Differential Response of Phaeodactylum tricornutum and Cylindrotheca fusiformis to High Concentrations of Cu2+ and Zn2+
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Scanning Electron Microscope (SEM) and Energy Dispersive Spectroscopy (EDS) Analysis
2.3. Transcriptomic Analysis
3. Results
3.1. Effects of Cu2+ and Zn2+ on Growth of P. tricornutum and C. fusiformis
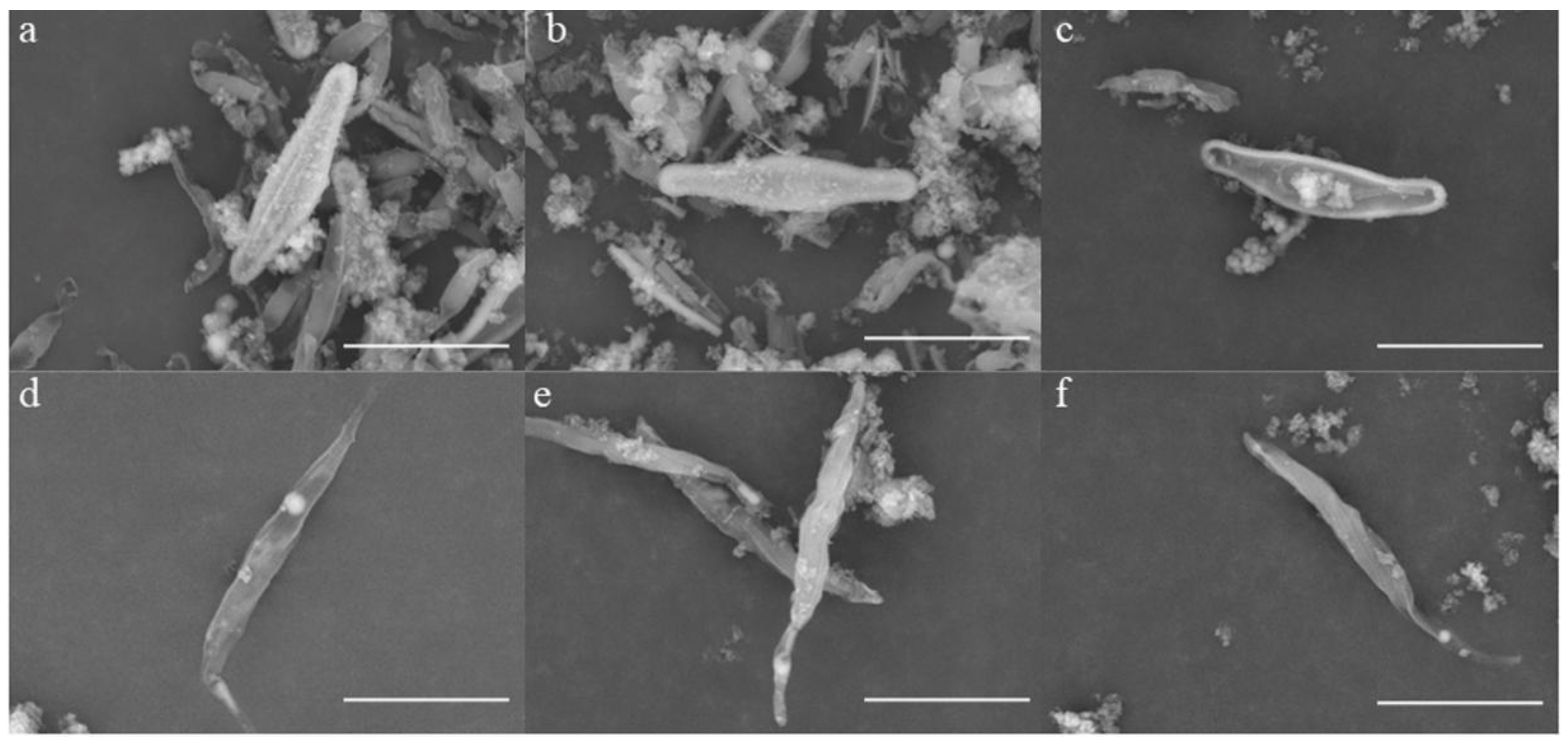
3.2. Effects of Cu2+ and Zn2+ on Cell Morphology of P. tricornutum and C. fusiformis
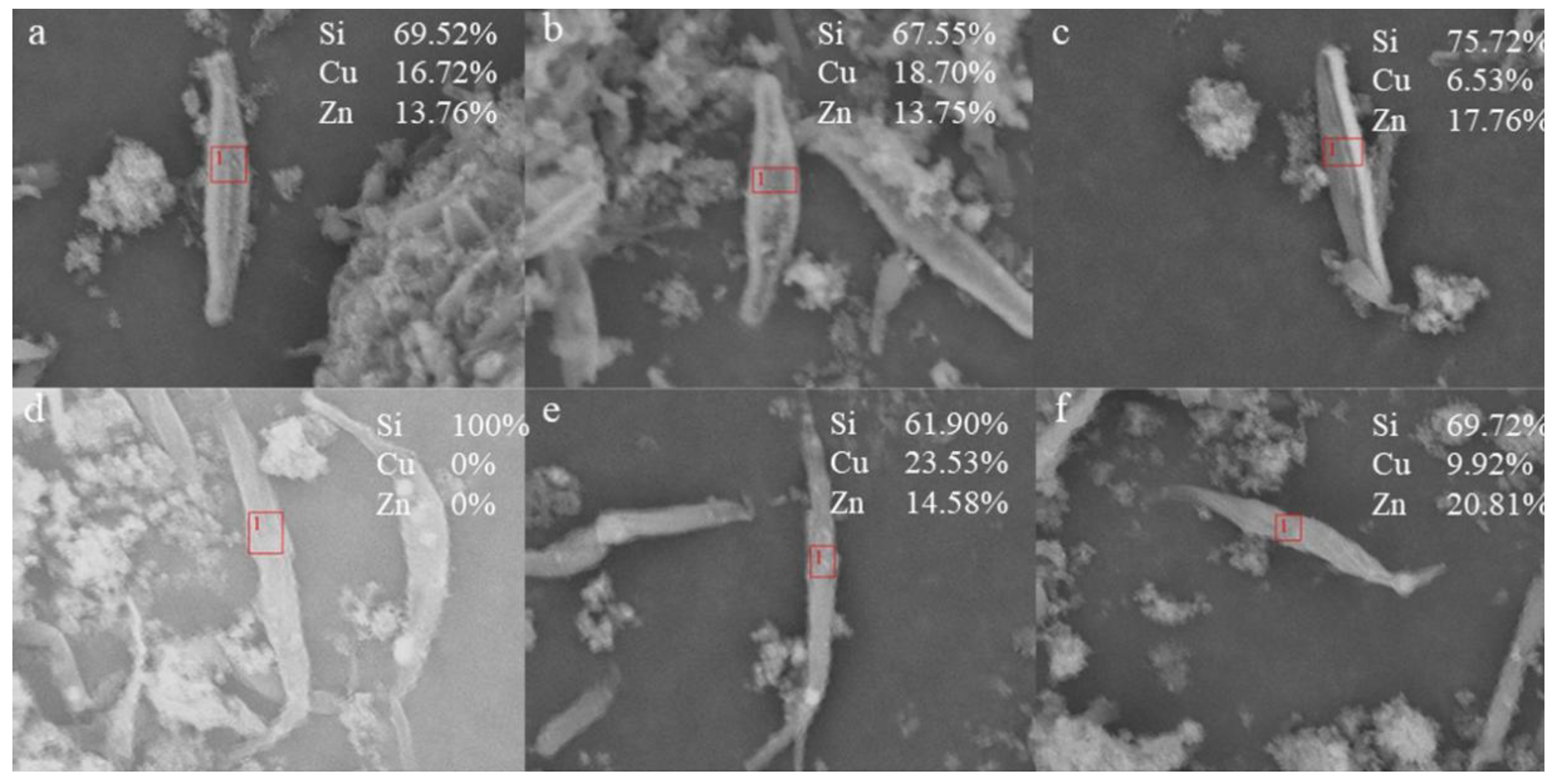
3.3. Accumulation of Cu2+ and Zn2+ on Biosilica Shell of P. tricornutum and C. fusiformis
3.4. Effects of Cu2+ and Zn2+ on Gene Transcription in P. tricornutum
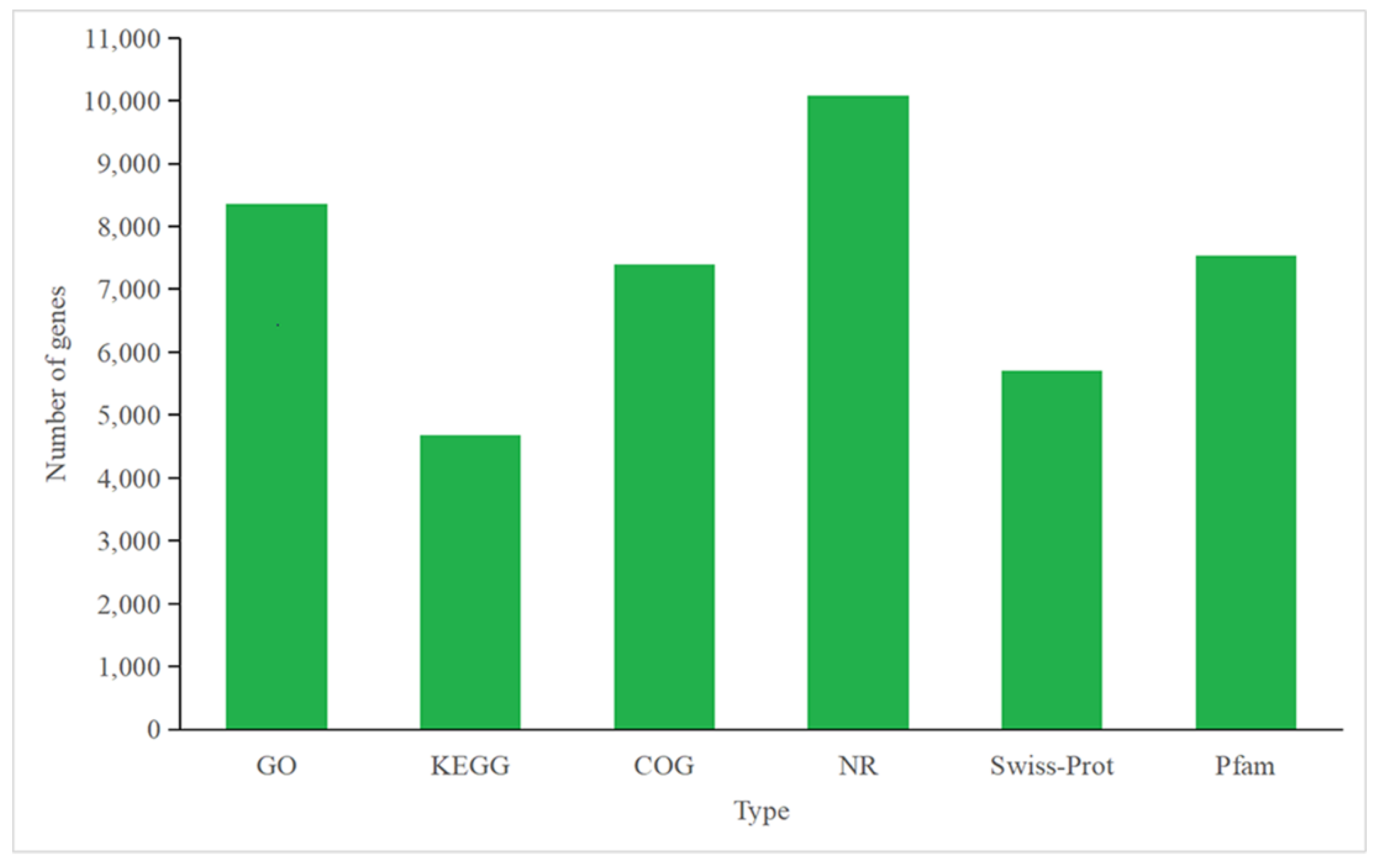
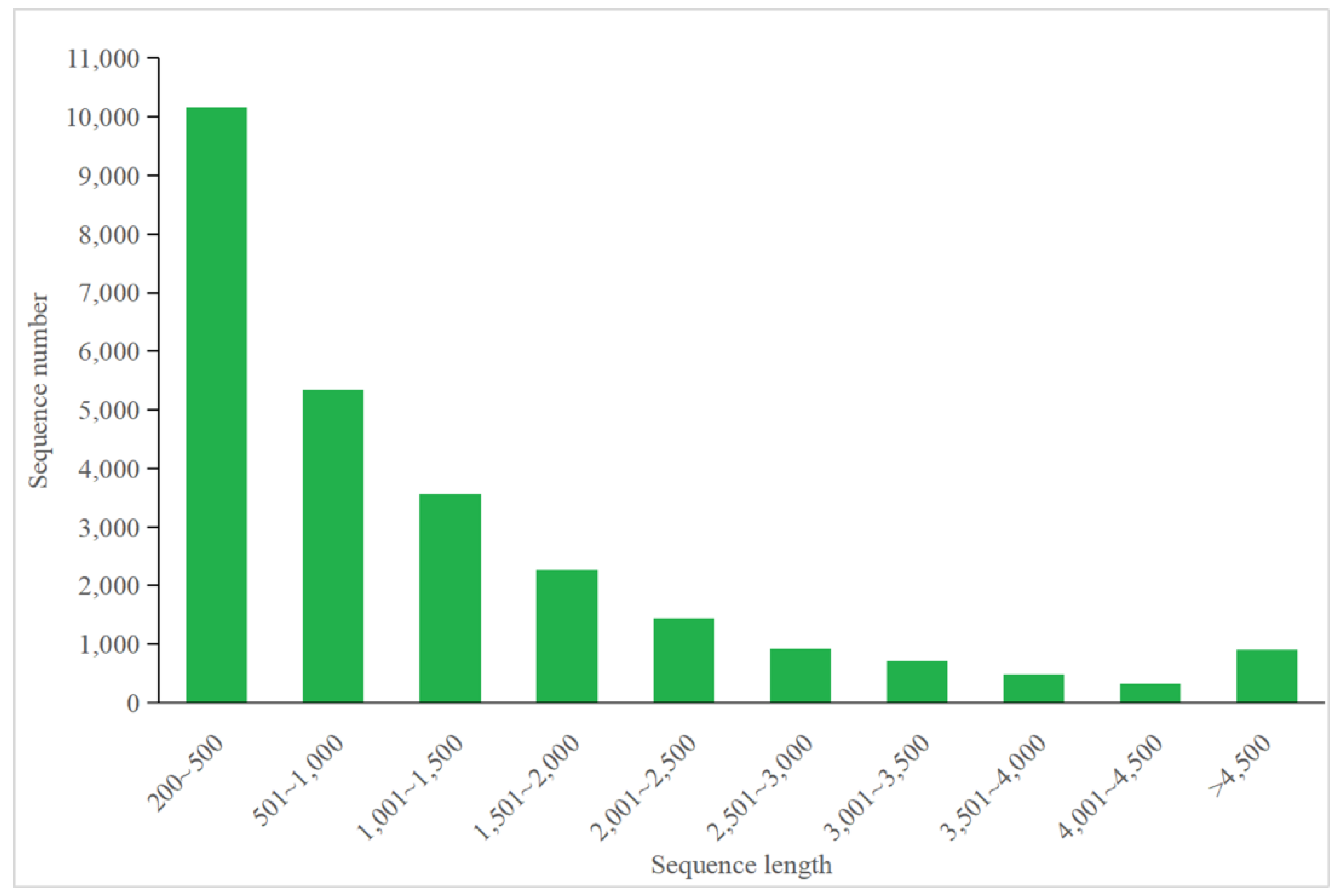
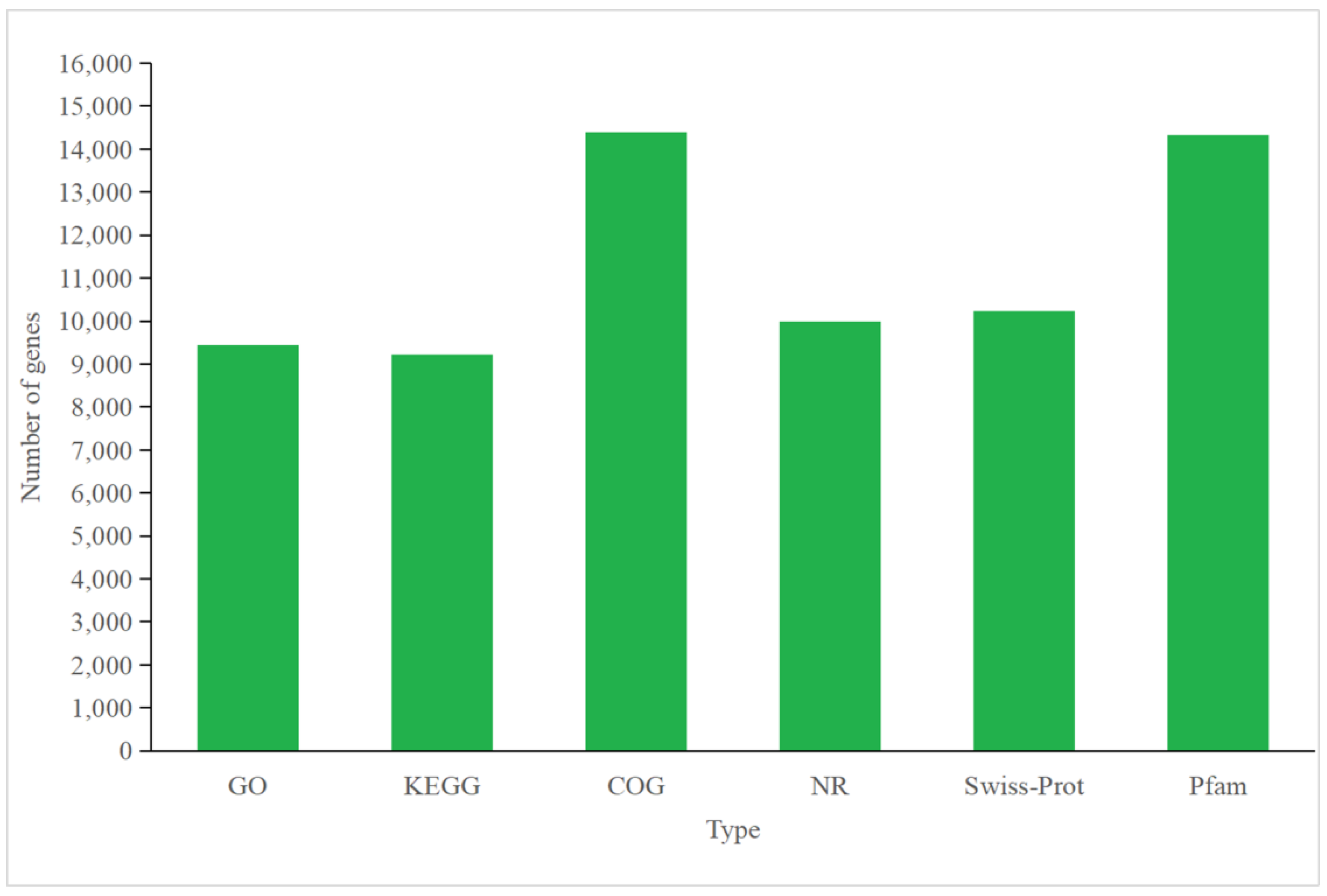
3.4.1. Annotation of P. tricornutum Transcriptome
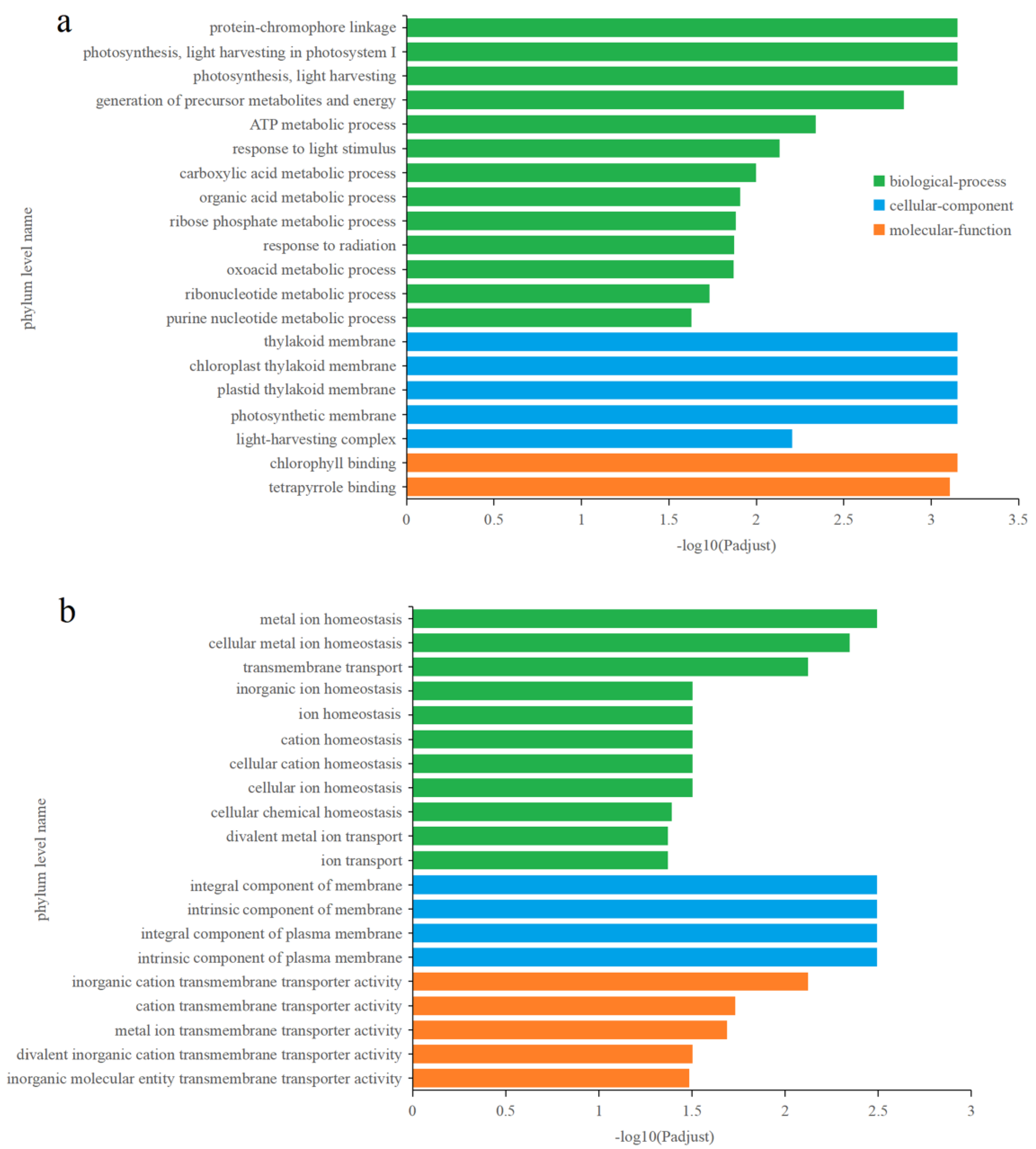
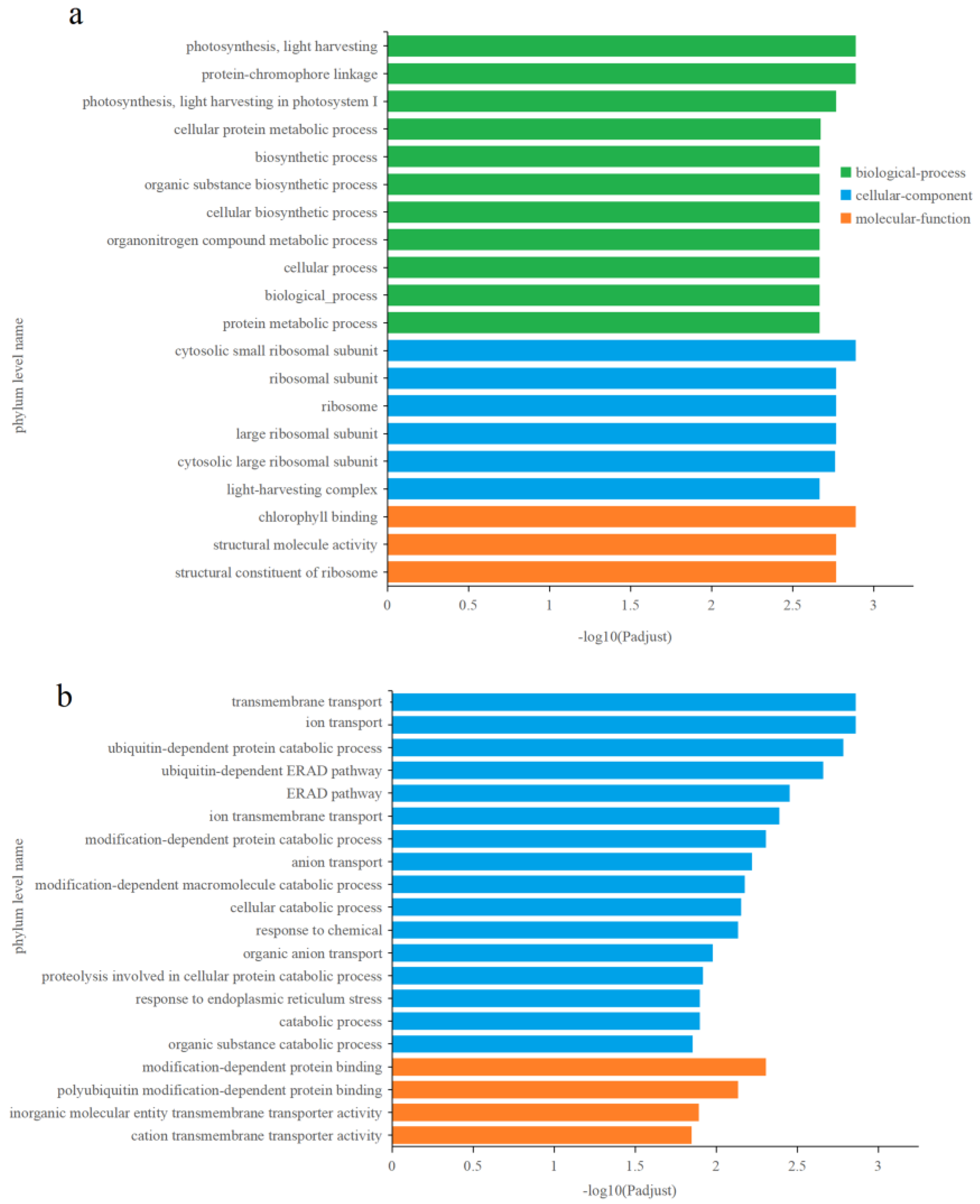
3.4.2. Identification and Functional Enrichment Analysis of Different Express Genes (DEGs) in P. tricornutum upon Cu2+ Treatment
3.4.3. Effects of Zn2+ on Gene Transcription in P. tricornutum
3.5. Effects of Cu2+ and Zn2+ on Gene Transcription in C. fusiformis
3.5.1. Annotation of C. fusiformis Transcriptome
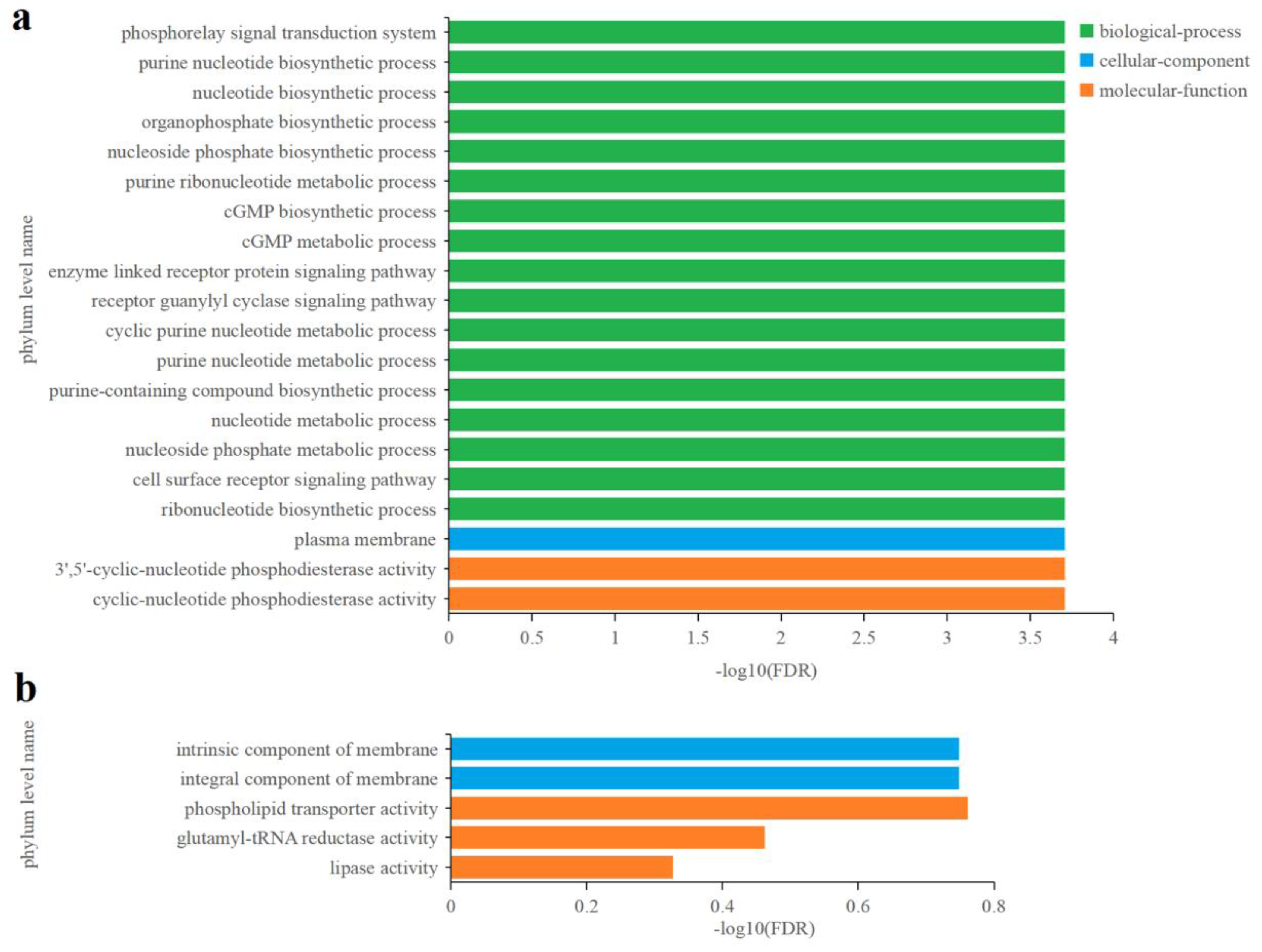
3.5.2. Effects of Cu2+ on Gene Transcription in C. fusiformis
3.5.3. Effects of Zn2+ on gene transcription in C. fusiformis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larned, S.T. A prospectus for periphyton: Recent and future ecological research. J. N. Am. Benthol. Soc. 2010, 29, 182–206. [Google Scholar] [CrossRef] [Green Version]
- Butcher, R.W. Studies in the Ecology of Rivers: VII. The Algae of Organically Enriched Waters. J. Ecol. 1947, 35, 186–191. [Google Scholar] [CrossRef]
- Tudesque, L.; Grenouillet, G.; Gevrey, M.; Khazraie, K.; Brosse, S. Influence of small-scale gold mining on French Guiana streams: Are diatom assemblages valid disturbance sensors? Ecol. Indic. 2012, 14, 100–106. [Google Scholar] [CrossRef]
