Increasing Hydrostatic Pressure Impacts the Prokaryotic Diversity during Emiliania huxleyi Aggregates Degradation
Abstract
1. Introduction
2. Materials and Methods
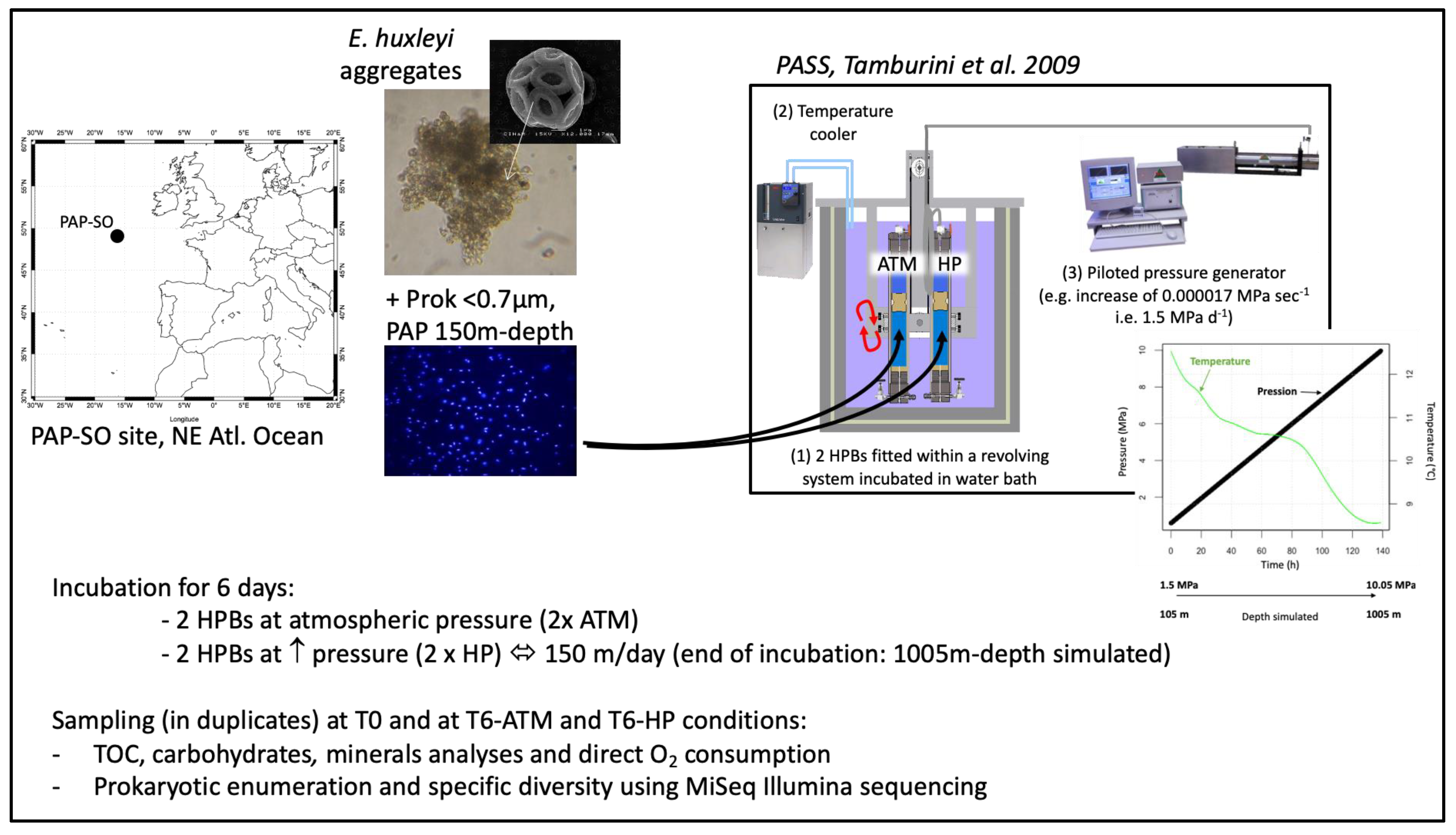
2.1. SINking PArticles Simulation Experiments (SINPAS Experiments)
2.2. Biogeochemical Analyses
2.3. 16S rRNA Extraction, PCR Amplification, and Sequencing
2.4. Analysis of the 16S rRNA-Based Community
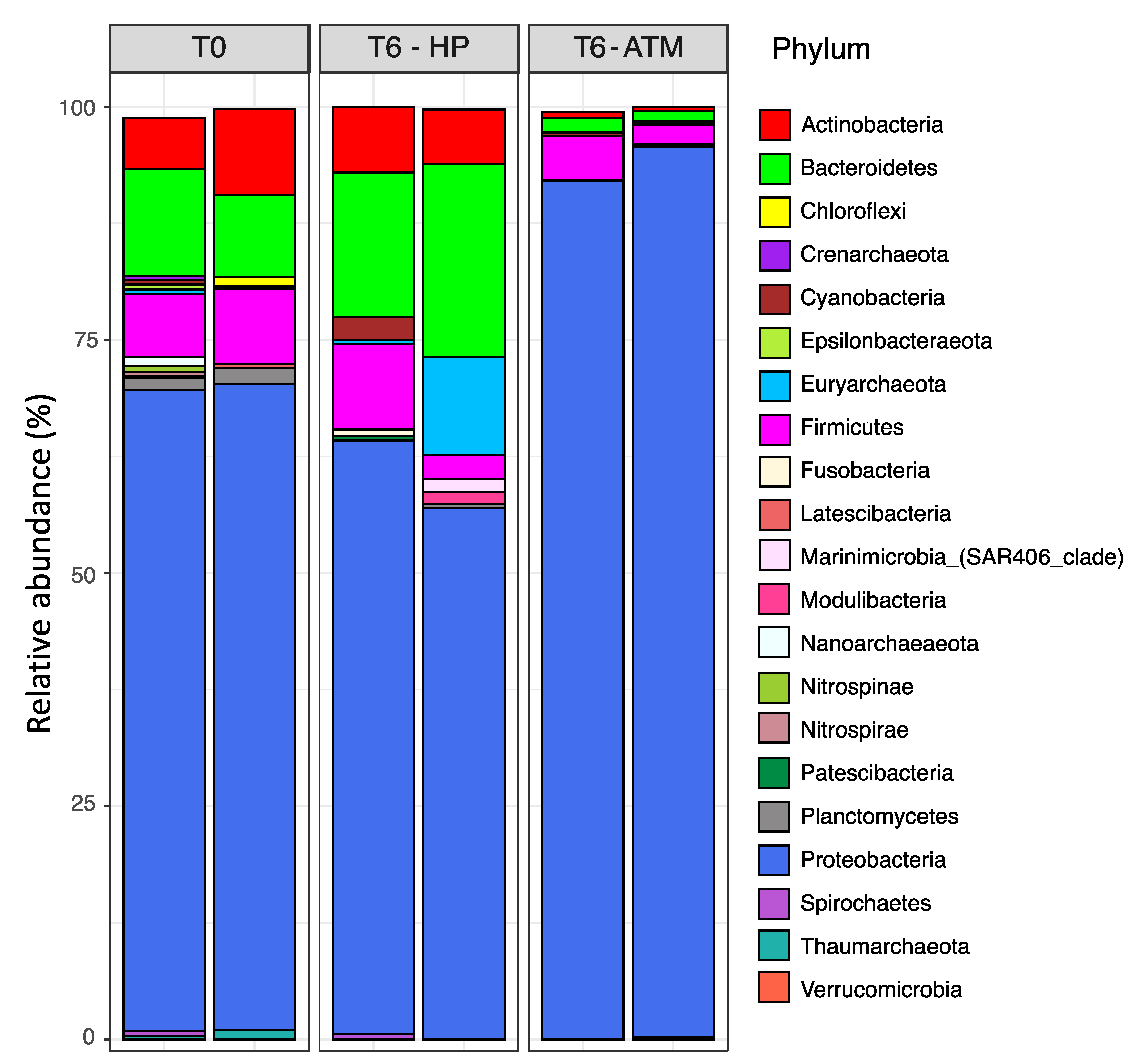
3. Results and Discussion
3.1. Biogeochemistry of E. huxleyi Particles in Sinking Particle Simulation Experiments
3.2. Prokaryotic Activity Associated with E. huxleyi Particles in Sinking Particles Simulation Experiments
| References | Aggregates | Pressure Simulated | Depth Simulated | Temperature | Sinking Rate | Dissolved O2 Measurement | Microbial Diversity | Main Results |
|---|---|---|---|---|---|---|---|---|
| [36] | Aggregate-forming diatom T. wessflogii | 2–14 MPa | 200–1400 m | 13 °C | Pressure increase of 1.5 MPa each day | ND | CARD-FISH | Increasing pressure decreased silica dissolution and aminopeptidase activity (relatively to constant ATM conditions) and increased Bacteroidetes abundance |
| [34] | Fecal pellets | 2–15 MPa | 200–1500 m | 13–13.4 °C | Continuously pressurized at a rate of 2 MPa d−1 | ND | CARD-FISH | Particulate carbohydrate, chloropigments and TEP decreased more slowly under HP than ATM condition |
| [35] | Aggregate-forming coccolithophorid E. huxleyii | 1–17 MPa | 100–1700 m | 13 °C | Continuously pressurized at a rate of 1.5 MPa d−1 | ND | CARD-FISH | Increasing pressure enhanced dissolution of calcite and particle aggregation (relatively to constant ATM conditions), and decreased 𝛼-Proteobacteria abundance |
| [67] | Synthetic inorganic Ca13CO3 (calcite) | 25 MPa | 2500 m | 21 °C | ND | ND | ND | Increasing pressure promoted calcite dissolution at 7 MPa |
| [38] | Culture of Thalassiosira weissflogii | 10 MPa | 1000 m | ND | Continuously pressurized at a rate of 3 MPa d−1 | ND | ND | Increasing pressure enhanced particulate inorganic carbon dissolution |
| [70] | No aggregates | 40 MPa | 4000 m | 20.5 °C | Pressure increase of 10 MPa each day | Yes | FISH * | Increasing pressure inhibited growth of surface-originated bacteria and decreased 𝛼-Proteobacteria et Bacteroidetes abundance |
| [71] | Aggregate-forming diatom Nannochloropsis and Tetraselmis algae | 30 MPa | 3000 m | 4 °C | Pressure increase of 2.5 MPa every 15 min | Yes | ND | Increasing pressure promoted particulate inorganic carbon dissolution |
