Untapped Potential of Moving Bed Biofilm Reactors with Different Biocarrier Types for Bilge Water Treatment: A Laboratory-Scale Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Analytical Methods
2.2. MBBR Setup and Operation
2.3. Microbial Profile Analysis
2.3.1. Sampling Campaigns
2.3.2. Next-Generation Sequencing
2.4. Scanning Electron Microscopy
2.5. Equations
3. Results
3.1. Performance of MBBRs
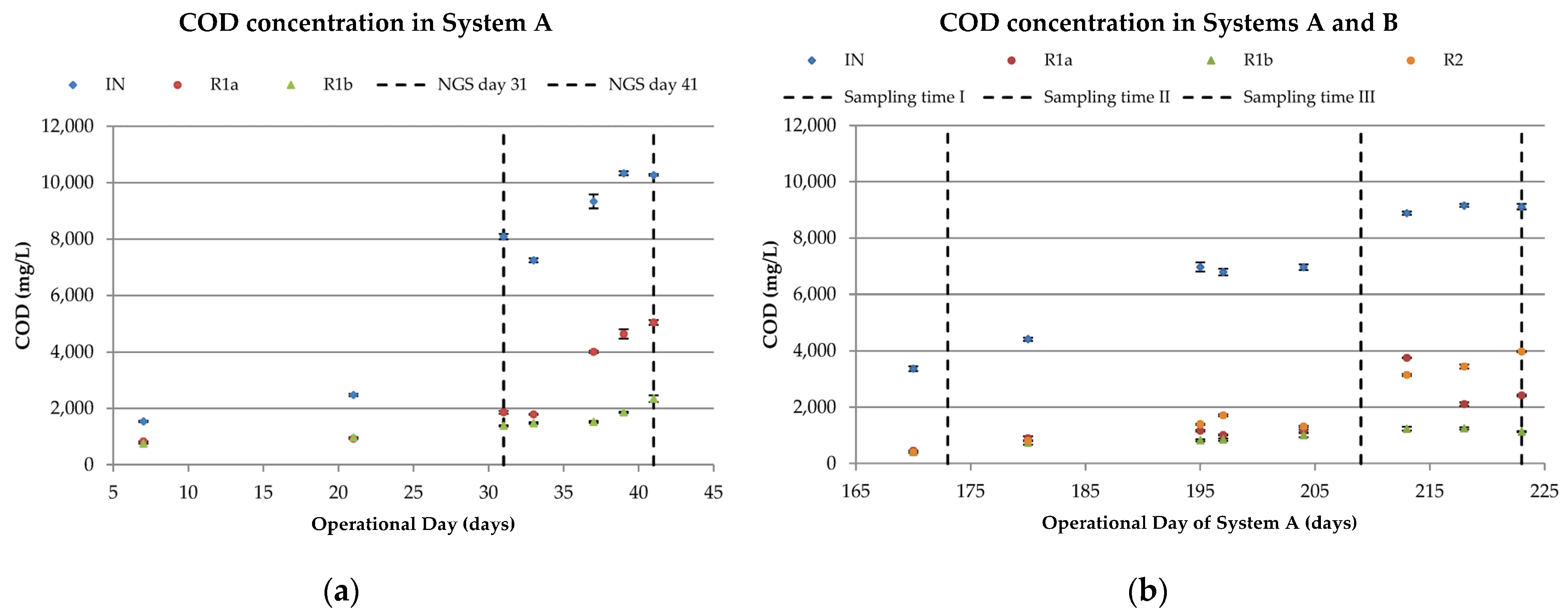
3.1.1. COD Removal
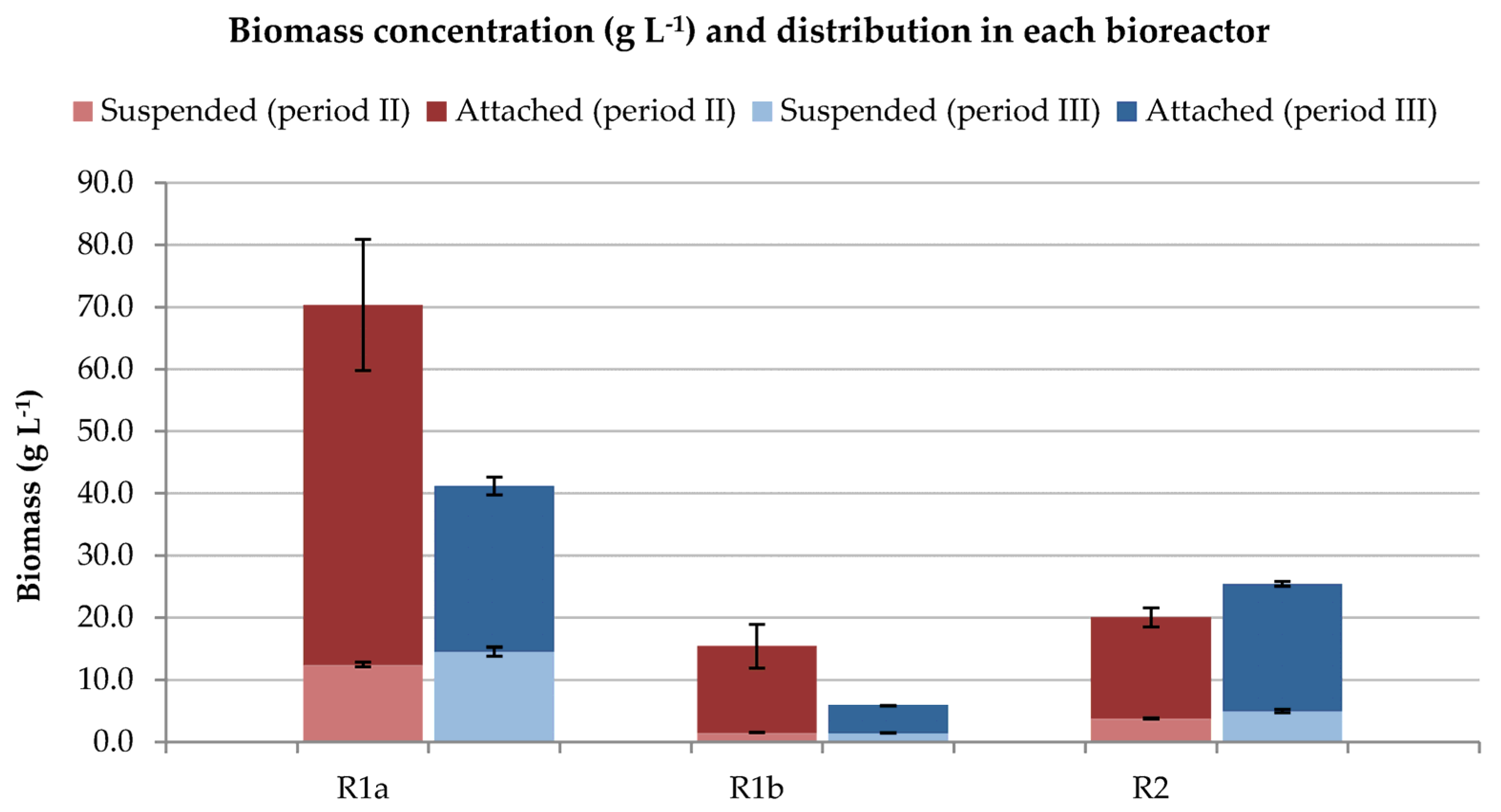
3.1.2. Biomass Concentration
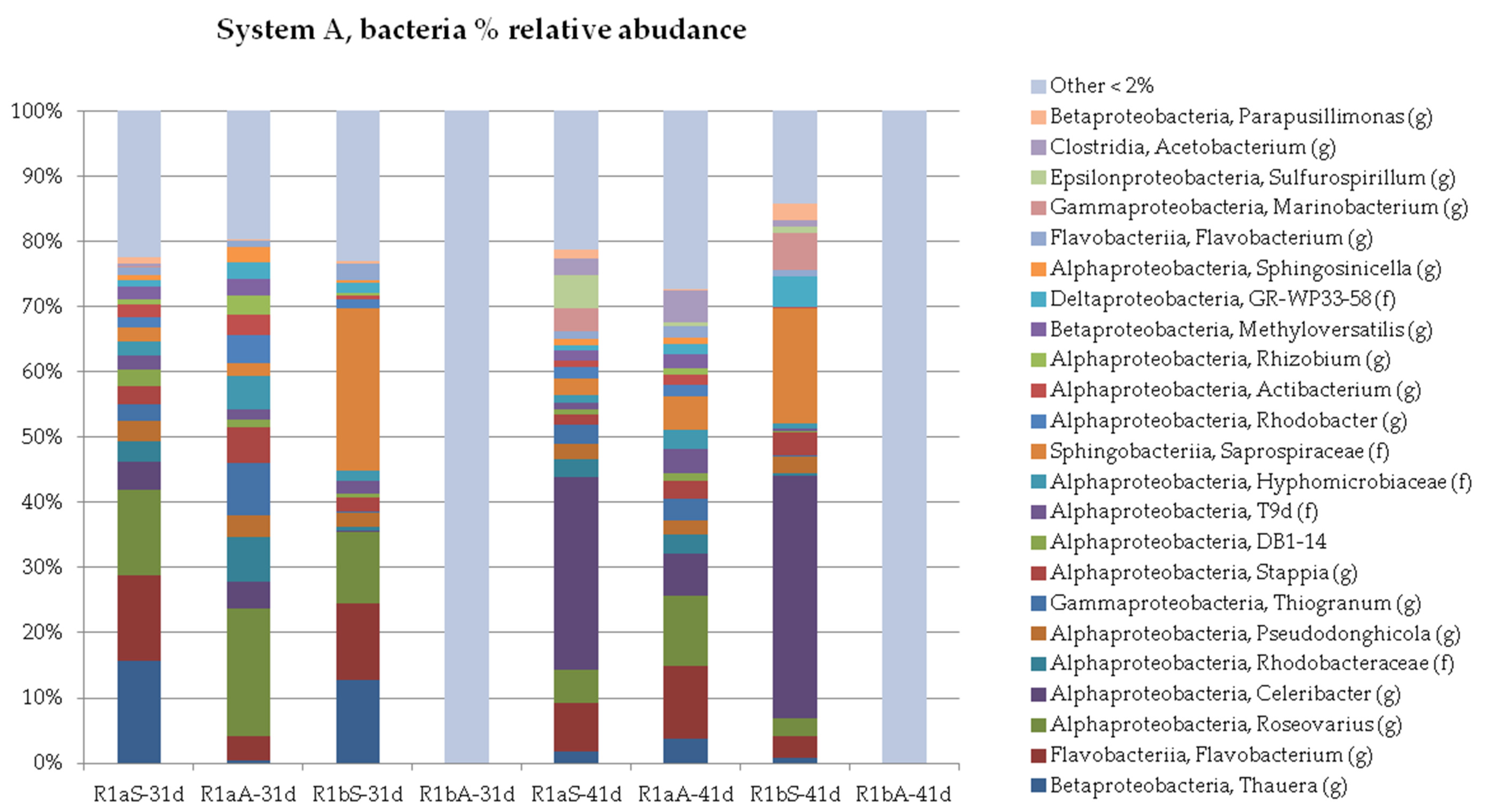
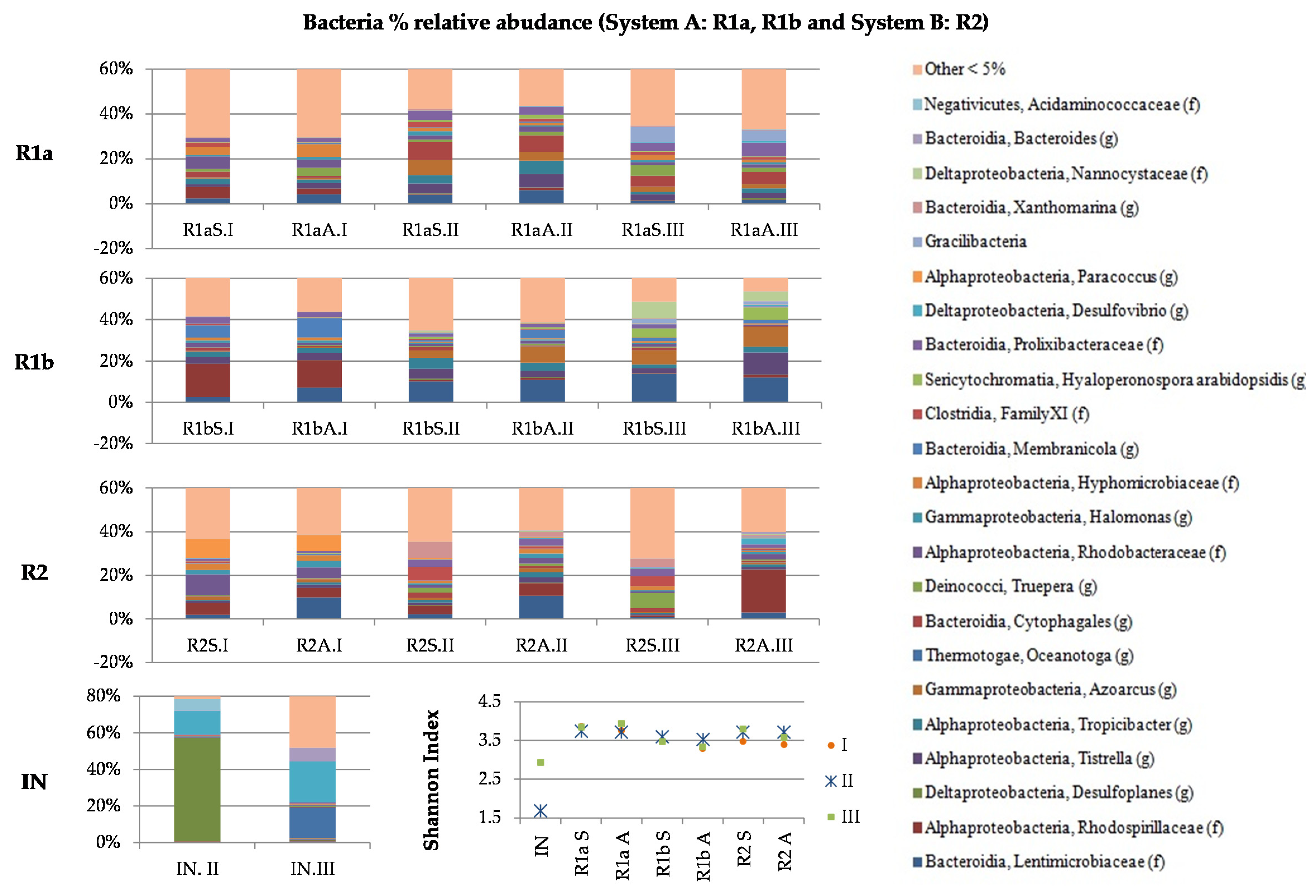
3.2. Microbial Profile
3.2.1. Organic and Hydraulic Shock Load (A)
3.2.2. Salinity Load Shock
3.2.3. Organic and Hydraulic Shock Load (B)
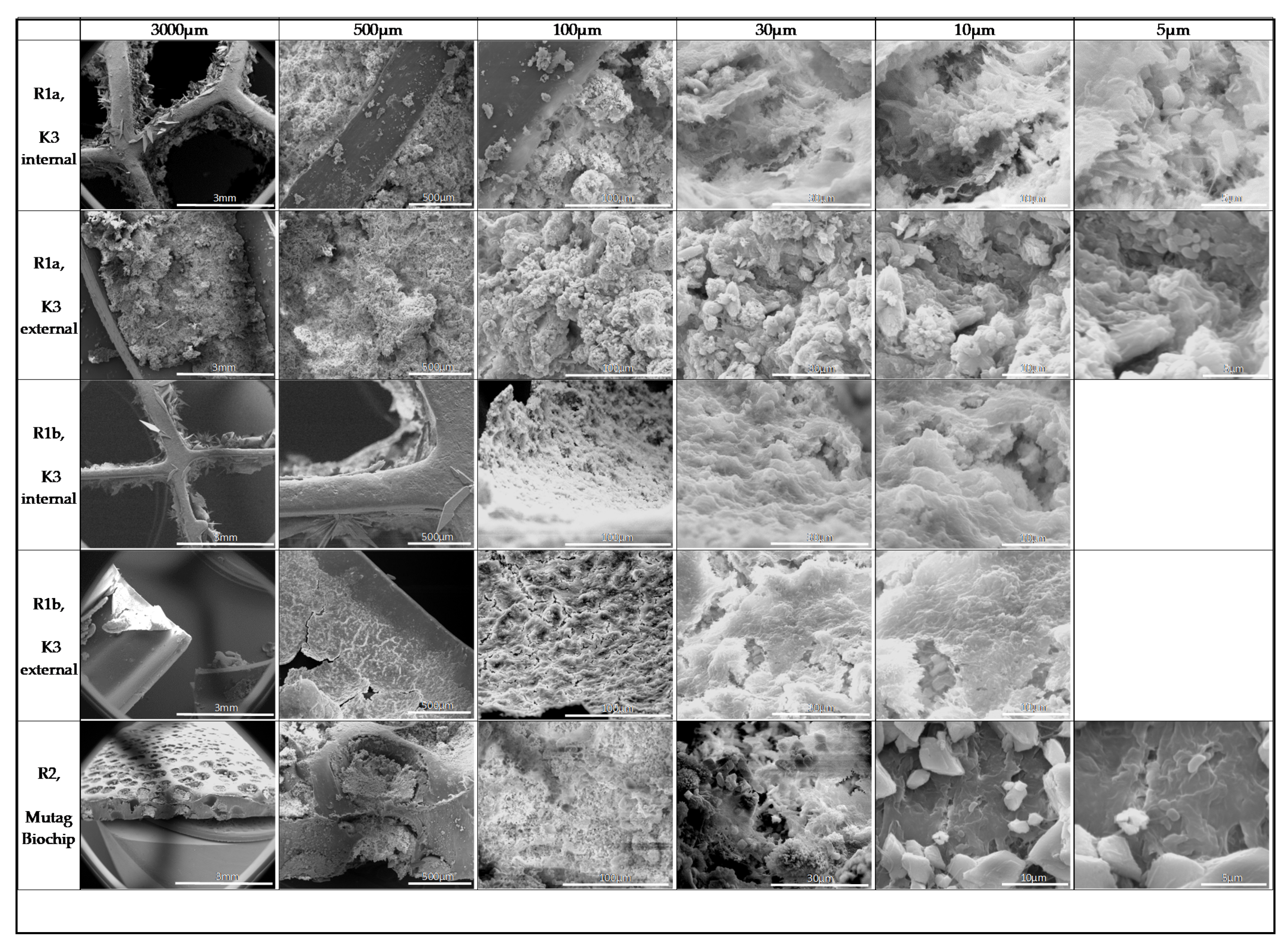
3.3. Biofilm Structure
4. Discussion
4.1. Comparison of the Two Biocarrier Types
4.2. Microbial Diversity in Relevant Studies
4.3. Biomineralization in Biofilms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, Y.H.; Su, J.F.; Shih, Y.; Wang, J.; Wang, P.Y.; Huang, C.P. Hazardous wastes treatment technologies. Water Environ. Res. 2020, 92, 1833–1860. [Google Scholar] [CrossRef]
- Tian, X.; Song, Y.; Shen, Z.; Zhou, Y.; Wang, K.; Jin, X.; Han, Z.; Liu, T. A comprehensive review on toxic petrochemical wastewater pretreatment and advanced treatment. J. Clean. Prod. 2020, 245, 118692. [Google Scholar] [CrossRef]
- Eusebi, A.L.; Massi, A.; Sablone, E.; Santinelli, M.; Battistoni, P. Industrial wastewater platform: Upgrading of the biological process and operative configurations for best performance. Water Sci. Technol. 2012, 65, 721–727. [Google Scholar] [CrossRef]
- Garner, E.; Davis, B.C.; Milligan, E.; Blair, M.F.; Keenum, I.; Maile-Moskowitz, A.; Pan, J.; Gnegy, M.; Liguori, K.; Gupta, S.; et al. Next generation sequencing approaches to evaluate water and wastewater quality. Water Res. 2021, 194, 116907. