Response of Algal–Bacterial Regrowth Characteristics to the Hypochlorite in Landscape Ponds Replenished with Reclaimed Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Basic Information of the Water for Experiments
2.2. Microcosmic Experiment
2.3. Chemical Analysis
2.4. Monitoring of the Algal Growth
2.5. Analysis of Microbial Community
2.5.1. Sample Preparation
2.5.2. 16S rDNA Sequencing and Data Processing
2.5.3. Downstream Analysis
3. Results
3.1. Raw Water for the Experiment
3.2. Microbial Community
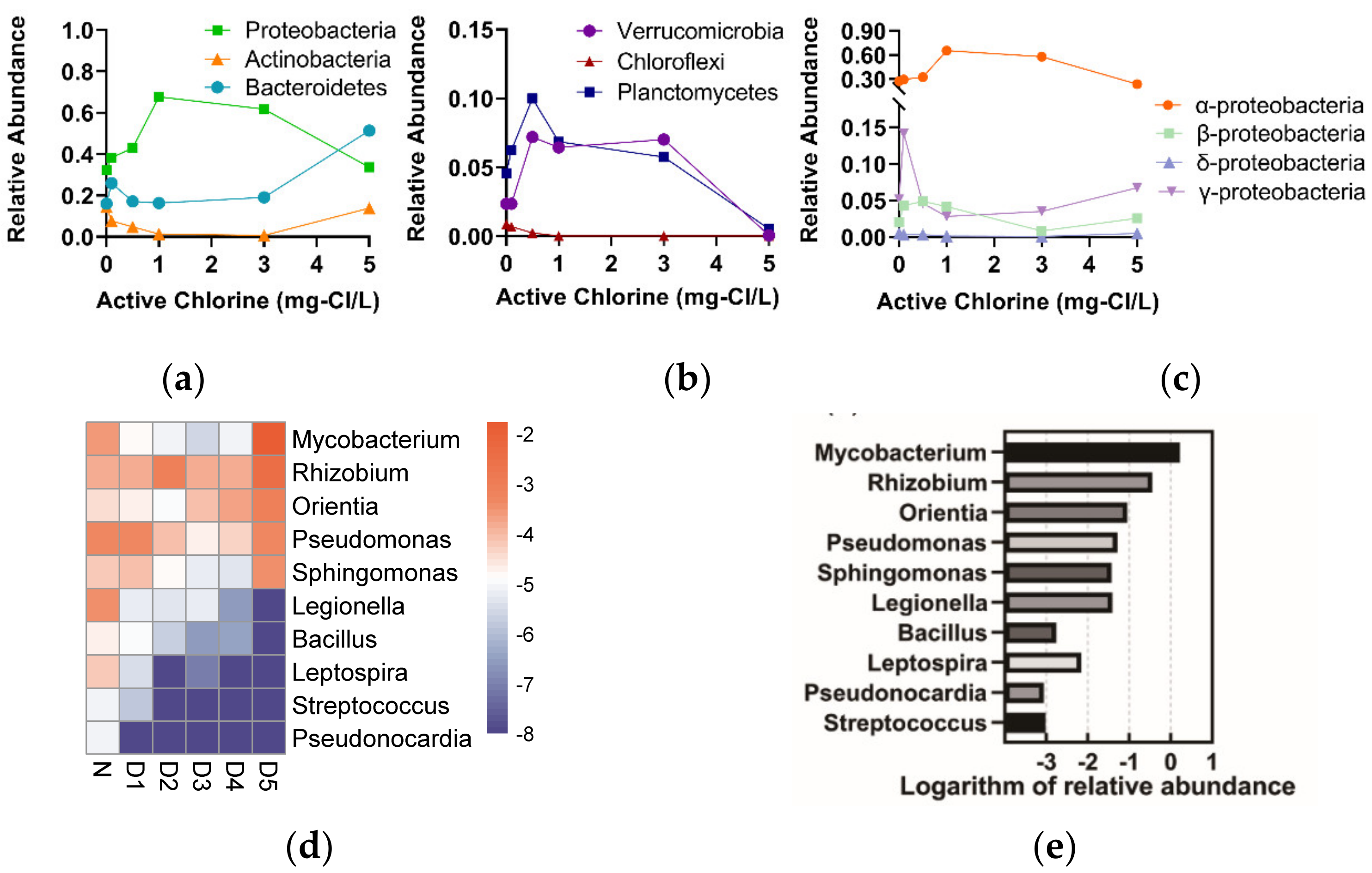
3.2.1. The Characteristics of Bacterial Composition and Abundance
3.2.2. The Responses of Microbial Community to the Chlorine Concentrations
3.3. Microbial Community
4. Discussion
4.1. The Algal and Bacterial Regrowth Potential with Chlorinated RW
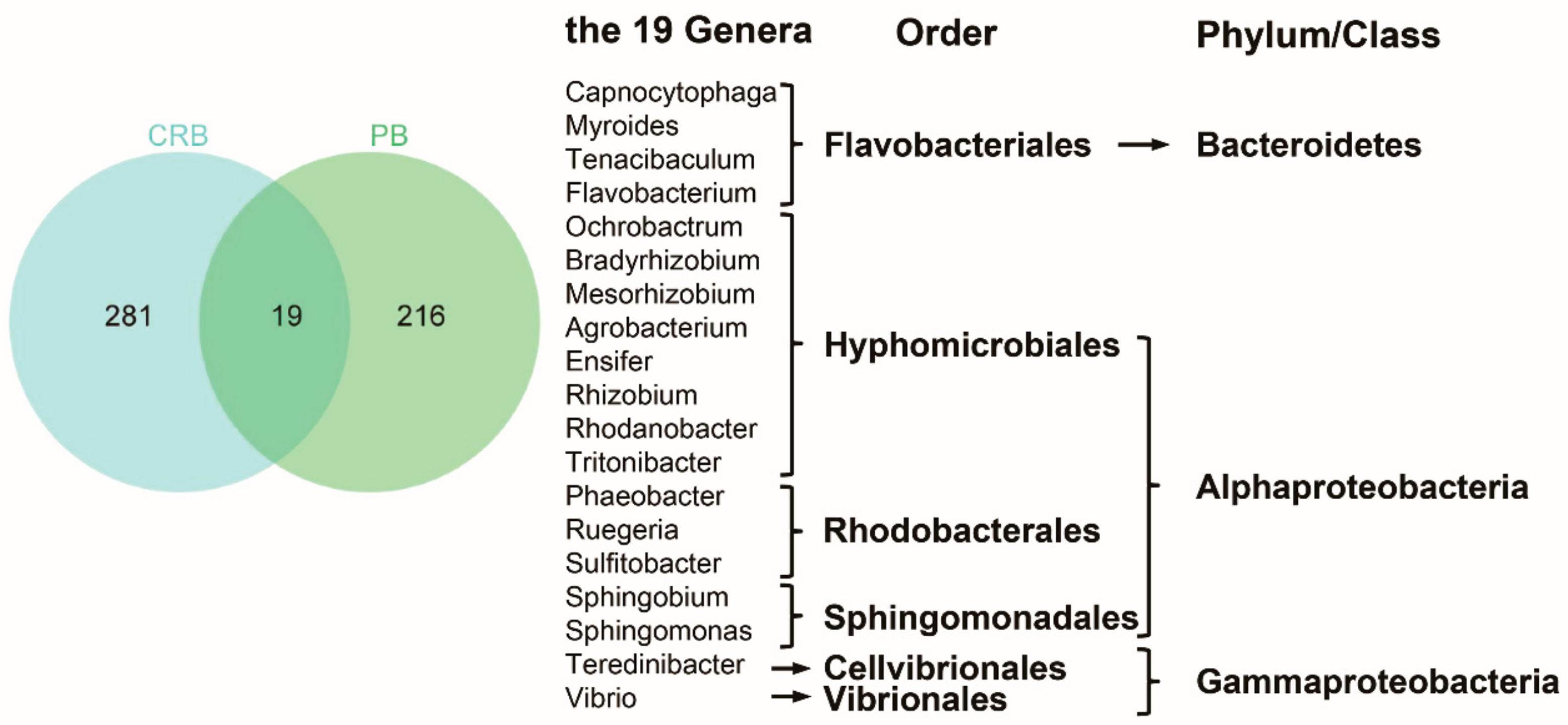
4.2. The Characteristics of Bacteria Regrew within Chlorinated RW
4.3. Ecological Risks Associated with Pathogenic Bacteria in Chlorinated RW
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Wang, Y.; Xiong, H.; Tan, Z.; Lv, Z.; Zheng, K.; Zou, L.; Luo, G.; Ye, L.; Zhang, Z.; et al. Investigation and Optimization Strategies on the Operation of Disinfection Facilities in Municipal WWTPs. China Water Wastewater 2020, 36, 7–19. (In Chinese) [Google Scholar]
- Zhang, T.; Wei, H.; Yao, J.; Chen, G.; Xu, B. Comparison of Disinfection Efficiency of Different UV/Chlorination Combined Processes in Wastewater Treatment Plant. China Water Wastewater 2021, 37, 19–24. (In Chinese) [Google Scholar]
- Li, Z.; Song, G.; Bi, Y.; Gao, W.; He, A.; Lu, Y.; Wang, Y.; Jiang, G. Occurrence and Distribution of Disinfection Byproducts in Domestic Wastewater Effluent, Tap Water, and Surface Water during the SARS-CoV-2 Pandemic in China. Environ. Sci. Technol. 2021, 55, 4103–4114. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.; Chen, S.; Meng, F.; Zhang, D.; Liu, Y.; Li, M.; Liu, X.; Huang, X.; Qu, J.H. Spatial variation of dissolved organic nitrogen in Wuhan surface waters: Correlation with the occurrence of disinfection byproducts during the COVID-19 pandemic. Water Res. 2021, 198, 117138. [Google Scholar] [CrossRef]