- Sbihi, K.; Cherifi, O.; Bertrand, M. Toxicity and biosorption of chromium from aqueous solutions by the diatom Planothidium lanceolatum (Brébisson) Lange-Bertalot. Am. J. Sci. 2012, 3, 27–38. [Google Scholar] [CrossRef]
- De Stefano, L.; Rotiroti, L.; De Stefano, M.; Lamberti, A.; Lettieri, S.; Setaro, A.; Maddalena, P. Marine diatoms as optical biosensors. Biosens. Bioelectron. 2009, 24, 1580–1584. [Google Scholar] [CrossRef]
- Marie, M.; Kirsten, H.; Pamela, Q.; Negri, A.P. Additive toxicity of herbicide mixtures and comparative sensitivity of tropical benthic microalgae. Mar. Pollut. Bull. 2010, 60, 1978–1987. [Google Scholar]
- Satoh, A.; Vudikaria, L.Q.; Kurano, N.; Miyachi, S. Evaluation of the sensitivity of marine microalgal strains to the heavy metals, Cu, As, Sb, Pb and Cd. Environ. Int. 2005, 31, 713–722. [Google Scholar] [CrossRef]
- Rimet, F. Recent views on river pollution and diatoms. Hydrobiologia 2012, 683, 1–24. [Google Scholar] [CrossRef]
- Masmoudi, S.; Nguyen-Deroche, N.; Caruso, A.; Ayadi, H.; Morant-Manceau, A.; Tremblin, G.; Bertrand, M.; Schoefs, B. Cadmium, copper, sodium and zinc effects on diatoms: From heaven to hell—A review. Cryptogam. Algol. 2013, 34, 185–225. [Google Scholar] [CrossRef]
- Owens, T.G.; Wold, E.R. Light-Harvesting Function in the Diatom Phaeodactylum tricornutum: I. Isolation and Characterization of Pigment-Protein Complexes. Plant Physiol. 1986, 80, 732. [Google Scholar] [CrossRef] [Green Version]
- Patil, V.; Reitan, K.I.; Knutsen, G.; Mortensen, L.M.; Källqvist, T.; Olsen, E.; Vogt, G.; Gislerød, H.R. Microalgae as source of polyunsaturated fatty acids for aquaculture. Curr. Top. Plant Biol. 2005, 6, 57–65. [Google Scholar]
- Alipanah, L.; Rohloff, J.; Winge, P.; Bones, A.M.; Brembu, T. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 2015, 66, 6281–6296. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef]
- De Risco, V.; Raniello, R.; Maumus, F.; Rogato, A.; Bowler, C.; Falciatore, A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009, 37, 96. [Google Scholar] [CrossRef] [Green Version]
- Stukenberg, D.; Zauner, S.; Dell’Aquila, G.; Maier, U.G. Optimizing CRISPR/Cas9 for the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2018, 9, 740. [Google Scholar] [CrossRef] [Green Version]
