Developing Indicators of Nutrient Pollution in Streams Using 16S rRNA Gene Metabarcoding of Periphyton-Associated Bacteria
Abstract
:1. Introduction
2. Materials and Methods
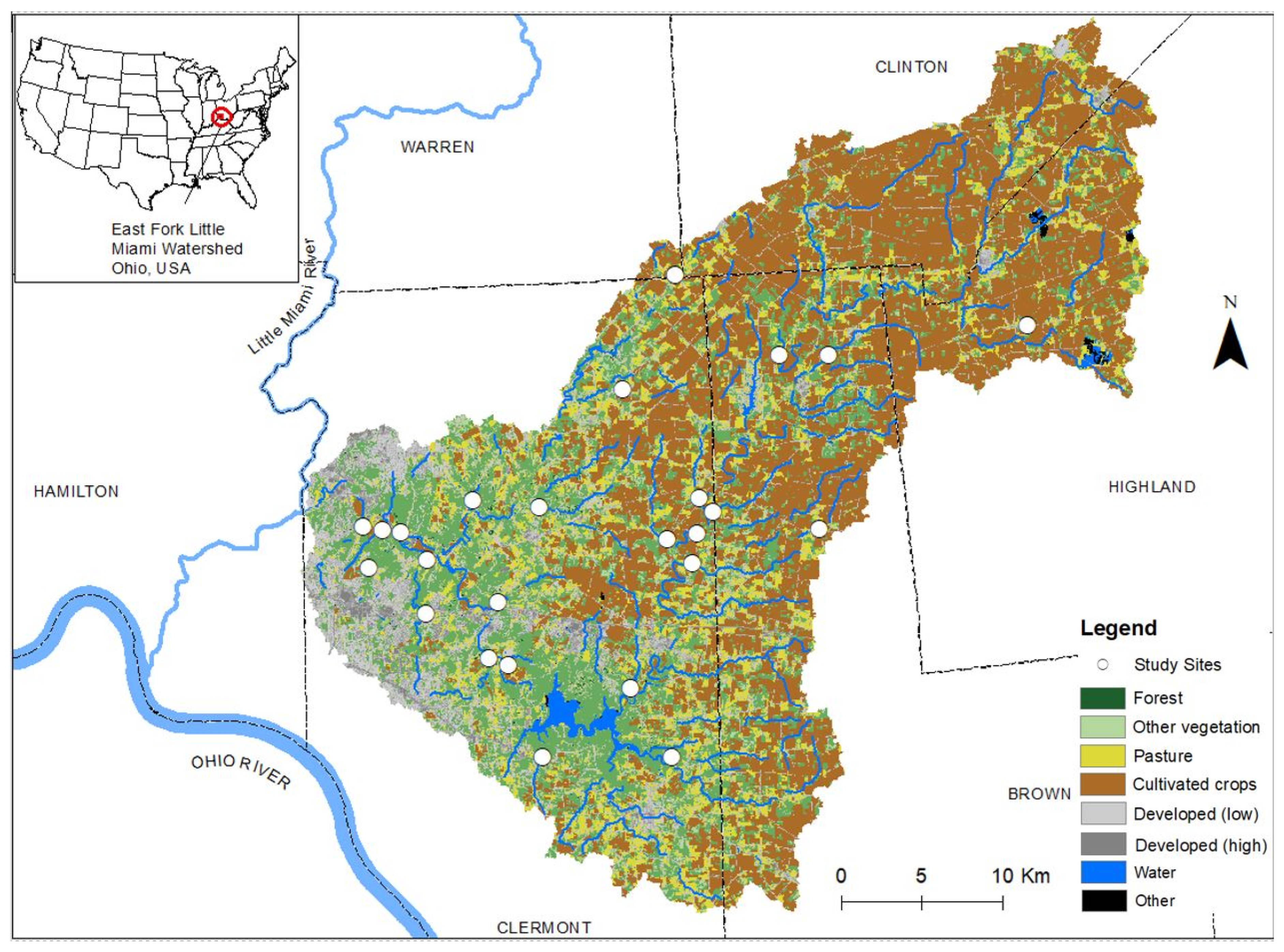
2.1. Site Selection and Sampling
2.2. DNA Metabarcoding Workflow
2.3. Bioinformatics
2.4. Statistical Analyses
2.5. Phylogenetic Analysis
3. Results
3.1. Descriptive Summary of Data
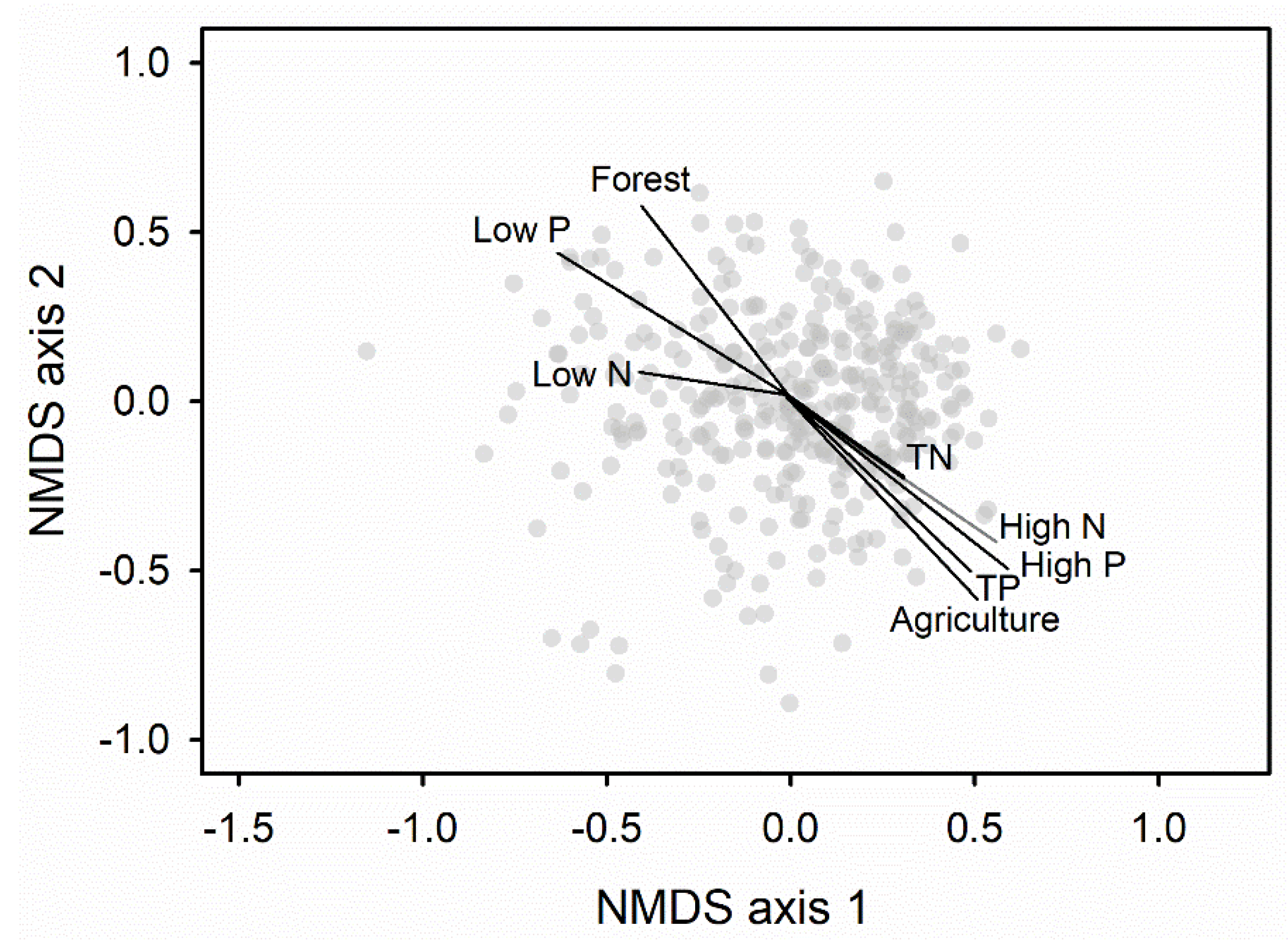
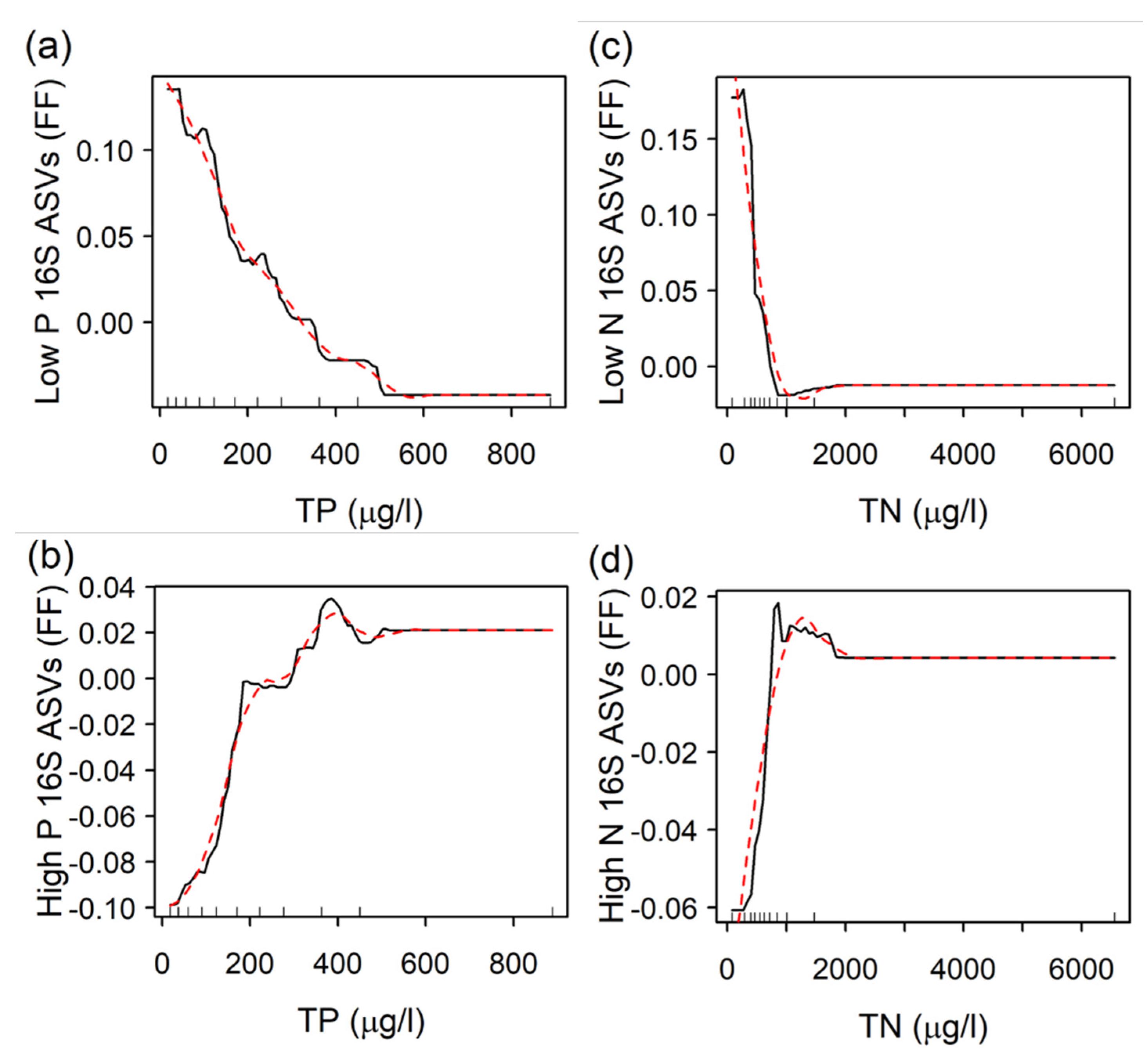
3.2. Bacterial Indicators and Relationships with TP and TN
3.3. Summary of Changes in Bacterial Assemblages
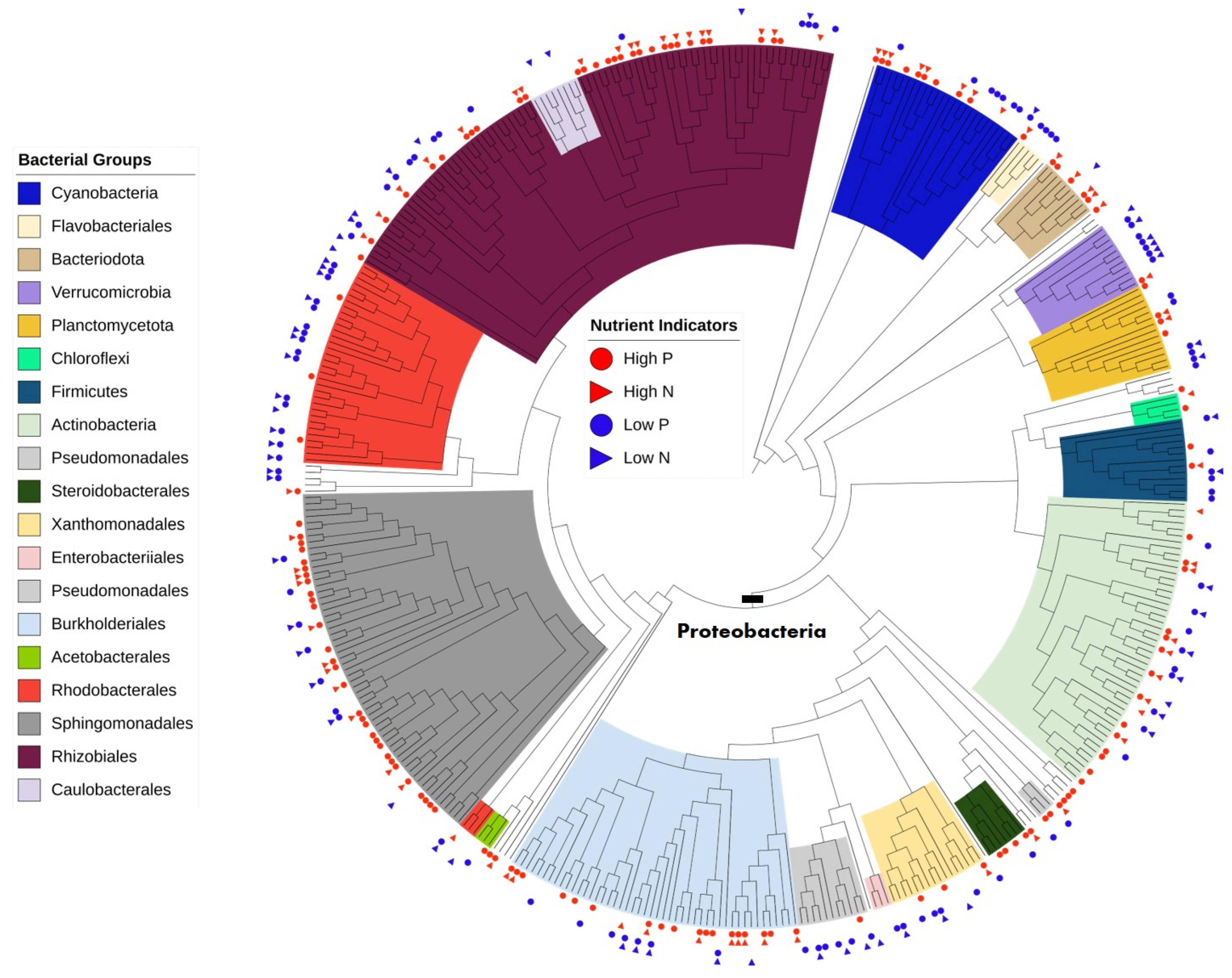
3.4. Taxonomy and Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beusen, A.H.W.; Bouwman, A.F.; Van Beek, L.P.H.; Mogollón, J.M.; Middelburg, J.J. Global riverine N and P transport to ocean increased during the 20th century despite increased retention along the aquatic continuum. Biogeosciences 2016, 13, 2441–2451. [Google Scholar] [CrossRef] [Green Version]
- Sabo, R.D.; Clark, C.M.; Bash, J.; Sobota, D.; Cooter, E.; Dobrowski, J.P.; Houlton, B.Z.; Rea, A.; Schwede, D.; Morford, S.L.; et al. Decadal shift in nitrogen inputs and fluxes across the contiguous United States: 2002–2012. J. Geophys. Res.-Biogeosci. 2019, 124, 3104–3124. [Google Scholar] [CrossRef]
- Sabo, R.D.; Clark, C.M.; Gibbs, D.A.; Metson, G.S.; Todd, M.J.; LeDuc, S.D.; Greiner, D.; Fry, J.A.; Polinsky, R.; Yang, Q.; et al. Phosphorus inventory for the conterminous United States (2002–2012). J. Geophys. Res.-Biogeosci. 2021, 126, e2020JG005684. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. National Rivers and Streams Assessment 2008–2009: A Collaborative Survey; EPA/841/R-16/007; Office of Water and Office of Research and Development: Washingon, DC, USA, 2016. [Google Scholar]
- Dodds, W.K.; Smith, V.H. Nitrogen, phosphorus, and eutrophication in streams. Inland Waters 2016, 6, 155–164. [Google Scholar] [CrossRef]
- Carvalho, L.; Mackay, E.B.; Cardoso, A.C.; Baattrup-Pedersen, A.; Birk, S.; Blackstock, K.L.; Borics, G.; Borja, A.; Feld, C.K.; Ferreira, M.T.; et al. Protecting and restoring Europe’s waters: An analysis of the future development needs of the Water Framework Directive. Sci. Total Environ. 2019, 658, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Poikane, S.; Várbíró, G.; Kelly, M.; Birk, S.; Phillips, G. Estimating river nutrient concentrations consistent with good ecological condition: More stringent nutrient thresholds needed. Ecol. Indic. 2021, 121, 107017. [Google Scholar] [CrossRef]
- Wang, L.; Robertson, D.M.; Garrison, P.L. Linkages between nutrients and assemblages of macroinvertebrates and fish in wadeable streams: Implication to nutrient criteria development. Environ. Manag. 2007, 39, 194–212. [Google Scholar] [CrossRef] [PubMed]
- Weijters, M.J.; Janse, J.H.; Alkemade, R.; Verhoeven, J.T.A. Quantifying the effect of catchment land use and water nutrient concentrations on freshwater river and stream biodiversity. Aquat. Conserv. 2009, 19, 104–112. [Google Scholar] [CrossRef]
- Stevenson, R.J.; Bennett, B.J.; Jordan, D.N.; French, R.D. Phosphorus regulates stream injury by filamentous green algae, DO, and pH with thresholds in responses. Hydrobiologia 2012, 695, 25–42. [Google Scholar] [CrossRef]
