
| Rainfall | Rainfall in weir catchment area | mm | |||
| Inflow | Weir inflow | m3 s−1 | |||
| Outflow | Weir outflow | m3 s−1 |
| Category | Feature | Mean | Minimum | Median | Maximum |
|---|---|---|---|---|---|
| Input features | Cyano(t) | 2976 | 0 | 165 | 112,735 |
| WT | 16.8 | 0.7 | 17.5 | 33.6 | |
| pH | 8.1 | 6.5 | 8.1 | 9.6 | |
| DO | 10.4 | 1.6 | 10.1 | 16.4 | |
| Chl-a | 20.1 | 2.3 | 15.45 | 87.2 | |
| BOD | 1.9 | 0.4 | 1.8 | 5.0 | |
| COD | 5.9 | 3.5 | 5.8 | 10.5 | |
| SS | 7.6 | 1.5 | 6.3 | 44.9 | |
| TN | 2.674 | 1.089 | 2.686 | 4.396 | |
| TP | 0.043 | 0.011 | 0.034 | 0.198 | |
| N/P | 81.5 | 12.7 | 76.6 | 255.5 | |
| TOC | 4.1 | 2.6 | 4.0 | 7.9 | |
| EC | 288 | 124 | 286 | 596 | |
| TotalColiform | 8219 | 2 | 264 | 340,000 | |
| TDN | 2.513 | 1.078 | 2.532 | 4.125 | |
| NH3-N | 0.113 | 0.003 | 0.091 | 0.790 | |
| NO3-N | 1.971 | 0.530 | 1.996 | 3.330 | |
| TDP | 0.024 | 0.003 | 0.018 | 0.125 | |
| PO4-P | 0.011 | 0.000 | 0.004 | 0.105 | |
| FecalColiform | 428 | 0 | 12 | 21,750 | |
| AT | 15.5 | −4.8 | 15.8 | 32.5 | |
| LT | 10.8 | −8.9 | 10.8 | 27.6 | |
| HT | 21.0 | −1.0 | 21.9 | 38.0 | |
| MaxSolarRad | 2.39 | 0.19 | 2.54 | 3.74 | |
| DaySolarRad | 15.56 | 0.69 | 15.465 | 31.02 | |
| WeirLevel | 25.52 | 25.02 | 25.56 | 25.86 | |
| StorageVolume | 75.321 | 68.181 | 75.930 | 79.005 | |
| Rainfall | 2.290 | 0.000 | 0.023 | 57.263 | |
| Inflow | 112.867 | 3.604 | 67.733 | 1147.669 | |
| Outflow | 113.379 | 8.098 | 69.004 | 1140.136 | |
| Target variable (Output feature) | Cyano(t+1) | 2903 | 0 | 165 | 112,735 |
| Group1 | Group2 | Group3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Class | Cyano(t+1) | Number | Class | Cyano(t+1) | Number | Class | Cyano(t+1) | Number |
| Normal | <1000 | 269 | Normal | <1000 | 269 | None | 0 | 136 |
| Caution | ≥1000 | 83 | Occurrence | ≥1000 | 109 | Normal | <1000 | 133 |
| Warning | ≥10,000 | 26 | - | Occurrence | ≥1000 | 109 | ||
| Outbreak | ≥1,000,000 | 0 | - | |||||
| Category | Feature | Mean | Minimum | Median | Maximum |
|---|---|---|---|---|---|
| Input features | logCyano | 0.000 | −1.200 | 0.224 | 2.046 |
| WT | −1.974 | 0.085 | 2.071 | ||
| pH | −3.340 | 0.032 | 3.193 | ||
| DO | −3.115 | −0.077 | 2.144 | ||
| Chl-a | −1.194 | −0.310 | 4.514 | ||
| BOD | −2.066 | −0.148 | 4.237 | ||
| COD | −2.316 | −0.100 | 4.430 | ||
| SS | −1.240 | −0.280 | 7.533 | ||
| TN | −2.451 | 0.018 | 2.661 | ||
| TP | −1.219 | −0.329 | 6.019 | ||
| N/P | −1.602 | −0.114 | 4.048 | ||
| TOC | −1.784 | −0.133 | 4.468 | ||
| EC | −2.312 | −0.041 | 4.324 | ||
| TotalColiform | −0.308 | −0.299 | 12.455 | ||