- Kawamura, T.; Roberts, R.D.; Nicholson, C.M. Factors affecting the food value of diatom strains for post-larval abalone Haliotis iris. Aquaculture 1998, 160, 81–88. [Google Scholar] [CrossRef]
- Gallardo, W.G.; Buen, S.M.A. Evaluation of mucus, Navicula, and mixed diatoms as larval settlement inducers for the tropical abalone Haliotis asinina. Aquaculture 2003, 221, 357–364. [Google Scholar] [CrossRef]
- Cid, A.; Torres, E.; Herrero, C.; Abalde, J.E. Disorders provoked by copper in the marine diatom Phaeodactylum tricornutum in short-time exposure assays. Cah. Biol. Mar. 1997, 38, 201–206. [Google Scholar]
- Guillard, R. Culture of Marine Invertebrate Animals, 1st ed.; Springer: New York, NY, USA, 1975; pp. 29–60. [Google Scholar]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hänsch, R.; Mende, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Peers, G.; Price, N.M. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature 2006, 441, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Peers, G.; Quesnel, S.A.; Price, N.M. Copper requirements for iron acquisition and growth of coastal and oceanic diatoms. Limnol. Oceanogr. 2005, 50, 1149–1158. [Google Scholar] [CrossRef]
- Maldonado, M.T.; Allen, A.E.; Chong, J.S.; Lin, K.; Leus, D.; Karpenko, N.; Harris, S.L. Copper-dependent iron transport in coastal and oceanic diatoms. Limnol. Oceanogr. 2006, 51, 1729–1743. [Google Scholar] [CrossRef] [Green Version]
- Cox, E.H.; McLendon, G.L.; Morel, F.M.M.; Lane, T.W.; Prince, R.C.; Pickering, I.J.; George, G.N. The Active Site Structure of Thalassiosira weissflogii Carbonic Anhydrase 1. Biochemistry 2000, 39, 12128–12130. [Google Scholar] [CrossRef]
- Rayko, E.; Maumus, F.; Maheswari, U.; Jabbari, K.; Bowler, C. Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 2010, 188, 52–66. [Google Scholar] [CrossRef]
- Rijstenbil, J.W.; Derksen, J.W.M.; Gerringa, L.J.A.; Poortvliet, T.C.W.; Sandee, A.; Berg, M.; Drie, J.; Wijnholds, J.A. Oxidative stress induced by copper: Defense and damage in the marine planktonic diatom Ditylum brightwellii, grown in continuous cultures with high and low zinc levels. Mar. Biol. 1994, 119, 583–590. [Google Scholar] [CrossRef] [Green Version]