- Wurtsbaugh, W.A.; Paerl, H.W.; Dodds, W.K. Nutrients, eutrophication and harmful algal blooms along the freshwater to marine continuum. WIREs Water 2019, 6, e1373. [Google Scholar] [CrossRef]
- Smucker, N.J.; Beaulieu, J.J.; Nietch, C.T.; Young, J.L. Increasingly severe cyanobacterial blooms and deep-water hypoxia coincide with warming temperatures in reservoirs. Glob. Chang. Biol. 2021, 27, 2507–2519. [Google Scholar] [CrossRef] [PubMed]
- Howarth, R.W.; Sharpley, A.; Walker, D. Sources of nutrient pollution to coastal waters in the United States: Implications for achieving coastal water quality goals. Estuaries 2002, 25, 656–676. [Google Scholar] [CrossRef]
- Vörösmarty, C.J.; McIntyre, P.B.; Gessner, M.O.; Dudgeon, D.; Prusevich, A.; Green, P.; Glidden, S.; Bunn, S.E.; Sullivan, C.A.; Reidy Liebermann, C.; et al. Global threats to human water security and river biodiversity. Nature 2010, 467, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Glibert, P.M.; Beusen, A.H.W.; Harrison, J.A.; Durr, H.H.; Bouwman, A.F.; Laruelle, G.G. Changing land-, sea-, and airscapes: Sources of nutrient pollution affecting habitat suitability for harmful algae. In Global Ecology and Oceanography of Harmful Algal Blooms; Ecological Studies (Analysis and Synthesis); Glibert, P., Berdalet, E., Burford, M., Pitcher, G., Zhou, M., Eds.; Springer: Cham, Switzerland, 2018; Volume 232. [Google Scholar] [CrossRef]
- Caruso, G.; La Ferla, R.; Azzaro, M.; Zoppini, A.; Marino, G.; Petochi, T.; Corinaldesi, C.; Leonarid, M.; Zaccone, R.; Fonda Umani, S.; et al. Microbial assemblages for environmental quality assessment: Knowledge, gaps and usefulness in the European marine strategy framework directive. Crit. Rev. Microbiol. 2016, 42, 883–904. [Google Scholar] [CrossRef]
- Sagova-Mareckova, M.; Boenigk, J.; Bouchez, A.; Cermakova, K.; Chonova, T.; Cordier, T.; Eisendle, U.; Elersek, T.; Fazi, S.; Fleituch, T.; et al. Expanding ecological assessment by integrating microorganisms into routine freshwater biomonitoring. Water Res. 2021, 191, 116767. [Google Scholar] [CrossRef]
- Mulholland, P.J.; Helton, A.M.; Poole, G.C.; Hall, R.O.; Hamilton, S.K.; Peterson, B.J.; Tank, J.L.; Ashkenas, L.R.; Cooper, L.W.; Dahm, C.N.; et al. Stream denitrification across biomes and its response to anthropogenic nitrate loading. Nature 2008, 452, 202–205. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Hill, B.H.; Follstad Shah, J.J. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 2009, 462, 795–798. [Google Scholar] [CrossRef]
- Besemer, K. Biodiversity, community structure and function of biofilms in stream ecosystems. Res. Microbiol. 2015, 166, 774–781. [Google Scholar] [CrossRef] [Green Version]
- Battin, T.J.; Besemer, K.; Bengtsson, M.M.; Romani, A.M.; Packmann, A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016, 14, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Antwis, R.E.; Griffiths, S.M.; Harrison, X.A.; Aranega-Bou, P.; Arce, A.; Bettridge, A.S.; Brailsford, F.L.; de Menezes, A.; Devaynes, A.; Forbes, K.M.; et al. Fifty important research questions in microbial ecology. FEMS Microbiol. Ecol. 2017, 93, fix044. [Google Scholar] [CrossRef]