| TDN | −2.357 | 0.031 | 2.646 | ||
| NH3-N | −1.213 | −0.243 | 7.458 | ||
| NO3-N | −2.425 | 0.042 | 2.288 | ||
| TDP | −1.045 | −0.307 | 4.957 | ||
| PO4-P | −0.570 | −0.355 | 5.068 | ||
| FecalColiform | −0.252 | −0.245 | 12.520 | ||
| AT | −2.286 | 0.024 | 1.907 | ||
| LT | −2.182 | 0.000 | 1.861 | ||
| HT | −2.372 | 0.088 | 1.826 | ||
| MaxSolarRad | −2.567 | 0.171 | 1.569 | ||
| DaySolarRad | −2.103 | −0.014 | 2.186 | ||
| WeirLevel | −3.042 | 0.219 | 2.030 | ||
| StorageVolume | −3.250 | 0.277 | 1.677 | ||
| Rainfall | −0.349 | −0.346 | 8.381 | ||
| Inflow | −0.751 | −0.310 | 7.111 | ||
| Outflow | −0.723 | −0.305 | 7.054 | ||
| Target variable | Each class of three groups (Group1, Group2, and Group3) based on Cyano(t+1) | ||||
| Algorithm | Parameter | Description |
|---|---|---|
| k-NN | n_neighbors | Number of neighbors |
| DT | max_depth | Maximum depth of the tree |
| LR | C | Regularization parameter |
| SVM | C | Regularization parameter |
| kernel | The kernel type to be used in the algorithm such as ‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, etc. |
| Feature | Group1 | Group2 | Group3 | |||
|---|---|---|---|---|---|---|
| F Value | p-Value | F Value | p-Value | F Value | p-Value | |
| logCyano | 132.367 | <0.001 | 256.089 | <0.001 | 270.917 | <0.001 |
| WT | 71.613 | <0.001 | 143.214 | <0.001 | 142.227 | <0.001 |
| pH | 0.545 | 0.580 | 0.313 | 0.576 | 6.180 | 0.002 |
| DO | 74.182 | <0.001 | 145.698 | <0.001 | 131.458 | <0.001 |
| Chl-a | 7.137 | 0.001 | 14.118 | <0.001 | 7.637 | 0.001 |
| BOD | 1.463 | 0.233 | 2.917 | 0.088 | 5.022 | 0.007 |
| COD | 2.599 | 0.076 | 5.186 | 0.023 | 18.898 | <0.001 |
| SS | 5.244 | 0.006 | 3.924 | 0.048 | 4.928 | 0.008 |
| TN | 63.964 | <0.001 | 123.352 | <0.001 | 108.115 | <0.001 |
| TP | 4.222 | 0.015 | 0.951 | 0.330 | 6.432 | 0.002 |
| N/P | 19.436 | <0.001 | 38.336 | <0.001 | 40.293 | <0.001 |
| TOC | 1.499 | 0.225 | 1.456 | 0.228 | 18.843 | <0.001 |
| EC | 6.176 | 0.002 | 0.170 | 0.680 | 8.701 | <0.001 |
| TotalColiform | 4.703 | 0.010 | 6.984 | 0.009 | 7.137 | 0.001 |
| TDN | 66.039 | <0.001 | 128.394 | <0.001 | 103.655 | <0.001 |
| NH3-N | 5.961 | 0.003 | 11.281 | 0.001 | 6.176 | 0.002 |
| NO3-N | 85.820 | <0.001 | 163.285 | <0.001 | 126.452 | <0.001 |
| TDP | 3.874 | 0.022 | 2.428 | 0.120 | 12.020 | <0.001 |
| PO4-P | 2.922 | 0.055 | 0.594 | 0.441 | 8.241 | <0.001 |
| FecalColiform | 1.754 | 0.175 | 3.176 | 0.076 | 5.414 | 0.005 |
| AT | 63.407 | <0.001 | 126.277 | <0.001 | 98.519 | <0.001 |
| LT | 66.861 | <0.001 | 133.669 | <0.001 | 103.961 | <0.001 |