- Buhmann, M.T.; Schulze, B.; Foerderer, A.; Schleheck, D.; Kroth, P.G. Bacteria may induce the secretion of mucin-like proteins by the diatom Phaeodactylum tricornutum. J. Phycol. 2016, 52, 463–474. [Google Scholar] [CrossRef] [Green Version]
- Willis, A.; Chiovitti, A.; Dugdale, T.M.; Wetherbee, R. Characterization of the extracellular matrix of Phaeodactylum tricornutum (Bacillariophyceae): Structure, composition, and adhesive characteristics. J. Phycol. 2013, 49, 937–949. [Google Scholar] [CrossRef]
- Tong, C.Y.; Derek, C.J.C. The role of substrates towards marine diatom Cylindrotheca fusiformis adhesion and biofilm development. J. Appl. Phycol 2021, 33, 2845–2862. [Google Scholar] [CrossRef]
- Tong, C.Y.; Derek, C.J.C. Biofilm formation of benthic diatoms on commercial polyvinylidene fluoride membrane. Algal Res. 2021, 55, 102260. [Google Scholar] [CrossRef]



| Gene_id | fc | Regulate | nr | Paths | Swissprot |
|---|---|---|---|---|---|
| Pt04g03550 | 5.7 | up | XP_002181744.1 (predicted protein) | map00480 (Glutathione metabolism); map00053 (Ascorbate and aldarate metabolism) | Probable L-ascorbate peroxidase 8 |
| Pt05g02260 | 2.1 | up | XP_002186090.1 (catalase-peroxidase) | map00360 (Phenylalanine metabolism); map00380 (Tryptophan metabolism) | Catalase-peroxidase |
| Pt08g02130 | 30.8 | up | XP_002179007.1 (predicted protein) | map00480 (Glutathione metabolism) | Probable cytosol aminopeptidase |
| Pt14g00980 | 0.3 | down | XP_002181057.1 (predicted protein) | ||
| Pt02g05550 | 3.3 | up | XP_002177701.1 (predicted protein) | map00480 (Glutathione metabolism) | |
| Pt03g03150 | 0.4 | down | XP_002185216.1 (glyoxalase) | map00620 (Pyruvate metabolism) | Hydroxyacylglutathione hydrolase |
| Pt02g03960 | 2.8 | up | XP_002177790.1 (predicted protein) | ||
| Pt07g01050 | 0.4 | down | XP_002185856.1 (predicted protein) | map00620 (Pyruvate metabolism) | |
| Pt15g02690 | 9.6 | up | XP_002182163.1 (predicted protein) | ||
| Pt12g00930 | 2.7 | up | XP_002180005.1 (predicted protein) | map00480 (Glutathione metabolism) | Glutathione S-transferase DHAR2 |