| [72] | Diatom-bacteria aggregates | 10 to 100 MPa | 100–10,000 m | 3 °C | Pressure increase or decrease of 10 MPa during 15–20 min | Yes | ND | Increasing pressure inhibited respiration on surface prokaryotic assemblage |
| [72] | Aggregate-forming diatom Skeletonema marinoi | 10 to 100 MPa | 100–10,000 m | 3 °C | Pressure increase or decrease of 10 MPa during 15–20 min | Yes | ND | Increasing pressure inhibited respiration on surface prokaryotic assemblage |
| [This study] | Aggregate-forming coccolithophorid E. huxleyii | 1.05–10.05 MPa | 105–1005 m | 13–8.5 °C | Continuously pressurized at a rate of 1.5 MPa d−1 | Yes | Metabarcoding | Increasing pressure inhibited respiration on surface prokaryotic assemblage, reduced both the phylogenetic diversity and the species richness (specifically Bacteroidetes) |
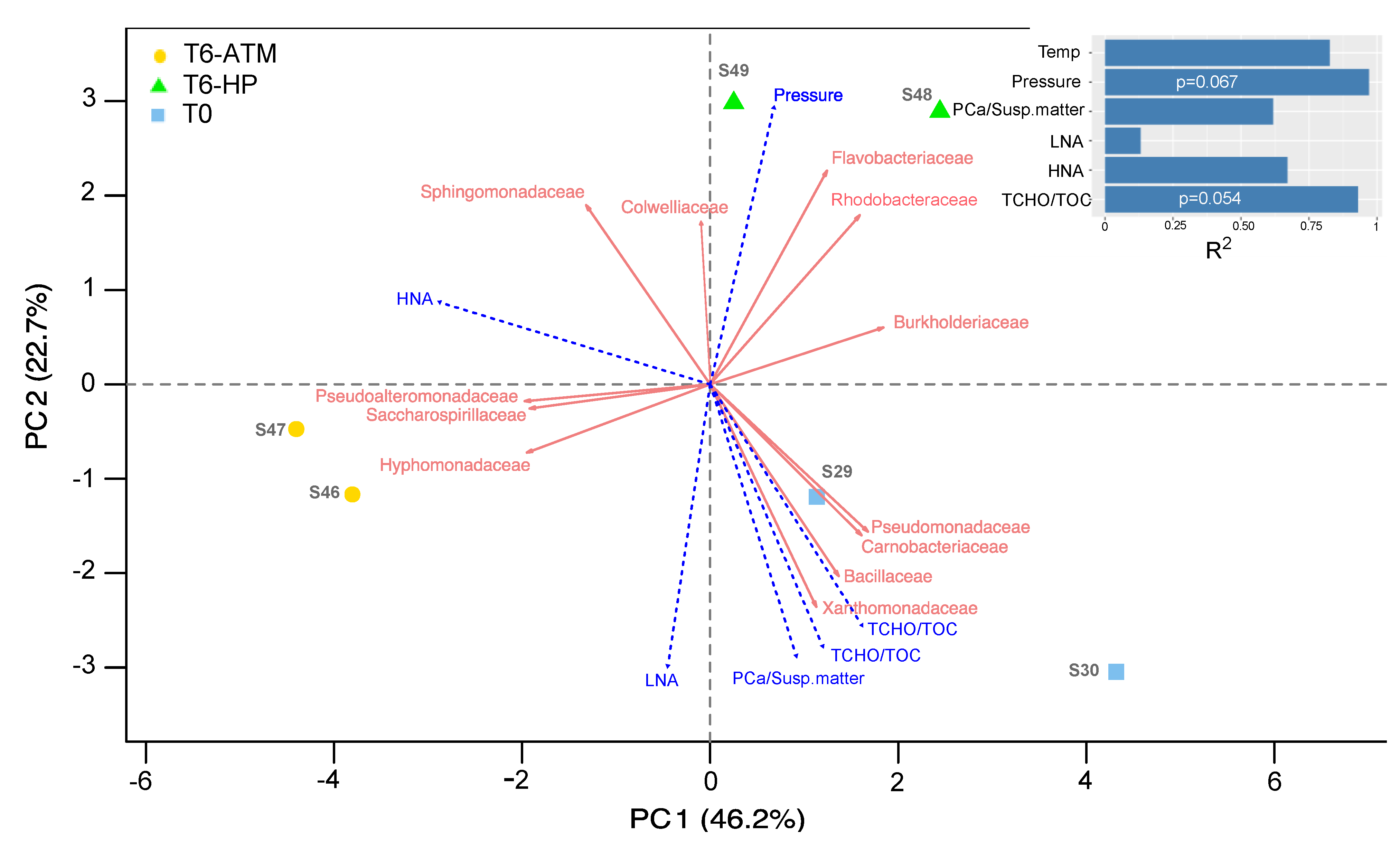
3.3. Effect of the Increasing Pressure on the Active Prokaryotic Communities
3.4. Factors Driving the Active Prokaryotic Communities
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eppley, R.W.; Peterson, B.J. Particulate organic matter flux and planktonic new production in the deep ocean. Nature 1979, 282, 677–680. [Google Scholar] [CrossRef]
- Siegel, D.A.; Buesseler, K.O.; Behrenfeld, M.J.; Benitez-Nelson, C.R.; Boss, E.; Brzezinski, M.A.; Burd, A.; Carlson, C.A.; D’Asaro, E.A.; Doney, S.C.; et al. Prediction of the Export and Fate of Global Ocean Net Primary Production: The EXPORTS Science Plan. Front. Mar. Sci. 2016, 3, 22. [Google Scholar] [CrossRef]
- Boyd, P.W.; Claustre, H.; Levy, M.; Siegel, D.A.; Weber, T. Multi-faceted particle pumps drive carbon sequestration in the ocean. Nature 2019, 568, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Le Moigne, F.A.C. Pathways of organic carbon downward transport by the oceanic biological carbon pump. Front. Mar. Sci. 2019, 6, 634. [Google Scholar] [CrossRef]
- Briggs, N.; Dall’Olmo, G.; Claustre, H. Major role of particle fragmentation in regulating biological sequestration of CO2 by the oceans. Science 2020, 367, 791–793. [Google Scholar] [CrossRef]
- Kiko, R.; Brandt, P.; Christiansen, S.; Faustmann, J.; Kriest, I.; Rodrigues, E.; Schütte, F.; Hauss, H. Zooplankton-mediated fluxes in the Eastern Tropical North Atlantic. Front. Mar. Sci. 2020, 7, 358. [Google Scholar] [CrossRef]
- Hernández-León, S.; Koppelmann, R.; Fraile-Nuez, E.; Bode, A.; Mompeán, C.; Irigoien, X.; Olivar, M.P.; Echevarría, F.; Fernández de Puelles, M.L.; González-Gordillo, J.I.; et al. Large deep-sea zooplankton biomass mirrors primary production in the global ocean. Nat. Commun. 2020, 11, 6048. [Google Scholar] [CrossRef]