- Abreu, C.I.; Friedman, J.; Andersen Woltz, V.L.; Gore, J. Mortality causes universal changes in microbial community composition. Nat. Commun. 2019, 10, 2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, F.; Acuña, V.; Font, C.; Freixa, A.; Sabater, S. Effects of multiple stressors on river biofilms depend on the time scale. Sci. Rep. 2019, 9, 15810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seviour, R.J.; Mino, T.; Onuki, M. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 2003, 27, 99–127. [Google Scholar] [CrossRef] [Green Version]
- Simonin, M.; Voss, K.A.; Hassett, B.A.; Rocca, J.D.; Wang, S.Y.; Bier, R.L.; Violin, C.R.; Wright, J.P.; Bernhardt, E.S. In search of microbial indicator taxa: Shifts in stream bacterial communities along an urbanization gradient. Environ. Microbiol. 2019, 21, 3652–3668. [Google Scholar] [CrossRef]
- Steen, A.D.; Crits-Christoph, A.; Carini, P.; DeAngelis, K.M.; Fierer, N.; Lloyd, K.G.; Thrash, J.C. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019, 13, 3126–3130. [Google Scholar] [CrossRef] [Green Version]
- Bodor, A.; Bounedjoun, N.; Vincze, G.E.; Kis, A.E.; Laczi, K.; Bende, G.; Szilagyi, A.; Kovacs, A.; Perei, K.; Rakhely, G. Challenges of unculturable bacteria: Environmental perspectives. Rev. Environ. Sci. Biotechnol. 2020, 19, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Lau, K.E.M.; Washington, V.J.; Fan, V.; Neale, M.W.; Lear, G.; Curran, J.; Lewis, G.D. A novel bacterial community index to assess stream ecological health. Freshwater Biol. 2015, 60, 1988–2002. [Google Scholar] [CrossRef]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A.; et al. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecol. Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef]
- Stein, E.D.; Martinez, M.C.; Stiles, S.; Miller, P.E.; Zakharov, E.V. Is DNA barcoding actually cheaper and faster than traditional morphological methods: Results from a survey of freshwater bioassessment efforts in the United States? PLoS ONE 2014, 9, e95525. [Google Scholar] [CrossRef]
- Keck, F.; Vasselon, V.; Tapolczai, K.; Rimet, F.; Bouchez, A. Freshwater biomonitoring in the information age. Front. Ecol. Environ. 2017, 15, 266–274. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apotheloz-Perret-Gentil, L.; Begja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Wetterstrand, K.A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available online: www.genome.gov/sequencingcostsdata (accessed on 15 November 2021).
- Bock, C.; Jensen, M.; Forster, D.; Marks, S.; Nuy, J.; Psenner, R.; Beisser, D.; Boenigk, J. Factors shaping community patterns of protists and bacteria on a European scale. Environ. Microbiol. 2020, 22, 2243–2260. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.A.; Biessy, L.; Pingram, M.A.; Hamer, M.P.; Wagenhoff, A.; Clapcott, J.; Pearman, J.K. Temporal and spatial variation in bacterial communities on uniform substrates in non-wadeable rivers. Environ. DNA 2021, 3, 1023–1034. [Google Scholar] [CrossRef]
- Juvigny-Khenafou, N.P.D.; Piggott, J.J.; Atkinson, D.; Zhang, Y.; Wu, N.; Matthaei, C.D. Fine sediment and flow velocity impact bacteria community and functional profile more than nutrient enrichment. Ecol. Appl. 2021, 31, e02212. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.E.; King, R.S. A new method for detecting and interpreting biodiversity and ecological community thresholds. Methods Ecol. Evol. 2010, 1, 25–37. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. Elements of Statistical Learning: Data Mining, Inference and Prediction; Springer: New York, NY, USA, 2009. [Google Scholar]