- Huang, G.; Ng, T.W.; Chen, H.; Chow, A.T.; Liu, S.; Wong, P.K. Formation of assimilable organic carbon (AOC) during drinking water disinfection: A microbiological prospect of disinfection byproducts. Environ. Int. 2020, 135, 105389. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, T.; Ngo, H.H.; Lu, Y.; Li, G.; Wu, Q.; Li, K.; Bai, Y.; Liu, S.; Hu, H.Y. Assimilable organic carbon (AOC) variation in reclaimed water: Insight on biological stability evaluation and control for sustainable water reuse. Bioresour. Technol. 2018, 254, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Elhadidy, A.M.; Van Dyke, M.I.; Peldszus, S.; Huck, P.M. Application of flow cytometry to monitor assimilable organic carbon (AOC) and microbial community changes in water. J. Microbiol. Methods 2016, 130, 154–163. [Google Scholar] [CrossRef]
- Munoz-Marin, M.C.; Gomez-Baena, G.; Lopez-Lozano, A.; Moreno-Cabezuelo, J.A.; Diez, J.; Garcia-Fernandez, J.M. Mixotrophy in marine picocyanobacteria: Use of organic compounds by Prochlorococcus and Synechococcus. ISME J. 2020, 14, 1065–1073. [Google Scholar] [CrossRef]
- Subashchandrabose, S.R.; Ramakrishnan, B.; Megharaj, M.; Venkateswarlu, K.; Naidu, R. Mixotrophic cyanobacteria and microalgae as distinctive biological agents for organic pollutant degradation. Environ. Int. 2013, 51, 59–72. [Google Scholar] [CrossRef]
- Ramanan, R.; Kim, B.H.; Cho, D.H.; Oh, H.M.; Kim, H.S. Algae-bacteria interactions: Evolution, ecology and emerging applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar] [CrossRef] [Green Version]
- Kazamia, E.; Helliwell, K.E.; Purton, S.; Smith, A.G. How mutualisms arise in phytoplankton communities: Building eco-evolutionary principles for aquatic microbes. Ecol. Lett. 2016, 19, 810–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benton, T.G.; Solan, M.; Travis, J.M.; Sait, S.M. Microcosm experiments can inform global ecological problems. TRENDS Ecol. Evol. 2007, 22, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.W.; Wu, Y.H.; Yu, T.; Wang, Y.H.; Chen, G.Q.; Tong, X.; Bai, Y.; Xu, C.; Wang, H.B.; Ikuno, N.; et al. Evaluating method and potential risks of chlorine-resistant bacteria (CRB): A review. Water Res. 2021, 188, 116474. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association. 4500-Cl chlorine (residual). In Standard Methods For the Examination of Water and Wastewater; APHA Press: Washington, DC, USA, 2018. [Google Scholar]
- Fichot, C.G.; Benner, R. The spectral slope coefficient of chromophoric dissolved organic matter (S275–295) as a tracer of terrigenous dissolved organic carbon in river-influenced ocean margins. Limnol. Oceanogr. 2012, 57, 1453–1466. [Google Scholar] [CrossRef] [Green Version]
- Helms, J.R.; Stubbins, A.; Ritchie, J.D.; Minor, E.C.; Kieber, D.J.; Mopper, K. Absorption spectral slopes and slope ratios as indicators of molecular weight, source, and photobleaching of chromophoric dissolved organic matter. Limnol. Oceanogr. 2008, 53, 955–969. [Google Scholar] [CrossRef] [Green Version]