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S. Microbial sequencing methods for monitoring of anaerobic treatment of antibiotics to optimize performance and prevent system failure. Appl. Microbiol. Biotechnol. 2016, 100, 5313–5321. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.; Brynolf, S.; Lindgren, J.F.; Wilewska-Bien, M. Shipping and the Environment, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 125–168. [Google Scholar] [CrossRef]
- McLaughlin, C.; Falatko, D.; Danesi, R.; Albert, R. Characterizing shipboard bilgewater effluent before and after treatment. Environ. Sci. Pollut. Res. 2014, 21, 5637–5652. [Google Scholar] [CrossRef] [PubMed]
- Mazioti, A.A.; Vasquez, M.I.; Vyrides, I. Comparison of different cultures and culturing conditions for the biological deterioration of organic load from real saline bilge wastewater: Microbial diversity insights and ecotoxicity assessment. Environ. Sci. Pollut. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Nievas, M.; Commendatore, M.; Olivera, N.; Esteves, J.; Bucalá, V. Biodegradation of bilge waste from Patagonia with an indigenous microbial community. Bioresour. Technol. 2006, 97, 2280–2290. [Google Scholar] [CrossRef]
- Cerqueira, V.S.; Hollenbach, E.B.; Maboni, F.; Vainstein, M.; Camargo, F.; Peralba, M.D.C.R.; Bento, F.M. Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour. Technol. 2011, 102, 11003–11010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappello, S.; Santisi, S.; Calogero, R.; Hassanshahian, M.; Yakimov, M. Characterisation of Oil-Degrading Bacteria Isolated from Bilge Water. Water Air Soil Pollut. 2012, 223, 3219–3226. [Google Scholar] [CrossRef]
- Feknous, N.; Branes, Z.; Rouabhia, K.; Batisson, I.; Amblard, C. Isolation characterization and growth of locally isolated hydrocarbonoclastic marine bacteria (eastern Algerian coast). Environ. Monit. Assess. 2017, 189, 49. [Google Scholar] [CrossRef]
- Uma, V.; Gandhimathi, R. Organic removal and synthesis of biopolymer from synthetic oily bilge water using the novel mixed bacterial consortium. Bioresour. Technol. 2019, 273, 169–176. [Google Scholar] [CrossRef]