- Ellis, N.; Smith, S.J.; Pitcher, C.R. Gradient forests: Calculating importance gradients on physical predictors. Ecology 2012, 93, 156–168. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency. Method 365.1, Revision 2.0: Determination of Phosphorus by Semi-Automated Colorimetry; EPA-600/R-93/100; United States Environmental Protection Agency: Washingon, DC, USA, 1993. [Google Scholar]
- Tucker, S. Determination of Orthophosphate in Waters by Flow Injection Analysis Colorimetry (High Throughput): QuikChem® Method 10-115-01-1-V; Lachat Instruments: Loveland, CO, USA, 2008. [Google Scholar]
- Patton, C.J.; Kryskalla, J.R. Methods of Analysis by the U.S. Geological Survey National Water Quality Laboratory: Evaluation of Alkaline Persulfate Digestion as an Alternative to Kjeldahl Digestion for Determination of Total and Dissolved Nitrogen and Phosphorus in Water; 03-4174:33; U.S. Department of the Interior, U.S. Geological Survey: Reston, VA, USA, 2003. [Google Scholar]
- Smith, P.; Bogren, K. Determination of Nitrate/Nitrite in Manual Persulfate Digestions: QuikChem® Method 10-107-04-4-A; Lachat Instruments: Loveland, CO, USA, 2003. [Google Scholar]
- Smucker, N.J.; Pilgrim, E.M.; Nietch, C.T.; Darling, J.A.; Johnson, B.R. DNA metabarcoding effectively quantifies diatom responses to nutrients in streams. Ecol. Appl. 2020, 30, e02205. [Google Scholar] [CrossRef]
- Werner, J.J.; Loren, O.; Hugenholtz, P.; DeSantis, T.Z.; Walters, W.A.; Caporaso, J.G.; Angenent, L.T.; Knight, R.; Ley, R.E. Impact of training sets on classification of high-throughput bacterial 16S rRNA gene surveys. ISME J. 2012, 6, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Hall, E.K.; Bernhardt, E.S.; Bier, R.L.; Bradford, M.A.; Boot, C.M.; Cotner, J.B.; del Giorgio, P.A.; Evans, S.E.; Graham, E.B.; Jones, S.E.; et al. Understanding how microbiomes influence the systems they inhabit. Nat. Microbiol. 2018, 3, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- McCune, B. ; Grace., J.B. Analysis of Ecological Communities; MjM Software Design: Gleneden Beach, OR, USA, 2002. [Google Scholar]
- Lavoie, I.; Dillon, P.J.; Campeau, S. The effect of excluding diatom taxa and reducing taxonomic resolution on multivariate analyses and stream bioassessment. Ecol. Indic. 2009, 9, 213–225. [Google Scholar] [CrossRef]
- Rimet, F.; Bouchez, A. Biomonitoring river diatoms: Implications of taxonomic resolution. Ecol. Indic. 2012, 15, 92–99. [Google Scholar] [CrossRef]
- Poos, M.S.; Jackson, D.A. Addressing the removal of rare species in multivariate bioassessments: The impact of methodological choices. Ecol. Indic. 2012, 18, 82–90. [Google Scholar] [CrossRef]
- Rothenberger, M.B.; Swaffield, T.; Calomeni, A.J.; Cabrey, C.D. Multivariate analysis of water quality and plankton assemblages in an urban estuary. Estuaries Coasts 2014, 37, 695–711. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. 2020. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 17 July 2020).