- Steinberg, D.K.; Van Mooy, B.A.S.; Buesseler, K.O.; Boyd, P.W.; Kobari, T.; Karl, D.M. Bacterial vs. zooplankton control of sinking particle flux in the ocean’s twilight zone. Limnol. Oceanogr. 2008, 53, 1327–1338. [Google Scholar] [CrossRef]
- Simon, M.; Grossart, H.; Schweitzer, B.; Ploug, H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 2002, 28, 175–211. [Google Scholar] [CrossRef]
- Giering, S.L.C.; Sanders, R.; Lampitt, R.S.; Anderson, T.R.; Tamburini, C.; Boutrif, M.; Zubkov, M.V.; Marsay, C.M.; Henson, S.A.; Saw, K.; et al. Reconciliation of the carbon budget in the ocean’s twilight zone. Nature 2014, 507, 480–483. [Google Scholar] [CrossRef]
- Martin, A.; Boyd, P.; Buesseler, K.; Cetinic, I.; Claustre, H.; Giering, S.; Henson, S.; Irigoien, X.; Kriest, I.; Memery, L.; et al. The oceans’ twilight zone must be studied now, before it is too late. Nature 2020, 580, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.K.; Fu, W.; Primeau, F.; Britten, G.L.; Lindsay, K.; Long, M.; Doney, S.C.; Mahowald, N.; Hoffman, F.; Randerson, J.T. Sustained climate warming drives declining marine biological productivity. Science 2018, 359, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Bopp, L.; Resplandy, L.; Orr, J.C.; Doney, S.C.; Dunne, J.P.; Gehlen, M.; Halloran, P.; Heinze, C.; Ilyina, T.; Séférian, R.; et al. Multiple stressors of ocean ecosystems in the 21st century: Projections with CMIP5 models. Biogeosciences 2013, 10, 6225–6245. [Google Scholar] [CrossRef]
- Ibarbalz, F.M.; Henry, N.; Brandão, M.C.; Martini, S.; Busseni, G.; Byrne, H.; Coelho, L.P.; Endo, H.; Gasol, J.M.; Gregory, A.C.; et al. Global Trends in Marine Plankton Diversity across Kingdoms of Life. Cell 2019, 179, 1084–1097.e21. [Google Scholar] [CrossRef]
- Baumas, C.M.J.; Le Moigne, F.A.C.; Garel, M.; Bhairy, N.; Guasco, S.; Riou, V.; Armougom, F.; Grossart, H.-P.; Tamburini, C. Mesopelagic microbial carbon production correlates with diversity across different marine particle fractions. ISME J. 2021, 15, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Henson, S.A.; Koski, M.; De La Rocha, C.L.; Painter, S.C.; Poulton, A.J.; Riley, J.; Salihoglu, B.; Visser, A.; Yool, A.; et al. The Biological Carbon Pump in the North Atlantic. Prog. Oceanogr. 2014, 129, 200–218. [Google Scholar] [CrossRef]
- Le Moigne, F.A.C.; Villa-Alfageme, M.; Sanders, R.J.; Marsay, C.; Henson, S.; García-Tenorio, R. Export of organic carbon and biominerals derived from 234Th and 210Po at the Porcupine Abyssal Plain. Deep. Res. Part. I Oceanogr. Res. Pap. 2013, 72, 88–101. [Google Scholar] [CrossRef]