| HT | 53.737 | <0.001 | 106.578 | <0.001 | 83.166 | <0.001 |
| MaxSolarRad | 5.712 | 0.004 | 9.368 | 0.002 | 6.774 | 0.001 |
| DaySolarRad | 4.996 | 0.007 | 7.154 | 0.008 | 5.754 | 0.003 |
| WeirLevel | 3.047 | 0.049 | 4.768 | 0.030 | 9.661 | <0.001 |
| StorageVolume | 2.737 | 0.066 | 4.370 | 0.037 | 9.695 | <0.001 |
| Rainfall | 2.256 | 0.106 | 0.370 | 0.543 | 0.327 | 0.721 |
| Inflow | 3.843 | 0.022 | 0.244 | 0.622 | 6.501 | 0.002 |
| Outflow | 3.649 | 0.027 | 0.148 | 0.701 | 6.543 | 0.002 |
| Modeling Case | Input Features |
|---|---|
| Case1 | logCyano, HT |
| Case2 | TN, AT |
| Case3 | TN, LT |
| Case4 | TN, HT |
| Case5 | TDN, HT |
| Case6 | NO3-N, AT |
| Case7 | NO3-N, LT |
| Case8 | NO3-N, HT |
| Algorithm (Parameter) | Group | Case1 | Case2 | Case3 | Case4 | Case5 | Case6 | Case7 | Case8 |
|---|---|---|---|---|---|---|---|---|---|
| k-NN (n_neighbors) | Group1 | 3 | 11 | 12 | 6 | 6 | 7 | 3 | 5 |
| Group2 | 3 | 16 | 13 | 13 | 9 | 5 | 5 | 3 | |
| Group3 | 19 | 16 | 19 | 17 | 16 | 10 | 14 | 14 | |
| DT (max_depth) | Group1 | 15 | 11 | 10 | 9 | 15 | 9 | 12 | 14 |
| Group2 | 1 | 3 | 4 | 14 | 14 | 10 | 8 | 6 | |
| Group3 | 3 | 4 | 3 | 3 | 3 | 4 | 3 | 2 | |
| LR (C) | Group1 | 0.01 | 1 | 0.001 | 0.1 | 1 | 0.01 | 0.01 | 0.001 |
| Group2 | 1 | 1 | 100 | 1 | 0.1 | 10 | 1 | 1 | |
| Group3 | 1 | 10 | 100 | 100 | 0.1 | 10 | 1 | 1 | |
| SVM (C/kernel) | Group1 | 1000/rbf | 1000/rbf | 1000/rbf | 1000/rbf | 1000/rbf | 1000/rbf | 1000/rbf | 1000/rbf |
| Group2 | 10/linear | 1/rbf | 1/rbf | 0.1/rbf | 1/rbf | 1/rbf | 100/rbf | 100/rbf | |
| Group3 | 10/rbf | 100/rbf | 10/linear | 1/linear | 0.1/rbf | 0.1/rbf | 1/linear | 0.1/rbf |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Jonoski, A.; Solomatine, D.P. A Classification-Based Machine Learning Approach to the Prediction of Cyanobacterial Blooms in Chilgok Weir, South Korea. Water 2022, 14, 542. https://doi.org/10.3390/w14040542
Kim J, Jonoski A, Solomatine DP. A Classification-Based Machine Learning Approach to the Prediction of Cyanobacterial Blooms in Chilgok Weir, South Korea. Water. 2022; 14(4):542. https://doi.org/10.3390/w14040542
Chicago/Turabian StyleKim, Jongchan, Andreja Jonoski, and Dimitri P. Solomatine. 2022. "A Classification-Based Machine Learning Approach to the Prediction of Cyanobacterial Blooms in Chilgok Weir, South Korea" Water 14, no. 4: 542. https://doi.org/10.3390/w14040542
APA StyleKim, J., Jonoski, A., & Solomatine, D. P. (2022). A Classification-Based Machine Learning Approach to the Prediction of Cyanobacterial Blooms in Chilgok Weir, South Korea. Water, 14(4), 542. https://doi.org/10.3390/w14040542