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org (accessed on 14 May 2021).
- King, R.S.; Baker, M.E. Use, misuse, and limitations of threshold indicator taxa analysis (TITAN) for natural resource management. In Application of Threshold Concepts in Natural Resource Decision Making; Guntenspergen, G.R., Ed.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Baker, M.E.; King, R.S.; Kahle, D. TITAN2: Threshold Indicator Taxa Analysis. 2015. Available online: https://cran.r-project.org/web/packages/TITAN2/TITAN2.pdf (accessed on 17 July 2020).
- Karr, J.R. Assessment of biotic integrity using fish communities. Fisheries 1981, 6, 21–27. [Google Scholar] [CrossRef]
- DeShon, J.E. Development and application of the invertebrate community index (ICI). In Biological Assessment and Criteria: Tools for Water Resource Planning and Decision Making; Davis, W.S., Simon, T.P., Eds.; Lewis: Boca Raton, FL, USA, 1995; pp. 217–244. [Google Scholar]
- Hill, B.H.; Herlihy, A.T.; Kaufmann, P.R.; Stevenson, R.J.; McCormick, F.H.; Johnson, C.B. Use of periphyton assemblage data as an index of biotic integrity. J. N. Am. Benthol. Soc. 2000, 19, 50–67. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Phillips, S.; Leathwick, J.; Elith, J. Dismo: Species Distribution Modeling. 2017. Available online: https://cran.r-project.org/web/packages/dismo/dismo.pdf (accessed on 17 July 2020).
- Elith, J.; Leathwick, J.R.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetic analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanish, L.F.; O’Neill, S.P.; Gonzalez, A.; Legg, T.M.; Knelman, J.; McKnight, D.M.; Spaulding, S.; Nemergut, D.R. Bacteria and diatom co-occurrence patterns in microbial mats from polar desert streams. Environ. Microbiol. 2012, 15, 1115–1131. [Google Scholar] [CrossRef]
- Wyatt, K.H.; Turetsky, M.R. Algae alleviate carbon limitation of heterotrophic bacteria in a boreal peatland. J. Ecol. 2015, 103, 1165–1171. [Google Scholar] [CrossRef] [Green Version]
- Koedooder, C.; Stock, W.; Willems, A.; Mangelinckx, S.; de Troch, M.; Vyverman, W.; Sabbe, K. Diatom-bacteria interactions modulate the composition and productivity of benthic diatom biofilms. Front. Microbiol. 2019, 10, 1255. [Google Scholar] [CrossRef] [Green Version]
- Beeckmans, S.; Xie, J.P. Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Espeland, E.M.; Francoeur, S.N.; Wetzel, R.G. Influence of algal photosynthesis on biofilm bacterial production and associated glucosidase and xylosidase activities. Microb. Ecol. 2001, 42, 524–530. [Google Scholar] [CrossRef]
- Rier, S.T.; Kuehn, K.A.; Francoeur, S.N. Algal regulation of extracellular enzyme activity in stream microbial communities associated with inert substrata and detritus. J. N. Am. Benthol. Soc. 2007, 26, 439–449. [Google Scholar] [CrossRef]
- Smucker, N.J.; Vis, M.L. Acid mine drainage affects the development and function of epilithic biofilms in streams. J. N. Am. Benthol. Soc. 2011, 30, 728–738. [Google Scholar] [CrossRef] [Green Version]
- Pope, C.A.; Halvorson, H.M.; Findlay, R.H.; Francoeur, S.N.; Kuehn, K.A. Light and temperature mediate algal stimulation of heterotrophic activity on decomposing leaf litter. Freshwater Biol. 2020, 65, 1210–1222. [Google Scholar] [CrossRef]
- Stein, E.D.; White, B.P.; Mazor, R.D.; Miller, P.E.; Pilgrim, E.M. Evaluating ethanol-based sample preservation to facilitate the use of DNA barcoding in routine freshwater biomonitoring programs using benthic macroinvertebrates. PLoS ONE 2013, 8, e51273. [Google Scholar] [CrossRef] [Green Version]
- Gaget, V.; Keulen, A.; Lau, M.; Monis, P.; Brookes, J.D. DNA extraction from benthic cyanobacteria: Comparative assessment and optimization. J. Appl. Microbiol. 2017, 122, 294–304. [Google Scholar] [CrossRef]
- Majaneva, M.; Diserud, O.H.; Eagle, S.H.C.; Hajibabaei, M.; Ekrem, T. Choice of DNA extraction method affects DNA metabarcoding of unsorted invertebrate bulk samples. Metabarcoding Metagenom. 2018, 2, e26664. [Google Scholar] [CrossRef]