- Ingalls, A.E.; Lee, C.; Wakeham, S.G.; Hedges, J.I. The role of biominerals in the sinking flux and preservation of amino acids in the Southern Ocean along 170[deg]W. Deep. Res. II 2003, 50, 713–738. [Google Scholar] [CrossRef]
- Hedges, J.I.; Baldock, J.A.; Gelinas, Y.; Lee, C.; Peterson, M.; Wakeham, S.G. Evidence for non-selective preservation of organic matter in sinking marine particles. Nature 2001, 409, 801–804. [Google Scholar] [CrossRef]
- Armstrong, R.A.; Lee, C.; Hedges, J.I.; Honjo, S.; Wakeham, S.G. A new, mechanistic model for organic carbon fluxes in the ocean based on the quantitative association of POC with ballast minerals. Deep. Res. II Top. Stud. Oceanogr. 2002, 49, 219–236. [Google Scholar] [CrossRef]
- Bressac, M.; Guieu, C.; Ellwood, M.J.; Tagliabue, A.; Wagener, T.; Laurenceau-Cornec, E.C.; Whitby, H.; Sarthou, G.; Boyd, P.W. Resupply of mesopelagic dissolved iron controlled by particulate iron composition. Nat. Geosci. 2019, 12, 995–1000. [Google Scholar] [CrossRef]
- Armstrong, R.A.; Peterson, M.L.; Lee, C.; Wakeham, S.G. Settling velocity spectra and the ballast ratio hypothesis. Deep Sea Res. Part. II Top. Stud. Oceanogr. 2009, 56, 1470–1478. [Google Scholar] [CrossRef]
- Panagiotopoulos, C.; Goutx, M.; Suroy, M.; Moriceau, B. Phosphorus limitation affects the molecular composition of Thalassiosira weissflogii leading to increased biogenic silica dissolution and high degradation rates of cellular carbohydrates. Org. Geochem. 2020, 148, 104068. [Google Scholar] [CrossRef]
- Bidle, K.D.; Azam, F. Accelerated dissolution of diatom silica by marine bacterial assemblages. Nature 1999, 397, 508–512. [Google Scholar] [CrossRef]
- Bidle, K.D.; Azam, F. Bacterial control of silicon regeneration from diatom detritus: Significance of bacterial ectohydrolases and species identity. Limnol. Oceanogr. 2001, 46, 1606–1623. [Google Scholar] [CrossRef]
- Goutx, M.; Wakeham, S.G.; Lee, C.; Duflos, M.; Guigue, C.; Liu, Z.; Moriceau, B.; Sempéré, R.; Tedetti, M.; Xue, J. Composition and degradation of sinking particles with different settling velocities. Limnol. Oceanogr. 2007, 52, 1645–1664. [Google Scholar] [CrossRef]
- Bidle, K.D.; Manganelli, M.; Azam, F. Regulation of oceanic silicon and carbon preservation by temperature control on bacteria. Science 2002, 298, 1980–1984. [Google Scholar] [CrossRef]