| Pt01g09200 | 0.5 | down | XP_002177254.1 (predicted protein, partial) | Glutathione gamma-glutamylcysteinyltransferase | |
| Pt11g01900 | 10.3 | up | XP_002182079.1 (predicted protein) | ||
| Pt14g03650 | 2.1 | up | XP_002180739.1 (glutathione peroxidase, partial) | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Phospholipid hydroperoxide glutathione peroxidase |
| Pt05g02470 | 2.2 | up | XP_002186390.1 (predicted protein) | ||
| Pt11g01090 | 5.9 | up | XP_002182079.1 (predicted protein) | ||
| Pt07g04170 | 5.1 | up | XP_002176312.1 (peroxidase domain-containing protein) | Putative heme-binding peroxidase | |
| Pt11g03130 | 17.5 | up | XP_002181851.1 (predicted protein) | ||
| Pt14g03650 | 2.1 | up | XP_002180739.1 (glutathione peroxidase, partial) | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Phospholipid hydroperoxide glutathione peroxidase |
| Pt21g01220 | 0.3 | down | XP_002183862.1 (predicted protein) |
| Gene_id | fc | Regulate | nr | Paths | Swissprot |
|---|---|---|---|---|---|
| Pt04g03550 | 6.91 | up | XP_002181744.1 (predicted protein) | map00480 (Glutathione metabolism); map00053 (Ascorbate and aldarate metabolism) | Probable L-ascorbate peroxidase 8 |
| Pt20g01650 | 2.55 | up | XP_002182954.1 (catalase) | map00630 (Glyoxylate and dicarboxylate metabolism); map00380 (Tryptophan metabolism); map04146 (Peroxisome) | Catalase |
| Pt13g01910 | 0.50 | down | XP_002180671.1 (predicted protein) | ||
| Pt23g00220 | 2.22 | up | XP_002184868.1 (predicted protein) | Peroxiredoxin-6 | |
| Pt10g01030 | 0.03 | down | XP_002179508.1 (predicted protein) | map00480 (Glutathione metabolism) | Glutathione S-transferase |
| Pt05g04280 | 9.78 | up | XP_002186195.1 (UDP-glucose 6-dehydrogenase) | map00520 (Amino sugar and nucleotide sugar metabolism); map00040 (Pentose and glucuronate interconversions); map00053 (Ascorbate and aldarate metabolism) | UDP-glucose 6-dehydrogenase 1 |
| Pt14g01270 | 0.24 | down | XP_002180872.1 (l-ascorbate peroxidase, partial) | map00480 (Glutathione metabolism); map00053 (Ascorbate and aldarate metabolism) | Putative heme-binding peroxidase |