- Al-Juboori, R.A.; Yusaf, T.; Pittaway, P.A. Exploring the correlations between common UV measurements and chemical fractionation for natural waters. Desalination Water Treat. 2015, 57, 16324–16335. [Google Scholar] [CrossRef]
- Weishaar, J.L.; Aiken, G.R.; Bergamaschi, B.A.; Fram, M.S.; Fujii, R.; Mopper, K. Evaluation of Specific Ultraviolet Absorbance as an Indicator of the Chemical Composition and Reactivity of Dissolved Organic Carbon. Environ. Sci. Technol. 2003, 37, 4702–4708. [Google Scholar] [CrossRef]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of Two 16S rRNA Primers (V3-V4 and V4-V5) for Studies of Arctic Microbial Communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef]
- Ueda, H.; Otsuka, S.; Senoo, K. Bacterial Communities Constructed in Artificial Consortia of Bacteria and Chlorella vulgaris. Microbes Environ. 2010, 25, 36–40. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.X.; Li, X.; Zhao, J.R.; Zhang, Z.X.; Fan, X.Y. Response of microbial communities based on full-scale classification and antibiotic resistance genes to azithromycin and copper combined pollution in activated sludge nitrification laboratory mesocosms at low temperature. Bioresour. Technol. 2021, 341, 125859. [Google Scholar] [CrossRef]
- Wei, M.; Li, C.; Dai, Y.; Zhou, H.; Cui, Y.; Zeng, Y.; Huang, Q.; Wang, Q. High-Throughput Absolute Quantification Sequencing Revealed Osteoporosis- Related Gut Microbiota Alterations in Han Chinese Elderly. Front. Cell Infect. Microbiol. 2021, 11, 630372. [Google Scholar] [CrossRef]
- Nyholm, N. Response variable in algal growth inhibition tests—Biomass or growth rate? Water Research 1985, 19, 273–279. [Google Scholar] [CrossRef]
- Shi, Z.; Xu, H.; Wang, Z.; Du, H.; Fu, X. Effects of co-exposure to copper and humic acids on microalga Chlorella vulgaris: Growth inhibition, oxidative stress, and extracellular secretion. Environ. Pollut. Bioavailab. 2021, 33, 415–424. [Google Scholar] [CrossRef]
- Zheng, Y.; Chi, Z.; Lucker, B.; Chen, S. Two-stage heterotrophic and phototrophic culture strategy for algal biomass and lipid production. Bioresour. Technol. 2012, 103, 484–488. [Google Scholar] [CrossRef]
- Lee, Y.-K. Algal Nutrition—Heterotrophic Carbon Nutrition. In Handbook of Microalgal Culture: Biotechnology and Applied Phycology; Richmond, A., Ed.; Blackwell Publishing Ltd.: Oxford, UK, 2004. [Google Scholar]