- Le Moigne, F.A.C.; Gallinari, M.; Laurenceau, E.; De La Rocha, C.L. Enhanced rates of particulate organic matter remineralization by microzooplankton are diminished by added ballast minerals. Biogeosciences 2013, 10, 5755–5765. [Google Scholar] [CrossRef]
- Engel, A.; Szlosek, J.; Abramson, L.; Liu, Z.; Lee, C. Investigating the effect of ballasting by CaCO3 in Emiliania huxleyi: I. Formation, settling velocities and physical properties of aggregates. Deep. Res. Part. II Top. Stud. Oceanogr. 2009, 56, 1396–1407. [Google Scholar] [CrossRef]
- Bissett, A.; Neu, T.R.; de Beer, D. Dissolution of Calcite in the Twilight Zone: Bacterial Control of Dissolution of Sinking Planktonic Carbonates Is Unlikely. PLoS ONE 2011, 6, e26404. [Google Scholar] [CrossRef]
- Wollast, R.; Chou, L. Distribution and fluxes of calcium carbonate along the continental margin in the Gulf of Biscay. Aquat. Geochem. 1998, 4, 369–393. [Google Scholar] [CrossRef]
- Wollast, R.; Chou, L. The carbon cycle at the ocean margin in the northern Gulf of Biscay. Deep Sea Res. Part. II Top. Stud. Oceanogr. 2001, 48, 3265–3293. [Google Scholar] [CrossRef][Green Version]
- Sulpis, O.; Jeansson, E.; Dinauer, A.; Lauvset, S.K.; Middelburg, J.J. Calcium carbonate dissolution patterns in the ocean. Nat. Geosci. 2021, 14, 423–428. [Google Scholar] [CrossRef]
- Tamburini, C.; Goutx, M.; Guigue, C.; Garel, M.; Lefèvre, D.; Charrière, B.; Sempéré, R.; Pepa, S.; Peterson, M.L.; Wakeham, S.; et al. Effects of hydrostatic pressure on microbial alteration of sinking fecal pellets. Deep. Res. II Top. Stud. Oceanogr. 2009, 56, 1533–1546. [Google Scholar] [CrossRef]
- Riou, V.; Para, J.; Garel, M.; Guigue, C.; Al Ali, B.; Santinelli, C.; Lefèvre, D.; Gattuso, J.-P.; Goutx, M.; Jacquet, S.; et al. Biodegradation of Emiliania huxleyi aggregates by a natural Mediterranean prokaryotic community under increasing hydrostatic pressure. Prog. Oceanogr. 2018, 163, 271–281. [Google Scholar] [CrossRef]
- Tamburini, C.; Garcin, J.; Grégori, G.; Leblanc, K.; Rimmelin, P.; Kirchman, D.L. Pressure effects on surface Mediterranean prokaryotes and biogenic silica dissolution during a diatom sinking experiment. Aquat. Microb. Ecol. 2006, 43, 267–276. [Google Scholar] [CrossRef]
- Bidle, K.D.; Brzezinski, M.A.; Long, R.A.; Jones, J.L.; Azam, F. Diminished efficiency in the oceanic silica pump caused by bacterially-mediated silica dissolution. Limnol. Oceanogr. 2003, 48, 1855–1868. [Google Scholar] [CrossRef]
- de Jesus Mendes, P.A.; Thomsen, L. Effects of Ocean Acidification on the Ballast of Surface Aggregates Sinking through the Twilight Zone. PLoS ONE 2012, 7, e50865. [Google Scholar] [CrossRef]