| Pt16g00880 | 6.11 | up | XP_002179589.1 (nad-dependent epimerase/dehydratase) | map00520 (Amino sugar and nucleotide sugar metabolism); map00053 (Ascorbate and aldarate metabolism) | GDP-mannose 3,5-epimerase |
| Pt08g03190 | 5.92 | up | XP_002178726.1 (predicted protein) | map00460 (Cyanoamino acid metabolism); map00480 (Glutathione metabolism); map00430 (Taurine and hypotaurine metabolism) | Glutathione hydrolase-like YwrD proenzyme |
| Pt04g01510 | 0.39 | down | XP_002183098.1 (glutathione peroxidase domain-containing protein) | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Probable phospholipid hydroperoxide glutathione peroxidase |
| Pt02g03960 | 2.85 | up | XP_002177790.1 (predicted protein) | ||
| Pt12g00930 | 4.58 | up | XP_002180005.1 (predicted protein) | map00480 (Glutathione metabolism) | Glutathione S-transferase DHAR2 |
| Pt21g02200 | 25.23 | up | XP_002183815.1 (predicted protein) | map02010 (ABC transporters) | Glutathione-binding protein GsiB |
| Pt18g02190 | 6.06 | up | XP_002185391.1 (predicted protein) | ||
| Pt05g02470 | 2.74 | up | XP_002186390.1 (predicted protein) | ||
| Pt23g01150 | 8.11 | up | XP_002184892.1 (predicted protein) | Glutathione gamma-glutamylcysteinyltransferase 2 | |
| Pt08g02730 | 0.47 | down | GAX19067.1 (hypothetical protein FisN_8Hh293 [Fistulifera solaris]) | ABC transporter G family member 1 | |
| Pt12g03160 | 0.48 | down | XP_002180322.1 (glutathione reductase) | map00480 (Glutathione metabolism) | Glutathione reductase |
| PtUn01s113 | 5.37 | up | XP_002177253.1 (mutase superoxide dismutase) | map04146 (Peroxisome) | Superoxide dismutase |
| Pt13g02930 | 12.70 | up | XP_002180497.1 (precursor of mutase superoxide dismutase [Fe/Mn], partial) | map04146 (Peroxisome) | Superoxide dismutase |
| Pt01g09190 | 7.47 | up | XP_002177253.1 (mutase superoxide dismutase | map04146 (Peroxisome) | Superoxide dismutase |
| Pt05g04470 | 0.40 | down | XP_002186201.1 (5′-Nucleotidase or metallophosphoesterase) | ||
| Pt07g04170 | 2.73 | up | XP_002176312.1 (peroxidase domain-containing protein) | Putative heme-binding peroxidase | |
| Pt20g01220 | 2.27 | up | XP_002182845.1 (predicted protein) | map04146 (Peroxisome) | Peroxiredoxin-2C |