- Sheppard, S.K. Strain wars and the evolution of opportunistic pathogens. Curr. Opin. Microbiol. 2022, 67, 102138. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Yang, J.; Jiang, H.; Deng, Y.; Lear, G. Distribution of potentially pathogenic bacteria in the groundwater of the Jianghan Plain, central China. Int. Biodeterior. Biodegrad. 2019, 143, 104711. [Google Scholar] [CrossRef]
- Fan, C.; Li, Y.; Liu, P.; Mu, F.; Xie, Z.; Lu, R.; Qi, Y.; Wang, B.; Jin, C. Characteristics of airborne opportunistic pathogenic bacteria during autumn and winter in Xi’an, China. Sci. Total Environ. 2019, 672, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Landi, P.; Minoarivelo, H.O.; Brännström, Å.; Hui, C.; Dieckmann, U. Complexity and stability of ecological networks: A review of the theory. Popul. Ecol. 2018, 60, 319–345. [Google Scholar] [CrossRef]
- Zabel, R.A.; Morrell, J.J. Wood Microbiology (Second Edition)-Decay and Its Prevention; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Ramanan, R.; Kang, Z.; Kim, B.H.; Cho, D.H.; Jin, L.; Oh, H.M.; Kim, H.S. Phycosphere bacterial diversity in green algae reveals an apparent similarity across habitats. Algal Res. Biomass Biofuels Bioprod. 2015, 8, 140–144. [Google Scholar] [CrossRef]
- Gomez-Pereira, P.R.; Fuchs, B.M.; Alonso, C.; Oliver, M.J.; Van Beusekom, J.E.E.; Amann, R. Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. Isme J. 2010, 4, 472–487. [Google Scholar] [CrossRef]
- Kim, B.H.; Ramanan, R.; Cho, D.H.; Oh, H.M.; Kim, H.S. Role of Rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass Bioenergy 2014, 69, 95–105. [Google Scholar] [CrossRef]
- Cho, D.H.; Ramanan, R.; Heo, J.; Lee, J.; Kim, B.H.; Oh, H.M.; Kim, H.S. Enhancing microalgal biomass productivity by engineering a microalgal-bacterial community. Bioresour. Technol. 2015, 175, 578–585. [Google Scholar] [CrossRef]
- Shi, M.; Li, J.; Zhou, Q.; Wang, G.; Zhang, W.; Zhang, Z.; Gao, Y.; Yan, S. Interactions between elevated CO2 levels and floating aquatic plants on the alteration of bacterial function in carbon assimilation and decomposition in eutrophic waters. Water Res. 2020, 171, 115398. [Google Scholar] [CrossRef]
- Munoz-Gomez, S.A.; Kreutz, M.; Hess, S. A microbial eukaryote with a unique combination of purple bacteria and green algae as endosymbionts. Sci. Adv. 2021, 7, eabg4102. [Google Scholar] [CrossRef]
- Garoma, T.; Yazdi, R.E. Investigation of the disruption of algal biomass with chlorine. BMC Plant Biol. 2019, 19, 18. [Google Scholar] [CrossRef]