- Turley, C.M. The effect of pressure on leucine and thymidine incorporation by free-living bacteria and by bacteria attached to sinking oceanic particles. Deep. Res. I 1993, 40, 2193–2206. [Google Scholar]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef]
- Santinelli, C.; Follett, C.; Retelletti Brogi, S.; Xu, L.; Repeta, D. Carbon isotope measurements reveal unexpected cycling of dissolved organic matter in the deep Mediterranean Sea. Mar. Chem. 2015, 177, 267–277. [Google Scholar] [CrossRef]
- Panagiotopoulos, C.; Sempéré, R.; Jacq, V.; Charrière, B. Composition and distribution of dissolved carbohydrates in the Beaufort Sea Mackenzie margin (Arctic Ocean). Mar. Chem. 2014, 166, 92–102. [Google Scholar] [CrossRef]
- Suroy, M.; Panagiotopoulos, C.; Boutorh, J.; Goutx, M.; Moriceau, B. Degradation of diatom carbohydrates: A case study with N- and Si-stressed Thalassiosira weissflogii. J. Exp. Mar. Bio. Ecol. 2015, 470, 1–11. [Google Scholar] [CrossRef]
- Jacquet, S.H.M.; Dehairs, F.; Lefèvre, D.; Cavagna, A.J.; Planchon, F.; Christaki, U.; Monin, L.; André, L.; Closset, I.; Cardinal, D. Early spring mesopelagic carbon remineralization and transfer efficiency in the naturally iron-fertilized Kerguelen area. Biogeosciences 2015, 12, 1713–1731. [Google Scholar] [CrossRef]
- Girault, M.; Arakawa, H.; Barani, A.; Ceccaldi, H.J.; Hashihama, F.; Gregori, G. Heterotrophic prokaryote distribution along a 2300 km transect in the North Pacific subtropical gyre during a strong la Niña conditions: Relationship between distribution and hydrological conditions. Biogeosciences 2015, 12, 3607–3621. [Google Scholar] [CrossRef]
- Garel, M.; Bonin, P.; Martini, S.; Guasco, S.; Roumagnac, M.; Bhairy, N.; Armougom, F.; Tamburini, C. Pressure-Retaining Sampler and High-Pressure Systems to Study Deep-Sea Microbes Under in situ Conditions. Front. Microbiol. 2019, 10, 453. [Google Scholar] [CrossRef]
- McNeil, C.L.; D’Asaro, E.A. A calibration equation for oxygen optodes based on physical properties of the sensing foil. Limnol. Oceanogr. Methods 2014, 12, 139–154. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Blazewicz, S.J.; Barnard, R.L.; Daly, R.A.; Firestone, M.K. Evaluating rRNA as an indicator of microbial activity in environmental communities: Limitations and uses. ISME J. 2013, 7, 2061–2068. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- Mcmurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package “Vegan” Title Community Ecology Package Version 2.5-7; 2020; Available online: https://cran.ism.ac.jp (accessed on 17 September 2020).