- Barwal, A.; Chaudhary, R. To study the performance of biocarriers in moving bed biofilm reactor (MBBR) technology and kinetics of biofilm for retrofitting the existing aerobic treatment systems: A review. Rev. Environ. Sci. Biotechnol. 2014, 13, 285–299. [Google Scholar] [CrossRef]
- Biswas, K.; Taylor, M.W.; Turner, S.J. Successional development of biofilms in moving bed biofilm reactor (MBBR) systems treating municipal wastewater. Appl. Microbiol. Biotechnol. 2014, 98, 1429–1440. [Google Scholar] [CrossRef]
- Huang, H.; Peng, C.; Peng, P.; Lin, Y.; Zhang, X.; Ren, H. Towards the biofilm characterization and regulation in biological wastewater treatment. Appl. Microbiol. Biotechnol. 2019, 103, 1115–1129. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Delatolla, R. Biofilm and microbiome response of attached growth nitrification systems across incremental decreases to low temperatures. J. Water Process Eng. 2021, 39, 101730. [Google Scholar] [CrossRef]
- Bouteh, E.; Ahmadi, N.; Abbasi, M.; Torabian, A.; van Loosdrecht, M.C.; Ducoste, J. Biodegradation of organophosphorus pesticides in moving bed biofilm reactors: Analysis of microbial community and biodegradation pathways. J. Hazard. Mater. 2021, 408, 124950. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, N.; Wei, Y.; Dang, K.; Li, M.; Li, Y.; Li, Q.; Mu, R. Pilot study on the upgrading configuration of UASB-MBBR with two carriers: Treatment effect, sludge reduction and functional microbial identification. Process Biochem. 2020, 99, 211–221. [Google Scholar] [CrossRef]
- Abu Bakar, S.N.H.; Abu Hasan, H.; Mohammad, A.W.; Abdullah, S.R.S.; Haan, T.Y.; Ngteni, R.; Yusof, K.M.M. A review of moving-bed biofilm reactor technology for palm oil mill effluent treatment. J. Clean. Prod. 2018, 171, 1532–1545. [Google Scholar] [CrossRef]
- Liang, C.; de Jonge, N.; Carvalho, P.N.; Nielsen, J.L.; Bester, K. Biodegradation kinetics of organic micropollutants and microbial community dynamics in a moving bed biofilm reactor. Chem. Eng. J. 2021, 415, 128963. [Google Scholar] [CrossRef]