| Gene_id | nr_Description | fc | Regulate | Paths | Swissprot |
|---|---|---|---|---|---|
| TRINITY_DN14518_c0_g1 | thioredoxin-like protein | 2.68 | up | map00940 (Phenylpropanoid biosynthesis) | 1-Cys peroxiredoxin A |
| TRINITY_DN1479_c0_g1 | glutathione synthetase | 0.28 | down | map00270 (Cysteine and methionine metabolism); map00480 (Glutathione metabolism) | Glutathione synthetase |
| TRINITY_DN495_c0_g2 | hypothetical protein | 0.19 | down | map00480 (Glutathione metabolism) | Glutathione S-transferase |
| TRINITY_DN7366_c1_g1 | glutathione peroxidase | 0.43 | down | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Hydroperoxy fatty acid reductase gpx1 |
| TRINITY_DN1758_c0_g1 | hydroxyacylglutathione hydrolase | 0.45 | down | map00620 (Pyruvate metabolism); map00790 (Folate biosynthesis) | Hydroxyacylglutathione hydrolase |
| TRINITY_DN3215_c0_g1 | hypothetical protein | 0.46 | down | ||
| TRINITY_DN2680_c0_g1 | hypothetical protein | 0.49 | down | map00270 (Cysteine and methionine metabolism); map00480 (Glutathione metabolism) | Glutamate--cysteine ligase catalytic subunit |
| TRINITY_DN6304_c0_g1 | hypothetical protein | 0.38 | down |
| Gene_id | nr_Description | fc | Significant | Regulate | Paths | Swissprot |
|---|---|---|---|---|---|---|
| TRINITY_DN14518_c0_g1 | thioredoxin-like protein | 3.17 | yes | up | map00940 (Phenylpropanoid biosynthesis) | 1-Cys peroxiredoxin A |
| TRINITY_DN1479_c0_g1 | glutathione synthetase | 0.33 | yes | down | map00270 (Cysteine and methionine metabolism); map00480 (Glutathione metabolism) | Glutathione synthetase |
| TRINITY_DN1711_c0_g1 | hypothetical protein | 2.81 | yes | up | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Glutathione S-transferase |
| TRINITY_DN1711_c0_g2 | hypothetical protein | 6.12 | yes | up | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Glutathione S-transferase 1 |
| TRINITY_DN2013_c0_g1 | glutathione-S-transferase | 0.44 | yes | down | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Glutathione S-transferase |