- Liu, C.; Shin, Y.H.; Wei, X.; Ersan, M.S.; Wagner, E.; Plewa, M.J.; Amy, G.; Karanfil, T. Preferential Halogenation of Algal Organic Matter by Iodine over Chlorine and Bromine: Formation of Disinfection Byproducts and Correlation with Toxicity of Disinfected Waters. Environ. Sci. Technol. 2022, 56, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Sathasivam, R.; Ebenezer, V.; Guo, R.Y.; Ki, J.S. Physiological and biochemical responses of the freshwater green algae Closterium ehrenbergii to the common disinfectant chlorine. Ecotoxicol. Environ. Saf. 2016, 133, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Gad, A.a.M.; El-Tawel, S. Effect of pre-oxidation by chlorine/permanganate on surface water characteristics and algal toxins. Desalin. Water Treat. 2016, 57, 17922–17934. [Google Scholar] [CrossRef]
- Chen, R.; Ao, D.; Ji, J.Y.; Wang, X.C.C.; Li, Y.Y.; Huang, Y.; Xue, T.; Guo, H.B.; Wang, N.; Zhang, L. Insight into the risk of replenishing urban landscape ponds with reclaimed wastewater. J. Hazard. Mater. 2017, 324, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, L.; Xu, K.; Huang, H.; Ren, H. Algae Biofilm Reduces Microbe-Derived Dissolved Organic Nitrogen Discharges: Performance and Mechanisms. Environ. Sci. Technol. 2021, 55, 6227–6238. [Google Scholar] [CrossRef] [PubMed]
- Bankston, E.; Wang, Q.; Higgins, B.T. Algae support populations of heterotrophic, nitrifying, and phosphate-accumulating bacteria in the treatment of poultry litter anaerobic digestate. Chem. Eng. J. 2020, 398, 122550. [Google Scholar] [CrossRef]
- Acharya, S.M.; Kurisu, F.; Kasuga, I.; Furumai, H. Chlorine Dose Determines Bacterial Community Structure of Subsequent Regrowth in Reclaimed Water. J. Water Environ. Technol. 2016, 14, 15–24. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Y.; Luo, L.; Yu, T.; Xu, A.; Xue, S.; Chen, G.; Ni, X.-Y.; Peng, L.; Chen, Z.; et al. Risks, characteristics, and control strategies of disinfection-residual-bacteria (DRB) from the perspective of microbial community structure. Water Res. 2021, 204, 117606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lyu, Y.H.; Richlen, M.L.; Anderson, D.M.; Cai, Z.H. Quorum Sensing Is a Language of Chemical Signals and Plays an Ecological Role in Algal-Bacterial Interactions. Crit. Rev. Plant Sci. 2016, 35, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.; Russo, J.; Fiegel, J.; Brogden, N. Antibiotic Delivery Strategies to Treat Skin Infections When Innate Antimicrobial Defense Fails. Antibiotics 2020, 9, 56. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.P.; Cornforth, D.M.; Mideo, N. Evolution of virulence in opportunistic pathogens: Generalism, plasticity, and control. Trends Microbiol. 2012, 20, 336–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anttila, J.; Kaitala, V.; Laakso, J.; Ruokolainen, L. Environmental Variation Generates Environmental Opportunist Pathogen Outbreaks. PLoS ONE 2015, 10, e0145511. [Google Scholar] [CrossRef]