- Wolff, D.; Krah, D.; Dötsch, A.; Ghattas, A.-K.; Wick, A.; Ternes, T.A. Insights into the variability of microbial community composition and micropollutant degradation in diverse biological wastewater treatment systems. Water Res. 2018, 143, 313–324. [Google Scholar] [CrossRef]
- Sun, C.; Leiknes, T.; Weitzenböck, J.; Thorstensen, B. Development of an integrated shipboard wastewater treatment system using biofilm-MBR. Sep. Purif. Technol. 2010, 75, 22–31. [Google Scholar] [CrossRef]
- Mancini, G.; Cappello, S.; Yakimov, M.; Polizzi, A.; Torregrossa, M. Biological Approaches to the Treatment of Saline Oily Waste(waters) Originated from Marine Transportation. Chem. Eng. Trans. 2012, 27, 37–42. [Google Scholar] [CrossRef]
- Vyrides, I.; Drakou, E.-M.; Ioannou, S.; Michael, F.; Gatidou, G.; Stasinakis, A.S. Biodegradation of bilge water: Batch test under anaerobic and aerobic conditions and performance of three pilot aerobic Moving Bed Biofilm Reactors (MBBRs) at different filling fractions. J. Environ. Manag. 2018, 217, 356–362. [Google Scholar] [CrossRef]
- Ahmadi, M.; Ahmadmoazzam, M.; Saeedi, R.; Abtahi, M.; Ghafari, S.; Jorfi, S. Biological treatment of a saline and recalcitrant petrochemical wastewater by using a newly isolated halo-tolerant bacterial consortium in MBBR. Desalination Water Treat. 2019, 167, 84–95. [Google Scholar] [CrossRef]
- Yu, Z.; Li, W.; Tan, S. Real-time monitoring of the membrane biofouling based on spectroscopic analysis in a marine MBBR-MBR (moving bed biofilm reactor-membrane bioreactor) for saline wastewater treatment. Chemosphere 2019, 235, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhou, W.; Chen, X.; Zhou, Y.; He, B.; Tan, S. Analysis of the biodegradation performance and biofouling in a halophilic MBBR-MBR to improve the treatment of disinfected saline wastewater. Chemosphere 2021, 269, 128716. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; American Public Health Association; American Water Works Association; Water Environment Federation: Washington, DC, USA, 2012. [Google Scholar]