- Lahti, L.; Shetty, S.; Blake, T. Tools for Microbiome Analysis in R; Microbiome Packag.; Version 0.99; 2017; Available online: http://microbiome.github.io/microbiome (accessed on 17 September 2020).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Bittinger, K. Abdiv: Alpha and Beta Diversity Measures; R Package Version 0.2.0; 2020; Available online: https://CRAN.R-project.org/package=abdiv (accessed on 17 September 2020).
- Lepš, J.; Šmilauer, P. Multivariate Analysis of Ecological Data Using CANOCO; Cambridge University Press: Cambridge, MA, USA, 2003. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation Statutes Computer: Vienna, Austria, 2013; ISBN 3-900051-07-0. Available online: http://www.R-project.org/ (accessed on 17 September 2020).
- Legendre, P.; Legendre, L. Numerical Ecology, 2nd ed.; ScienceDirect; Elsevier: New York, NY, USA, 1998. [Google Scholar]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and Community Ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 370. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, J.; Jia, Z.; Chen, S.; Zhang, L.; Gao, W. DNA stable-isotope probing reveals potential key players for microbial decomposition and degradation of diatom-derived marine particulate matter. Microbiologyopen 2020, 9, e1013. [Google Scholar] [CrossRef]
- Dong, S.; Subhas, A.V.; Rollins, N.E.; Naviaux, J.D.; Adkins, J.F.; Berelson, W.M. A kinetic pressure effect on calcite dissolution in seawater. Geochim. Cosmochim. Acta 2018, 238, 411–423. [Google Scholar] [CrossRef]
- Bouvier, T.; Del Giorgio, P.A.; Gasol, J.M. A comparative study of the cytometric characteristics of High and Low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 2007, 9, 2050–2066. [Google Scholar] [CrossRef]
- Tamburini, C.; Boutrif, M.; Garel, M.; Colwell, R.R.; Deming, J.W. Prokaryotic responses to hydrostatic pressure in the ocean—A review. Environ. Microbiol. 2013, 15, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.-P.; Gust, G. Hydrostatic pressure affects physiology and community structure of marine bacteria during settling to 4000 m: An experimental approach. Mar. Ecol. Ser. 2009, 390, 97–104. [Google Scholar] [CrossRef]
- de Jesus Mendes, P.A.; Thomsen, L.; Holscher, B.; de Stigter, H.C.; Gust, G. Pressure effects on the biological degradation of organo-mineral aggregates in submarine canyons. Mar. Geol. 2007, 246, 165–175. [Google Scholar] [CrossRef]
- Stief, P.; Elvert, M.; Glud, R.N. Respiration by “marine snow” at high hydrostatic pressure: Insights from continuous oxygen measurements in a rotating pressure tank. Limnol. Oceanogr. 2021, 66, 2797–2809. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Cottrell, M.T.; Kirchman, D.L. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 2000, 66, 5116–5122. [Google Scholar] [CrossRef]
- Kirchman, D.L. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 2002, 39, 91–100. [Google Scholar] [CrossRef]
- Zabeti, N.; Bonin, P.; Volkman, J.K.; Jameson, I.D.; Guasco, S.; Rontani, J.-F. Potential alteration of U 37K′ paleothermometer due to selective degradation of alkenones by marine bacteria isolated from the haptophyte Emiliania huxleyi. FEMS Microbiol. Ecol. 2010, 73, 83–94. [Google Scholar] [CrossRef]
- Enke, T.N.; Leventhal, G.E.; Metzger, M.; Saavedra, J.T.; Cordero, O.X. Microscale ecology regulates particulate organic matter turnover in model marine microbial communities. Nat. Commun. 2018, 9, 2743. [Google Scholar] [CrossRef] [PubMed]
- Dini-Andreote, F.; Stegen, J.C.; Van Elsas, J.D.; Salles, J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial Surface Colonization and Biofilm Development in Marine Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Gómez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedrós-Alió, C. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef] [PubMed]
- Elifantz, H.; Horn, G.; Ayon, M.; Cohen, Y.; Minz, D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013, 85, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Peoples, L.M.; Bartlett, D.H. Ecogenomics of Deep-Ocean Microbial Bathytypes. In Microbial Ecology of Extreme Environments; Springer International Publishing: Cham, Switzerland, 2017; pp. 7–50. [Google Scholar] [CrossRef]