- Fang, T.; Cui, Q.; Huang, Y.; Dong, P.; Wang, H.; Liu, W.T.; Ye, Q. Distribution comparison and risk assessment of free-floating and particle-attached bacterial pathogens in urban recreational water: Implications for water quality management. Sci. Total Environ. 2018, 613–614, 428–438. [Google Scholar] [CrossRef]
- Gueretz, J.S.; Da Silva, F.A.; Simionatto, E.L.; Ferard, J.F.; Radetski, C.M.; Somensi, C.A. A multi-parametric study of the interaction between the Parati river and Babitonga Bay in terms of water quality. J. Env. Sci. Health B 2020, 55, 257–264. [Google Scholar] [CrossRef]
- Park, B.S.; Choi, W.J.; Guo, R.Y.; Kim, H.; Ki, J.S. Changes in Free-Living and Particle-Associated Bacterial Communities Depending on the Growth Phases of Marine Green Algae, Tetraselmis suecica. J. Mar. Sci. Eng. 2021, 9, 171. [Google Scholar] [CrossRef]
- Qi, F.; Jia, Y.T.; Mu, R.M.; Ma, G.X.; Guo, Q.Y.; Meng, Q.Y.; Yu, G.J.; Xie, J. Convergent community structure of algal-bacterial consortia and its effects on advanced wastewater treatment and biomass production. Sci. Rep. 2021, 11, 21118. [Google Scholar] [CrossRef] [PubMed]
- Hattenrath-Lehmann, T.K.; Gobler, C.J. Identification of unique microbiomes associated with harmful algal blooms caused by Alexandrium fundyense and Dinophysis acuminata. Harmful Algae 2017, 68, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Y.; Zhou, Y.; Zheng, W.; Tian, Y.; Zheng, T. Bacterial community dynamics during a bloom caused by Akashiwo sanguinea in the Xiamen sea area, China. Harmful Algae 2012, 20, 132–141. [Google Scholar] [CrossRef]
- Fulbright, S.P.; Robbins-Pianka, A.; Berg-Lyons, D.; Knight, R.; Reardon, K.F.; Chisholm, S.T. Bacterial community changes in an industrial algae production system. Algal Res. Biomass Biofuels Bioprod. 2018, 31, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Cantera, S.; Fischer, P.Q.; Sanchez-Andrea, I.; Marin, D.; Sousa, D.Z.; Munoz, R. Impact of the algal-bacterial community structure, physio-types and biological and environmental interactions on the performance of a high rate algal pond treating biogas and wastewater. Fuel 2021, 302, 121148. [Google Scholar] [CrossRef]