- Mazioti, A.-A.; Stasinakis, A.S.; Pantazi, Y.; Andersen, H.R. Biodegradation of benzotriazoles and hydroxy-benzothiazole in wastewater by activated sludge and moving bed biofilm reactor systems. Bioresour. Technol. 2015, 192, 627–635. [Google Scholar] [CrossRef] [Green Version]
- Falås, P.; Baillon-Dhumez, A.; Andersen, H.; Ledin, A.; Jansen, J.L.C. Suspended biofilm carrier and activated sludge removal of acidic pharmaceuticals. Water Res. 2012, 46, 1167–1175. [Google Scholar] [CrossRef]
- Fonseca, D.L.; Bassin, J.P. Investigating the most appropriate methods for attached solids determination in moving-bed biofilm reactors. Bioprocess Biosyst. Eng. 2019, 42, 1867–1878. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Kleerebezem, R.; Beckers, J.; Pol, L.W.H.; Lettinga, G. High rate treatment of terephthalic acid production wastewater in a two-stage anaerobic bioreactor. Biotechnol. Bioeng. 2005, 91, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.-R.; Zhang, L.-M.; He, J.-Z. Sorption mechanism and distribution of cadmium by different microbial species. J. Environ. Manag. 2019, 237, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, J.; Xu, N.; He, A.; Xin, F.; Ma, J.; Fang, Y.; Zhang, W.; Liu, S.; Jiang, M.; et al. Performance evaluation of a lab-scale moving bed biofilm reactor (MBBR) using polyethylene as support material in the treatment of wastewater contaminated with terephthalic acid. Chemosphere 2019, 227, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Nisenbaum, M.; Corti-Monzón, G.; Villegas-Plazas, M.; Junca, H.; Mangani, A.; Patat, M.L.; González, J.F.; Murialdo, S.E. Enrichment and key features of a robust and consistent indigenous marine-cognate microbial consortium growing on oily bilge wastewaters. Biodegradation 2020, 31, 91–108. [Google Scholar] [CrossRef]