- Gibson, J.F.; Shokralla, S.; Curry, C.; Baird, D.J.; Monk, W.A.; King, I.; Hajibabaei, M. Large-scale biomonitoring of remote and threatened ecosystems via high-throughput sequencing. PLoS ONE 2015, 10, e0138432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbrecht, V.; Vamos, E.E.; Meissner, K.; Aroviita, J.; Leese, F. Assessing strengths and weaknesses of DNA metabarcoding-based macroinvertebrate identification for routine stream monitoring. Methods Ecol. Evol. 2017, 8, 1265–1275. [Google Scholar] [CrossRef] [Green Version]
- Vasselon, V.; Bouchez, A.; Rimet, F.; Jacquet, S.; Trobajo, R.; Corniquel, M.; Tapolczai, K.; Domaizon, I. Avoiding quantification bias in metabarcoding: Application of a cell biovolume correction factor in diatom molecular biomonitoring. Methods Ecol. Evol. 2018, 9, 1060–1069. [Google Scholar] [CrossRef] [Green Version]
- Mathon, L.; Valentini, A.; Guerin, P.-E.; Normandeau, E.; Noel, C.; Lionnet, C.; Boulanger, E.; Thuiller, W.; Bernatchez, L.; Mouillot, D.; et al. Benchmarking bioinformatic tools for fast and accurate eDNA metabarcoding species identification. Mol. Ecol. Resour. 2021, 21, 2565–2579. [Google Scholar] [CrossRef]
- Trebitz, A.S.; Hoffman, J.C.; Grant, G.W.; Billehaus, T.M.; Pilgrim, E.M. Potential for DNA-based identification of Great Lakes fauna: Match and mismatch between taxa inventories and DNA barcode libraries. Sci. Rep. 2015, 5, 12162. [Google Scholar] [CrossRef] [Green Version]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Handelsman, J. Status of the microbial census. Microbiol. Mol. Biol. Rev. 2004, 68, 686–691. [Google Scholar] [CrossRef] [Green Version]
- Cordier, T.; Lanzen, A.; Apotheloz-Perret-Gentil, L.; Stoeck, T.; Pawlowski, J. Embracing environmental genomics and machine learning for routine biomonitoring. Trends Microbiol. 2019, 27, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Zolkefli, N.; Sharuddin, S.S.; Yusoff, M.Z.M.; Hassan, M.A.; Maeda, T.; Ramli, N. A review of current and emerging approaches for water pollution monitoring. Water 2020, 12, 3417. [Google Scholar] [CrossRef]
- Michan, C.; Blasco, J.; Alhama, J. High-throughput molecular analyses of microbiomes as a tool to monitor the wellbeing of aquatic environments. Microb. Biotechnol. 2021, 14, 870–885. [Google Scholar] [CrossRef]
- Stoeck, T.; Frühe, L.; Forster, D.; Cordier, T.; Martins, C.I.M.; Pawlowski, J. Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Mar. Pollut. Bull. 2018, 127, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Keeley, N.; Wood, S.A.; Pochon, X. Development and preliminary validation of a multi-trophic metabarcoding biotic index for monitoring benthic organic enrichment. Ecol. Indic. 2018, 85, 1044–1057. [Google Scholar] [CrossRef]
- Dully, V.; Balliet, H.; Frühe, L.; Däumer, M.; Thielen, A.; Gallie, S.; Berrill, I.; Stoeck, T. Robustness, sensitivity and reproducibility of eDNA metabarcoding as an environmental biomonitoring tool in coastal salmon aquaculture—An inter-laboratory study. Ecol. Indic. 2021, 121, 107049. [Google Scholar] [CrossRef]
- Aylagas, E.; Atalah, J.; Sanchez-Jerez, P.; Pearman, J.K.; Casado, N.; Asensi, J.; Toledo-Guedes, K.; Carvalho, S. A step towards the validation of bacteria biotic indices using DNA metabarcoding for benthic monitoring. Mol. Ecol. Resour. 2021, 21, 1889–1903. [Google Scholar] [CrossRef]
- Smucker, N.J.; Pilgrim, E.M.; Wu, H.; Nietch, C.T.; Darling, J.A.; Molina, M.; Johnson, B.R.; Yuan, L.L. Characterizing temporal variability in streams supports nutrient indicator development using diatom and bacterial DNA metabarcoding. Sci. Total Environ. 2022, 154960. [Google Scholar] [CrossRef] [PubMed]