- Kruger, K.; Chafee, M.; Ben Francis, T.; Glavina Del Rio, T.; Becher, D.; Schweder, T.; Amann, R.I.; Teeling, H. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019, 13, 2800–2816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, T.B.; Kruger, K.; Fuchs, B.M.; Teeling, H.; Amann, R.I. Candidatus Prosiliicoccus vernus, a spring phytoplankton bloom associated member of the Flavobacteriaceae. Syst. Appl. Microbiol. 2019, 42, 41–53. [Google Scholar] [CrossRef]
- Balleza, D.; Quinto, C.; Elias, D.; Gomez-Lagunas, F. A high-conductance cation channel from the inner membrane of the free-living soil bacteria Rhizobium etli. Arch. Microbiol. 2010, 192, 595–602. [Google Scholar] [CrossRef]
- Shao, K.Q.; Zhang, L.; Wang, Y.P.; Yao, X.; Tang, X.M.; Qin, B.Q.; Gao, G. The responses of the taxa composition of particle-attached bacterial community to the decomposition of Microcystis blooms. Sci. Total Environ. 2014, 488, 236–242. [Google Scholar] [CrossRef]
- Chakraborty, D.; Sharma, G.D.; Deb, B. Diversity of Free Living Nitrogen Fixing Bacteria in Sugarcane Rhizosphere of Barak Valley, Assam. J. Pure Appl. Microbiol. 2012, 6, 1351–1355. [Google Scholar]
- Islam, M.R.; Madhaiyan, M.; Boruah, H.P.D.; Yim, W.; Lee, G.; Saravanan, V.S.; Fu, Q.L.; Hu, H.Q.; Sa, T. Characterization of Plant Growth-Promoting Traits of Free-Living Diazotrophic Bacteria and Their Inoculation Effects on Growth and Nitrogen Uptake of Crop Plants. J. Microbiol. Biotechnol. 2009, 19, 1213–1222. [Google Scholar] [CrossRef] [PubMed]



| Groups | Index | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TN a (mg/L) | −N a (mg/L) | −N (mg/L) | TP a (mg/L) | SRP b (mg/L) | CODcr (mg/L) | Chlorine (mg-Cl/L) | pH | TDS (mg/L) | ORP (mV) | |
| NC | 2.13 ± 0.04 | 2.04 ± 0.02 | <0.1 | <0.01 | <0.01 | N.D. | 0 | 7.5~7.8 | 1200~1400 | 195~215 |
| N1 | 5.91 ± 0.10 | 4.43 ± 0.05 | 0.61 ± 0.11 | 0.15 ± 0.01 | 0.05 ± 0.01 | 21.3 ± 1.0 | ||||
| N2 | 10.6 ± 0.13 | 9.33 ± 0.09 | ||||||||
| N3 | 14.3 ± 0.22 | 13.1 ± 0.11 | ||||||||
| N4 | 5.79 ± 0.08 | 4.52 ± 0.07 | 0.30 ± 0.02 | 0.18 ± 0.02 | ||||||
| N5 | 10.3 ± 0.15 | 9.55 ± 0.09 | ||||||||
| N6 | 14.2 ± 0.28 | 12.9 ± 0.15 | ||||||||
| D1 | 10.4 ± 0.20 | 9.43 ± 0.11 | 0.61 ± 0.11 | 0.30 ± 0.02 | 0.18 ± 0.02 | 19.2 ± 1.5 | 0.1 | |||
| D2 | 0.5 | |||||||||
| D3 | 1.0 | |||||||||
| D4 | 3.0 | |||||||||
| D5 | 5.0 | |||||||||
| Indicators | Quantification Methods | Environmental Meanings | Reference |
|---|---|---|---|
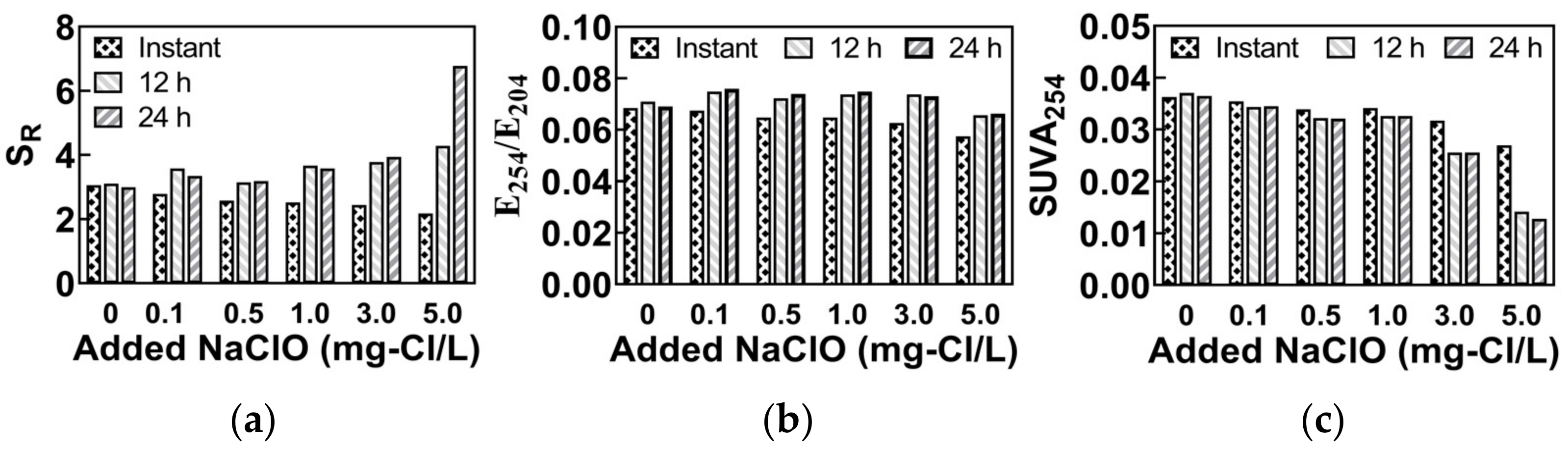