- Cao, J.; Lai, Q.; Yuan, J.; Shao, Z. Genomic and metabolic analysis of fluoranthene degradation pathway in Celeribacter indicus P73T. Sci. Rep. 2015, 5, 7741. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.-A.; Kang, I.; Moon, M.; Ryu, U.-C.; Kwon, K.K.; Cho, J.-C.; Oh, H.-M. Complete genome sequence of Celeribacter marinus IMCC12053T, the host strain of marine bacteriophage P12053L. Mar. Genom. 2016, 26, 5–7. [Google Scholar] [CrossRef]
- Procópio, L. Changes in microbial community in the presence of oil and chemical dispersant and their effects on the corrosion of API 5L steel coupons in a marine-simulated microcosm. Appl. Microbiol. Biotechnol. 2020, 104, 6397–6411. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Paul, B.G.; Castelle, C.J.; Hu, P.; Tringe, S.G.; Valentine, D.L.; Andersen, G.L.; Banfield, J.F. Unusual Metabolism and Hypervariation in the Genome of a Gracilibacterium (BD1-5) from an Oil-Degrading Community. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; Zhu, J.; Liang, X.; Zhu, Y.; Sand, W.; Yang, J.; Li, F.; Ma, C.; Liu, Y.; Yang, B. Microbial uniqueness of architecture modified loofah sponge as biological filler for efficient nitrogen removal. Bioresour. Technol. Rep. 2018, 3, 95–101. [Google Scholar] [CrossRef]
- Zhao, H.-P.; Wang, L.; Ren, J.-R.; Li, Z.; Li, M.; Gao, H.-W. Isolation and characterization of phenanthrene-degrading strains Sphingomonas sp. ZP1 and Tistrella sp. ZP5. J. Hazard. Mater. 2008, 152, 1293–1300. [Google Scholar] [CrossRef]
- Xue, J.; Zhang, Y.; Liu, Y.; El-Din, M.G. Treatment of oil sands process-affected water (OSPW) using a membrane bioreactor with a submerged flat-sheet ceramic microfiltration membrane. Water Res. 2016, 88, 1–11. [Google Scholar] [CrossRef]
- Denaro, R.; Aulenta, F.; Crisafi, F.; Di Pippo, F.; Viggi, C.C.; Matturro, B.; Tomei, P.; Smedile, F.; Martinelli, A.; Di Lisio, V.; et al. Marine hydrocarbon-degrading bacteria breakdown poly(ethylene terephthalate) (PET). Sci. Total Environ. 2020, 749, 141608. [Google Scholar] [CrossRef] [PubMed]