- Laroche, O.; Wood, S.A.; Tremblay, L.A.; Ellis, J.I.; Lear, G.; Pochon, X. A cross-taxa study using environmental DNA/RNA metabarcoding to measure biological impacts of offshore oil and gas drilling and production operations. Mar. Pollut. Bull. 2018, 127, 97–107. [Google Scholar] [CrossRef]
- Torres, G.G.; Figueroa-Galvis, I.; Munoz-Garcia, A.; Polania, J.; Vanegas, J. Potential bacterial bioindicators of urban pollution in mangroves. Environ. Pollut. 2019, 255 Pt 2, 113293. [Google Scholar] [CrossRef]
- da Costa Silva, T.A.; de Paula, M., Jr.; Silva, W.S.; Lacorte, G.A. Can moderate heavy metal soil contaminations due to cement production influence the surrounding soil bacterial communities? Ecotoxicology 2022, 31, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Pin, L.; Eiler, A.; Fazi, S.; Friberg, N. Two different approaches of microbial community structure characterization in riverine epilithic biofilms under multiple stressors conditions: Developing molecular indicators. Mol. Ecol. Resour. 2021, 21, 1200–1215. [Google Scholar] [CrossRef] [PubMed]
- Graves, C.J.; Makrides, E.J.; Schmidt, V.T.; Giblin, A.E.; Cardon, Z.G.; Rand, D.M. Functional response of salt marsh microbial communities to long-term nutrient enrichment. Appl. Environ. Microbiol. 2016, 82, 2862–2871. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Liu, G.; Chen, H.; Chen, C.; Wang, J.; Ai, S.; Wei, D.; Li, D.; Ma, B.; Tang, C.; et al. Long-term nutrient inputs shift soil microbial functional profiles of phosphors cycling in diverse agroecosystems. ISME J. 2020, 14, 757–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Barberan, A.; Borer, E.T.; Firn, J.L.; Harpole, W.S.; Hobbie, S.E.; Hofmockel, K.S.; Knops, J.M.H.; et al. Consistent response of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA 2015, 112, 10967–10972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Jing, H.; Xia, X.; Cheung, S.; Suzuki, K.; Liu, H. Metagenomic insights into the microbial community and nutrient cycling in the western subarctic Pacific Ocean. Front. Microbiol. 2018, 9, 623. [Google Scholar] [CrossRef] [PubMed]
- Bailet, B.; Bouchez, A.; Franc, A.; Frigerio, J.M.; Keck, F.; Karjalainen, S.M.; Rimet, F.; Schneider, S.; Kahler, M. Molecular versus morphological data for benthic diatoms biomonitoring in Northern Europe freshwater and consequences for ecological status. Metabarcoding Metagenom. 2019, 3, e34002. [Google Scholar] [CrossRef]


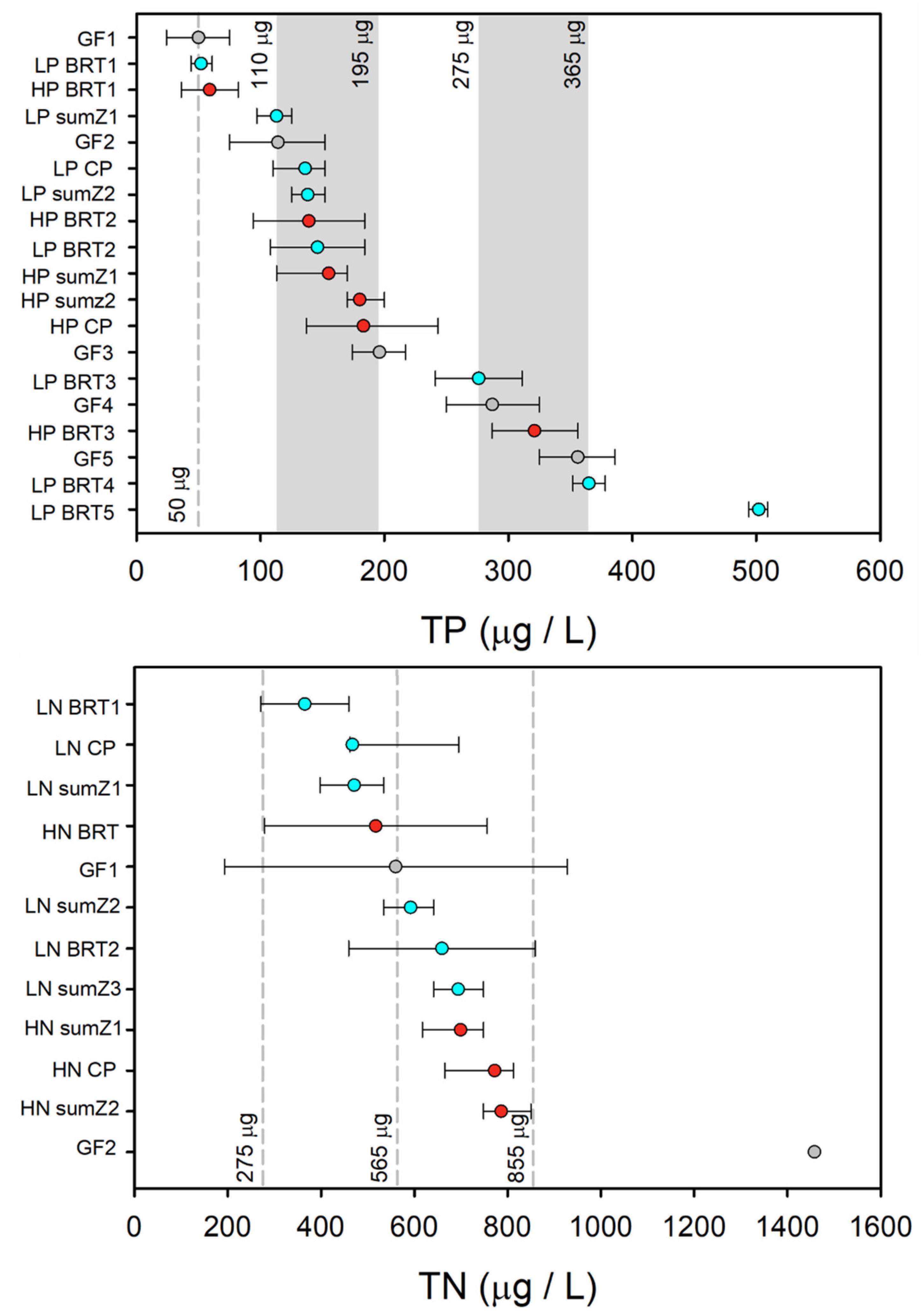
| Start of Response | Mid-Response | End of Response | |
|---|---|---|---|
| Response | (µg/L) | (µg/L) | (µg/L) |
| GF TP1 | 24 | 50 | 75 |
| BRT LP1 | 44 | 52 | 61 |
| BRT HP1 | 36 | 59 | 82 |
| LP sumz1 | 97 | 113 | 125 |
| GF TP2 | 75 | 114 | 152 |
| LP CP | 110 | 136 | 152 |
| LP sumz2 | 125 | 138 | 152 |
| BRT HP2 | 94 | 139 | 184 |
| BRT LP2 | 108 | 146 | 184 |
| HP sumz1 | 113 | 155 | 170 |
| HP sumz2 | 170 | 180 | 200 |
| HP CP | 137 | 183 | 243 |
| GF TP3 | 174 | 196 | 217 |
| BRT LP3 | 241 | 276 | 311 |
| GF TP4 | 250 | 287 | 325 |
| BRT HP3 | 287 | 321 | 356 |
| GF TP5 | 325 | 356 | 386 |
| BRT LP4 | 352 | 365 | 378 |
| BRT LP5 | 494 | 502 | 509 |
| Start of Response | Mid-Response | End of Response | |
|---|---|---|---|
| Response | (µg/L) | (µg/L) | (µg/L) |
| BRT LN1 | 271 | 365 | 459 |
| LN CP | 462 | 467 | 695 |
| LN sumz1 | 398 | 471 | 534 |
| BRT HN | 279 | 517 | 756 |
| GF1 | 193 | 560 | 928 |
| LN sumz2 | 534 | 592 | 641 |
| BRT LN2 | 459 | 659 | 859 |
| LN sumz3 | 641 | 694 | 748 |
| HN sumz1 | 617 | 699 | 748 |
| HN CP | 665 | 772 | 812 |
| HN sumz2 | 748 | 786 | 850 |
| GF2 | - | 1458 | - |
| % ag | TN | TP | |
|---|---|---|---|
| Low P | −0.84 | −0.59 | −0.68 |
| High P | 0.86 | 0.62 | 0.71 |
| Low N | −0.77 | −0.64 | −0.47 |
| High N | 0.88 | 0.64 | 0.71 |
| TP | 0.85 | 0.66 | |
| TN | 0.65 |
| Relative Importance (%) | ||||||
|---|---|---|---|---|---|---|
| Observed Deviance Explained (%) | CV Deviance Explained (%) | CV Correlation | TP | TN | EC | |
| Low P ASVs | 0.59 | 0.45 ± 0.05 | 0.69 ± 0.02 | 55.0 | 25.1 | 19.9 |