| SR | The ratio of spectral slopes in the band of 275~295 nm and 350~400 nm. | To characterize the molecular weight of DOM; A higher value indicates a smaller molecular weight. | [15,16] |
| E254/E204 | The ratio of α254 to α204; α254 and α204 are the absorption coefficients of the sample at 254 nm and 204 nm, respectively. | To characterize the hydrophobic property of DOM; A higher value indicates a higher hydrophobicity. | [17] |
| SUVA254 | The ratio of α254 to DOC; α254 is the absorption coefficients of the sample at 254 nm, and the DOC is the concentration of dissolved organic carbons. | To characterize the aromaticity of DOM; A higher value indicates a higher aromaticity. | [18] |
| Genus | Taxonomic Annotation | Putative Eco-Functions | ||
|---|---|---|---|---|
| Order | Class | Phylum | ||
| Algoriphagus | Cytophagales | Cytophagia | Bacteroidetes | Degradation of high molecular organic matters |
| Mycobacterium | Corynebacteriales | Actinomycetia | Actinobacteria | Unclear |
| Gemmobacter | Rhodobacterales | Alphaproteobacteria | Proteobacteria | Dissimilatory nitrate reduction |
| Flavobacterium | Flavobacteriales | Flavobacteriia | Bacteroidetes | Degradation of complex organic matters |
| Fluviicola | Flavobacteriales | Flavobacteriia | Bacteroidetes | |
| Rhizobium | Hyphomicrobiales | Alphaproteobacteria | Proteobacteria | lysis of algal cells and its toxin degradation |
| Devosia | Hyphomicrobiales | Alphaproteobacteria | Proteobacteria | Unclear. Probably Nitrogen-fixing |
| Rheinheimera | Chromatiales | Gammaproteobacteria | Proteobacteria | Anoxygenic photosynthesis and symbionts with algae |
| Alishewanella | Chromatiales | Gammaproteobacteria | Proteobacteria | |
| Orientia | Rickettsiales | Alphaproteobacteria | Proteobacteria | Unclear |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Liu, J.; Zhang, C.; Wang, J.; Li, P.; Sun, J.; Sun, Y. Response of Algal–Bacterial Regrowth Characteristics to the Hypochlorite in Landscape Ponds Replenished with Reclaimed Water. Water 2022, 14, 3893. https://doi.org/10.3390/w14233893
Li M, Liu J, Zhang C, Wang J, Li P, Sun J, Sun Y. Response of Algal–Bacterial Regrowth Characteristics to the Hypochlorite in Landscape Ponds Replenished with Reclaimed Water. Water. 2022; 14(23):3893. https://doi.org/10.3390/w14233893
Chicago/Turabian StyleLi, Meng, Jiaheng Liu, Chao Zhang, Jinli Wang, Pengfeng Li, Jingmei Sun, and Yongli Sun. 2022. "Response of Algal–Bacterial Regrowth Characteristics to the Hypochlorite in Landscape Ponds Replenished with Reclaimed Water" Water 14, no. 23: 3893. https://doi.org/10.3390/w14233893