- Djahnit, N.; Chernai, S.; Catania, V.; Hamdi, B.; China, B.; Cappello, S.; Quatrini, P. Isolation, characterization and determination of biotechnological potential of oil-degrading bacteria from Algerian centre coast. J. Appl. Microbiol. 2019, 126, 780–795. [Google Scholar] [CrossRef]
- Yao, J.; Li, W.; Ou, D.; Lei, L.; Asif, M.; Liu, Y. Performance and granular characteristics of salt-tolerant aerobic granular reactors response to multiple hypersaline wastewater. Chemosphere 2021, 265, 129170. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, J.; Liberto, T.; Barentin, C.; Dysthe, D.K. Mechanisms of Phase Transformation and Creating Mechanical Strength in a Sustainable Calcium Carbonate Cement. Preprints 2018. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, J.; Southam, G. Advanced biofilm staining techniques for TEM and SEM in geomicrobiology: Implications for visualizing EPS architecture, mineral nucleation, and microfossil generation. Chem. Geol. 2018, 498, 115–127. [Google Scholar] [CrossRef]
- Keren-Paz, A.; Kolodkin-Gal, I. A brick in the wall: Discovering a novel mineral component of the biofilm extracellular matrix. New Biotechnol. 2020, 56, 9–15. [Google Scholar] [CrossRef]
- Yang, T.-M.; Su, C.-S.; Li, J.-S.; Lu, K.-T.; Yeh, T.-F. Recrystallization and Micronization of p-Toluenesulfonamide Using the Rapid Expansion of Supercritical Solution (RESS) Process. Crystals 2019, 9, 449. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Martinez, A.; Leyva-Díaz, J.; Rodriguez-Sanchez, A.; Palazon, B.M.; Rivadeneyra, M.A.; Poyatos, J.M.; Martinez-Toledo, M. Isolation and metagenomic characterization of bacteria associated with calcium carbonate and struvite precipitation in a pure moving bed biofilm reactor-membrane bioreactor. Biofouling 2015, 31, 333–348. [Google Scholar] [CrossRef] [PubMed]
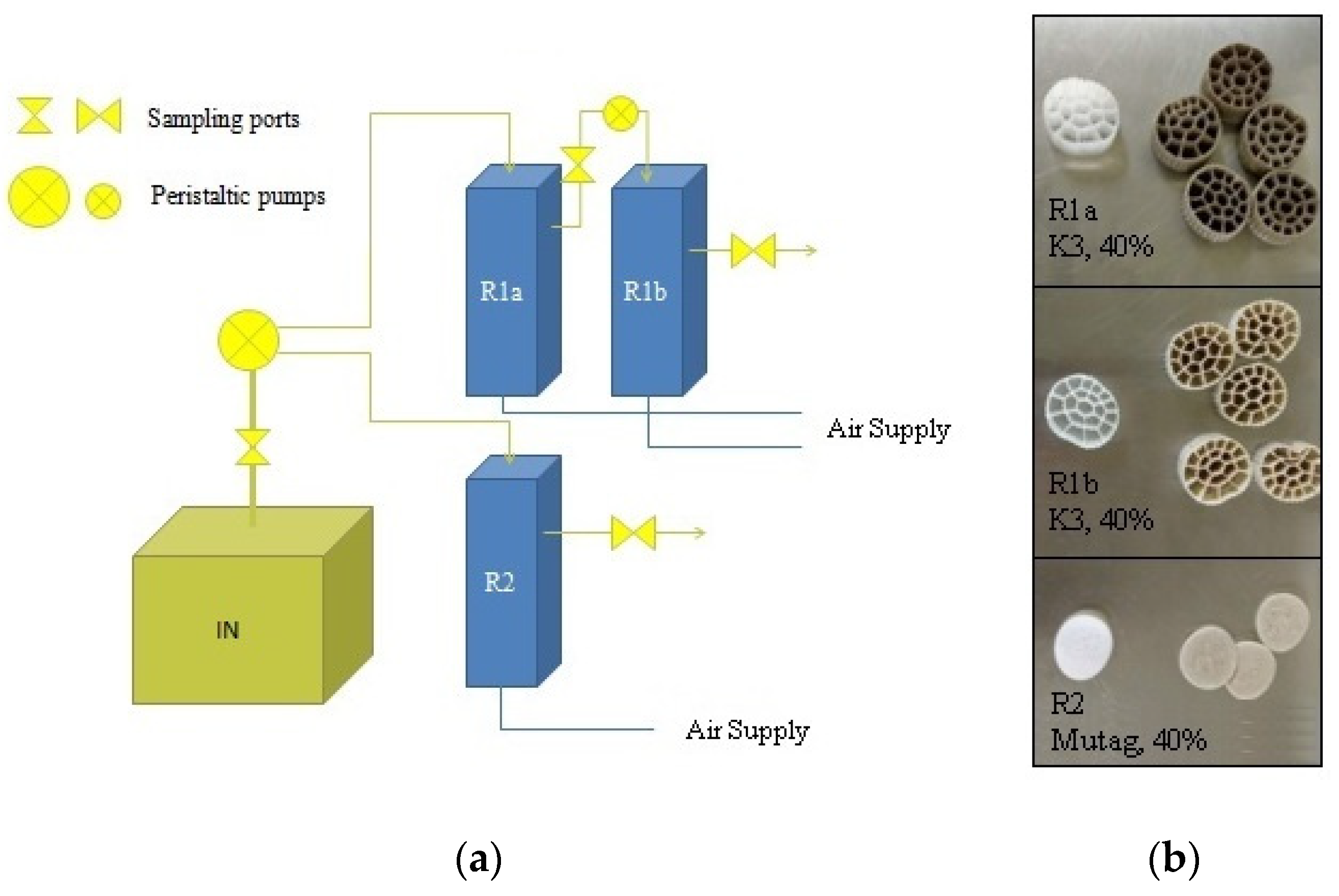