- Boeuf, D.; Edwards, B.R.; Eppley, J.M.; Hu, S.K.; Poff, K.E.; Romano, A.E.; Caron, D.A.; Karl, D.M.; DeLong, E.F. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc. Natl. Acad. Sci. USA 2019, 116, 11824–11832. [Google Scholar] [CrossRef] [PubMed]
- Wannicke, N.; Frindte, K.; Gust, G.; Liskow, I.; Wacker, A.; Meyer, A.; Grossart, H.-P. Measuring bacterial activity and community composition at high hydrostatic pressure using a novel experimental approach: A pilot study. FEMS Microbiol. Ecol. 2015, 91, fiv036. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F.; Yayanos, A.A. Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science 1985, 228, 1101–1113. [Google Scholar] [CrossRef]
- Burd, A.B.; Hansell, D.A.; Steinberg, D.K.; Anderson, T.R.; Arístegui, J.; Baltar, F.; Beaupré, S.R.; Buesseler, K.O.; DeHairs, F.; Jackson, G.A.; et al. Assessing the Apparent Imbalance Between Geochemical and Biochemical Indicators of Meso- and Bathypelagic Biological Activity: What the @$#! is wrong with present calculations of carbon budgets? Deep Sea Res. II Top. Stud.Oceanogr. 2010, 57, 1557–1571. [Google Scholar] [CrossRef]
- Bižić-Ionescu, M.; Ionescu, D.; Grossart, H.-P. Organic Particles: Heterogeneous Hubs for Microbial Interactions in Aquatic Ecosystems. Front. Microbiol. 2018, 9, 2569. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, D.; Bizic-Ionescu, M.; Khalili, A.; Malekmohammadi, R.; Morad, M.R.; de Beer, D.; Grossart, H.-P. A new tool for long-term studies of POM-bacteria interactions: Overcoming the century-old Bottle Effect. Sci. Rep. 2015, 5, 14706. [Google Scholar] [CrossRef] [PubMed]
- Pelve, E.A.; Fontanez, K.M.; DeLong, E.F. Bacterial Succession on Sinking Particles in the Ocean’s Interior. Front. Microbiol. 2017, 8, 2269. [Google Scholar] [CrossRef] [PubMed]


| Conditions | Samples | Input | Filtered | Denoised | Merged | Non-Chimeric | Final Retained (%) | Global Richness | Shannon | Chao1 | PD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T0 | T01 | 59,218 | 43,696 | 43,524 | 42,323 | 42,242 | 71 | 106 | 4.13 | 106 | 10.47 |
| T02 | 71,661 | 54,986 | 54,827 | 53,562 | 53,412 | 75 | 96 | 3.86 | 96 | 8.45 | |
| T6-ATM | ATM1 | 50,037 | 36,087 | 35,891 | 34,808 | 34,746 | 69 | 90 | 2.55 | 104 | 8.80 |
| ATM2 | 98,599 | 72,212 | 71,853 | 70,168 | 69,053 | 70 | 98 | 2.14 | 99 | 9.21 | |
| T6-HP | HP1 | 94,669 | 72,866 | 72,701 | 71,523 | 71,500 | 76 | 76 | 3.83 | 76 | 6.20 |
| HP2 | 71,436 | 53,275 | 53,177 | 52,386 | 52,386 | 73 | 54 | 3.50 | 54 | 5.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamburini, C.; Garel, M.; Barani, A.; Boeuf, D.; Bonin, P.; Bhairy, N.; Guasco, S.; Jacquet, S.; Le Moigne, F.A.C.; Panagiotopoulos, C.; et al. Increasing Hydrostatic Pressure Impacts the Prokaryotic Diversity during Emiliania huxleyi Aggregates Degradation. Water 2021, 13, 2616. https://doi.org/10.3390/w13192616
Tamburini C, Garel M, Barani A, Boeuf D, Bonin P, Bhairy N, Guasco S, Jacquet S, Le Moigne FAC, Panagiotopoulos C, et al. Increasing Hydrostatic Pressure Impacts the Prokaryotic Diversity during Emiliania huxleyi Aggregates Degradation. Water. 2021; 13(19):2616. https://doi.org/10.3390/w13192616
Chicago/Turabian StyleTamburini, Christian, Marc Garel, Aude Barani, Dominique Boeuf, Patricia Bonin, Nagib Bhairy, Sophie Guasco, Stéphanie Jacquet, Frédéric A. C. Le Moigne, Christos Panagiotopoulos, and et al. 2021. "Increasing Hydrostatic Pressure Impacts the Prokaryotic Diversity during Emiliania huxleyi Aggregates Degradation" Water 13, no. 19: 2616. https://doi.org/10.3390/w13192616
APA StyleTamburini, C., Garel, M., Barani, A., Boeuf, D., Bonin, P., Bhairy, N., Guasco, S., Jacquet, S., Le Moigne, F. A. C., Panagiotopoulos, C., Riou, V., Veloso, S., Santinelli, C., & Armougom, F. (2021). Increasing Hydrostatic Pressure Impacts the Prokaryotic Diversity during Emiliania huxleyi Aggregates Degradation. Water, 13(19), 2616. https://doi.org/10.3390/w13192616