| TRINITY_DN327_c0_g2 | glutathione S-transferase | 2.73 | yes | up | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Glutathione S-transferase 1 |
| TRINITY_DN6449_c0_g1 | hypothetical protein | 2.06 | yes | up | Glutathionyl-hydroquinone reductase | |
| TRINITY_DN7366_c1_g1 | glutathione peroxidase | 0.37 | yes | down | map00590 (Arachidonic acid metabolism); map00480 (Glutathione metabolism) | Hydroperoxy fatty acid reductase |
| TRINITY_DN2338_c0_g3 | hypothetical protein | 0.40 | yes | down | DEP domain-containing mTOR-interacting protein | |
| TRINITY_DN3_c0_g4 | mercuric reductase | 0.43 | yes | down | General L-amino acid-binding periplasmic protein Aap | |
| TRINITY_DN17353_c0_g1 | LhcSR | 2.98 | yes | up | map00196 (Photosynthesis—Antenna proteins) | Light-harvesting complex stress-related protein |
| TRINITY_DN1775_c0_g1 | hypothetical protein | 0.30 | yes | down | Thyroid peroxidase | |
| TRINITY_DN319_c0_g1 | oxidative stress-related Abc1-like protein | 2.10 | yes | up | Protein ACTIVITY OF BC1 COMPLEX KINASE 8 | |
| TRINITY_DN3894_c1_g1 | catalase peroxidase | 2.40 | yes | up | map00940 (Phenylpropanoid biosynthesis); map00380 (Tryptophan metabolism); map00360 (Phenylalanine metabolism) | Catalase-peroxidase |
| TRINITY_DN5279_c0_g2 | methionine sulfoxide reductase B | 0.45 | yes | down | Peptide methionine sulfoxide reductase | |
| TRINITY_DN6304_c0_g1 | hypothetical protein | 0.39 | yes | down | Peroxinectin A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, A.; Wang, Y.; Duan, J.; Guo, S.; Xie, Z. Differential Response of Phaeodactylum tricornutum and Cylindrotheca fusiformis to High Concentrations of Cu2+ and Zn2+. Water 2022, 14, 3305. https://doi.org/10.3390/w14203305
Huang A, Wang Y, Duan J, Guo S, Xie Z. Differential Response of Phaeodactylum tricornutum and Cylindrotheca fusiformis to High Concentrations of Cu2+ and Zn2+. Water. 2022; 14(20):3305. https://doi.org/10.3390/w14203305
Chicago/Turabian StyleHuang, Aiyou, Yujue Wang, Jiawen Duan, Shiyi Guo, and Zhenyu Xie. 2022. "Differential Response of Phaeodactylum tricornutum and Cylindrotheca fusiformis to High Concentrations of Cu2+ and Zn2+" Water 14, no. 20: 3305. https://doi.org/10.3390/w14203305