| System A (Reactors R1a and R1b Connected in Series) | System B (Reactor R2) | Ιnflow Wastewater Parameters (Mean, SE) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Operational period description | Duration (days) | Operational day at the end of the period | HRT of each reactor (days) | Operational period description | Duration (days) | Operational day at the end of the period | HRT (days) | pH | Salinity (ppt) | COD (mg L−1) |
| Startup | 31 | 31 | 5 | No operation | 7.1 | 20.5 | 2111 ± 501 | |||
| Organic/hydraulic load shock (A) | 10 | 41 | 2 | 7.2 | 22.4 | 9054 ± 608 | ||||
| Maintenance | 132 | 173 * | 7 | Startup | 61 | 61 * | 7 | 7.0 | 27.6 | 4036 ± 909 |
| Salinity load shock | 36 | 209 ** | 5 | Salinity load shock | 36 | 96 ** | 5 | 6.2 | 40.0 | 6286 ± 626 |
| Organic/hydraulic load shock (B) | 13 | 223 *** | 3 | Organic/hydraulic load shock (B) | 13 | 109 *** | 3 | 5.8 | 19.7 | 9051 ± 83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazioti, A.A.; Koutsokeras, L.E.; Constantinides, G.; Vyrides, I. Untapped Potential of Moving Bed Biofilm Reactors with Different Biocarrier Types for Bilge Water Treatment: A Laboratory-Scale Study. Water 2021, 13, 1810. https://doi.org/10.3390/w13131810
Mazioti AA, Koutsokeras LE, Constantinides G, Vyrides I. Untapped Potential of Moving Bed Biofilm Reactors with Different Biocarrier Types for Bilge Water Treatment: A Laboratory-Scale Study. Water. 2021; 13(13):1810. https://doi.org/10.3390/w13131810
Chicago/Turabian StyleMazioti, Aikaterini A., Loukas E. Koutsokeras, Georgios Constantinides, and Ioannis Vyrides. 2021. "Untapped Potential of Moving Bed Biofilm Reactors with Different Biocarrier Types for Bilge Water Treatment: A Laboratory-Scale Study" Water 13, no. 13: 1810. https://doi.org/10.3390/w13131810
APA StyleMazioti, A. A., Koutsokeras, L. E., Constantinides, G., & Vyrides, I. (2021). Untapped Potential of Moving Bed Biofilm Reactors with Different Biocarrier Types for Bilge Water Treatment: A Laboratory-Scale Study. Water, 13(13), 1810. https://doi.org/10.3390/w13131810