| High P ASVs | 0.67 | 0.56 ± 0.06 | 0.77 ± 0.02 | 46.2 | 28.8 | 25 |
| Low N ASVs | 0.54 | 0.46 ± 0.08 | 0.70 ± 0.04 | 13.7 | 76.2 | 10.1 |
| High N ASVs | 0.60 | 0.50 ± 0.10 | 0.79 ± 0.03 | 40.5 | 24.6 | 34.9 |
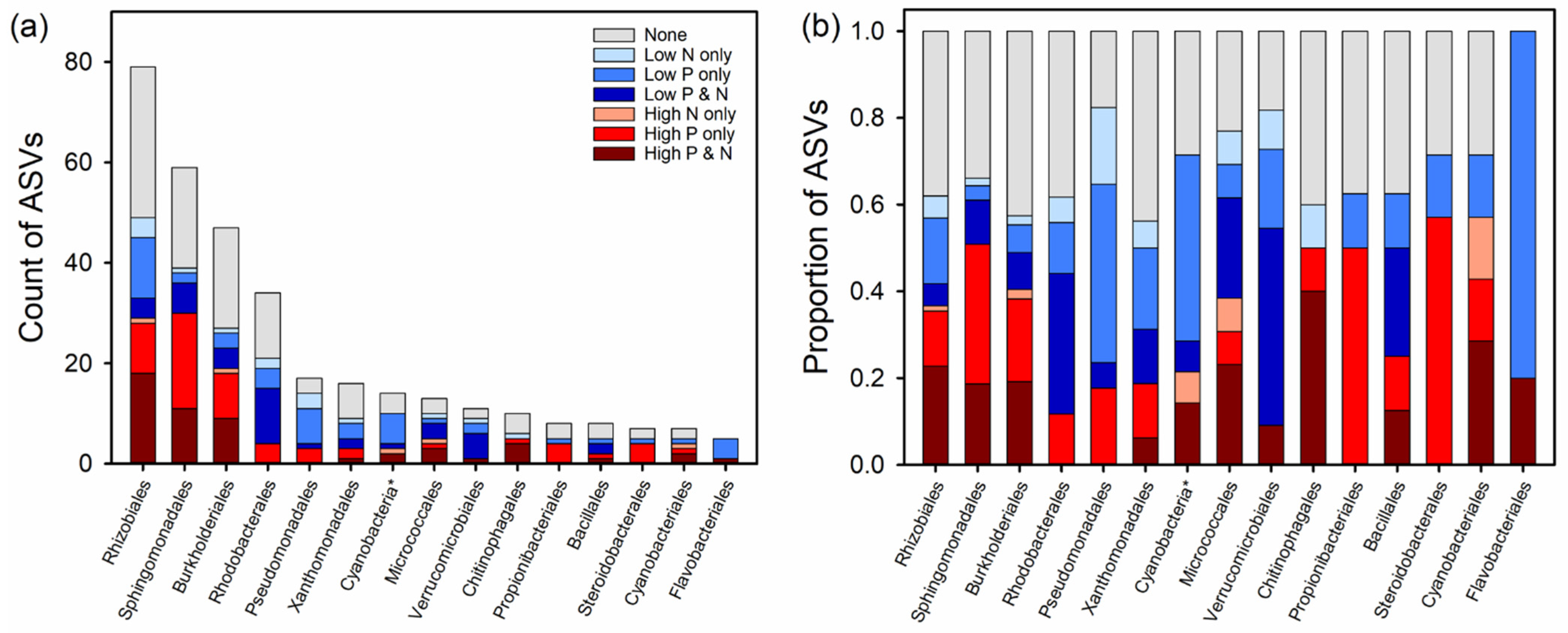
| Phylum | Order | Total | No. Indicators | High P and N | High P Only | High N Only | Low P and N | Low P Only | Low N Only |
|---|---|---|---|---|---|---|---|---|---|
| Proteo- | Rhizobiales | 79 | 49 | 18 | 10 | 1 | 4 | 12 | 4 |
| bacteria | Sphingomonadales | 59 | 39 | 11 | 19 | 6 | 2 | 1 | |
| Burkholderiales | 47 | 27 | 9 | 9 | 1 | 4 | 3 | 1 | |
| Rhodobacterales | 34 | 21 | 4 | 11 | 4 | 2 | |||
| Pseudomonadales | 16 | 13 | 3 ** | 1 | 7 | 3 ** | |||
| Xanthomonadales | 16 | 9 | 1 | 2 | 2 | 3 | 1 | ||
| Caulobacterales | 12 | 4 | 1 | 3 | |||||
| Steroidobacterales | 7 | 5 | 4 | 1 | |||||
| Acetobacterales | 3 | 2 | 1 | 1 | |||||
| Enterobacterales | 3 | 1 | 1 | ||||||
| Gammaproteobacteria * | 2 | 1 | 1 | ||||||
| Azospirillales | 1 | 1 | 1 | ||||||
| Reyranellales | 1 | 1 | 1 | ||||||
| Rickettsiales | 1 | 1 | 1 | ||||||
| Alphaproteobacteria * | 1 | 1 | 1 | ||||||
| “PLTA13” | 1 | 1 | 1 | ||||||
| “PHOS-HD29” | 1 | 1 | 1 | ||||||
| Tistrellales | 1 | 0 | |||||||
| Actino- | Micrococcales | 13 | 10 | 3 | 1 | 1 | 3 | 1 | 1 |
| bacteriota | Propionibacteriales | 8 | 5 | 4 | 1 | ||||
| Microtrichales | 8 | 4 | 2 | 1 | 1 | ||||
| Corynebacteriales | 5 | 3 | 1 | 1 | 1 | ||||
| “PeM15” | 3 | 1 | 1 | ||||||
| Gaiellales | 3 | 1 | 1 | ||||||
| Solirubrobacterales | 3 | 1 | 1 | ||||||
| Actinobacteriota * | 3 | 0 | |||||||
| Frankiales | 1 | 1 | 1 | ||||||
| Kineosporiales | 1 | 1 | 1 | ||||||
| Cyano- | Cyanobacteria * | 14 | 10 | 2 | 1 | 1 | 6 | ||
| bacteria | Cyanobacteriales | 7 | 5 | 2 | 1 | 1 | 1 | ||
| “SepB-3” | 4 | 2 | 1 | 1 | |||||
| Chroococcales | 1 | 1 | 1 | ||||||
| Bactero- | Chitinophagales | 10 | 6 | 4 | 1 | 1 | |||
| idota | Flavobacteriales | 5 | 5 | 1 | 4 | ||||
| Cytophagales | 1 | 0 | |||||||
| Firmicutes | Bacillales | 8 | 5 | 1 | 1 | 2 | 1 | ||
| Exiguobacterales | 4 | 3 | 3 | ||||||
| Alicyclobacillales | 1 | 1 | 1 | ||||||
| Lactobacillales | 1 | 0 | |||||||
| Plancto- | Pirellulales | 7 | 4 | 1 | 2 | 1 | |||
| mycetota | Gemmatales | 5 | 3 | 1 | 2 | ||||
| Isosphaerales | 1 | 1 | 1 | ||||||
| Planctomycetales | 1 | 1 | 1 | ||||||
| Verruco-microbiota | Verrucomicrobiales | 11 | 9 | 1 | 5 | 2 | 1 | ||
| Chloroflexi | Chloroflexi * | 4 | 2 | 1 | 1 | ||||
| Acido- | Vicinamibacterales | 2 | 2 | 2 | |||||
| bacteriota | Blastocatellales | 2 | 0 | ||||||
| Gemmati-monadota | Gemmatimonadales | 2 | 1 | 1 | |||||
| Desulfo-bacterota | Desulfobulbales | 1 | 1 | 1 | |||||
| Thermi/Deinococci | Deinococcales | 1 | 1 | 1 | |||||
| Fuso-bacteriota | Fusobacteriales | 1 | 0 | ||||||
| Myxococcota | Myxococcota * | 1 | 0 | ||||||
| Nitrospirota | Nitrospirales | 1 | 0 | ||||||
| 429 | 267 | 67 | 71 | 7 | 45 | 60 | 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilgrim, E.M.; Smucker, N.J.; Wu, H.; Martinson, J.; Nietch, C.T.; Molina, M.; Darling, J.A.; Johnson, B.R. Developing Indicators of Nutrient Pollution in Streams Using 16S rRNA Gene Metabarcoding of Periphyton-Associated Bacteria. Water 2022, 14, 2361. https://doi.org/10.3390/w14152361
Pilgrim EM, Smucker NJ, Wu H, Martinson J, Nietch CT, Molina M, Darling JA, Johnson BR. Developing Indicators of Nutrient Pollution in Streams Using 16S rRNA Gene Metabarcoding of Periphyton-Associated Bacteria. Water. 2022; 14(15):2361. https://doi.org/10.3390/w14152361
Chicago/Turabian StylePilgrim, Erik M., Nathan J. Smucker, Huiyun Wu, John Martinson, Christopher T. Nietch, Marirosa Molina, John A. Darling, and Brent R. Johnson. 2022. "Developing Indicators of Nutrient Pollution in Streams Using 16S rRNA Gene Metabarcoding of Periphyton-Associated Bacteria" Water 14, no. 15: 2361. https://doi.org/10.3390/w14152361






