Seasonal Water Level Fluctuation and Concomitant Change of Nutrients Shift Microeukaryotic Communities in a Shallow Lake
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. DNA Extraction, PCR, and Sequencing
2.3. Analyses
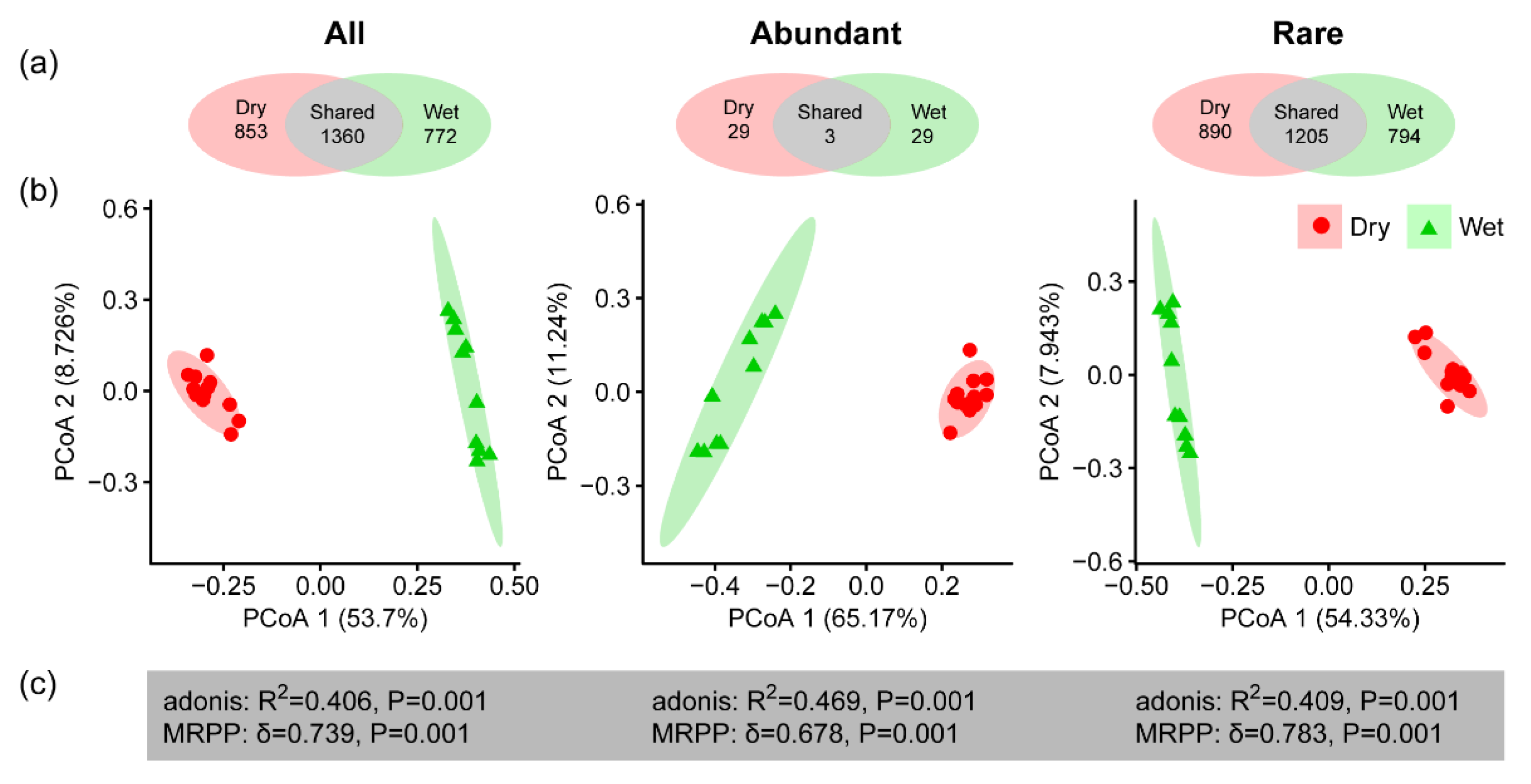
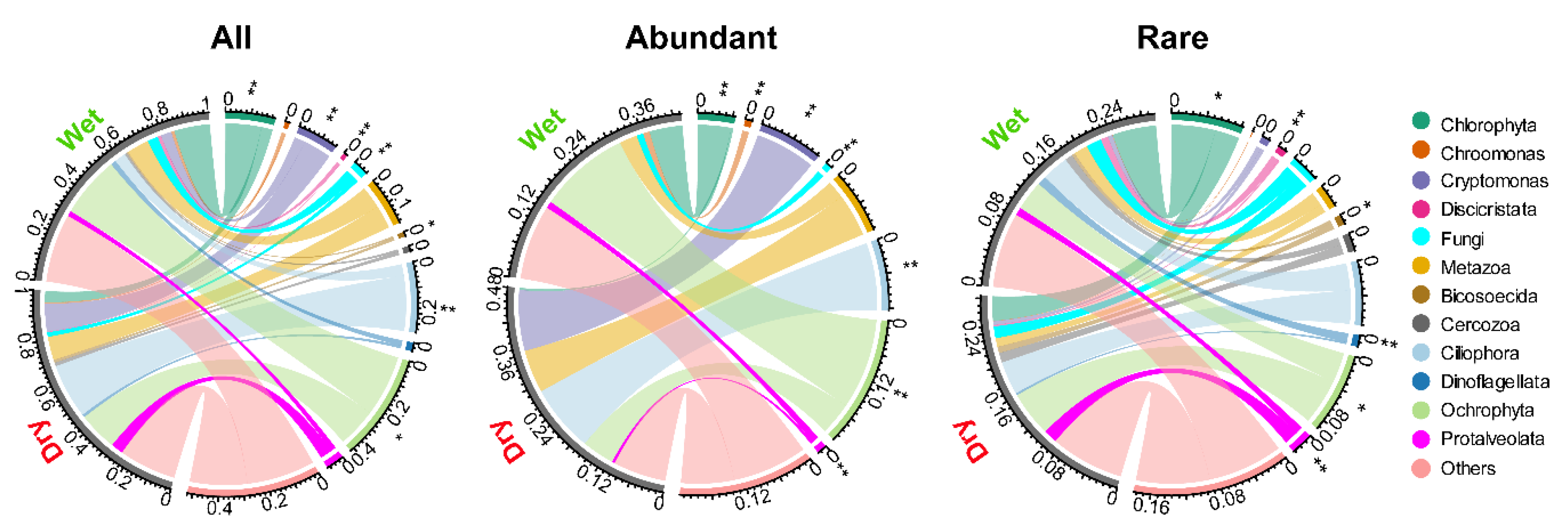
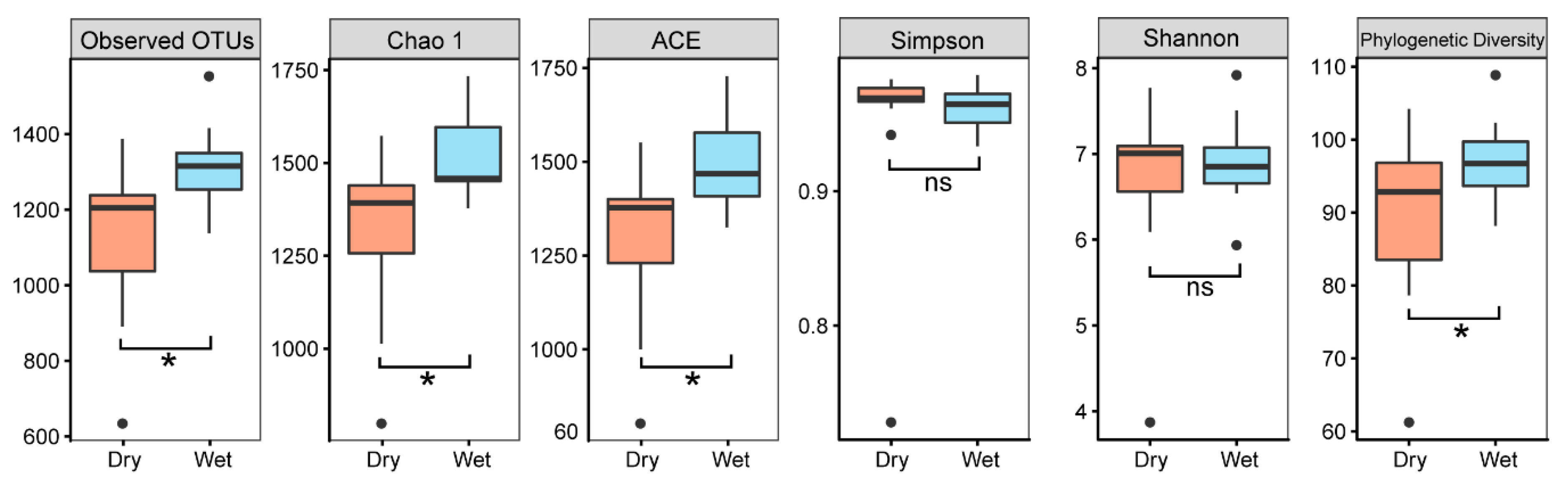
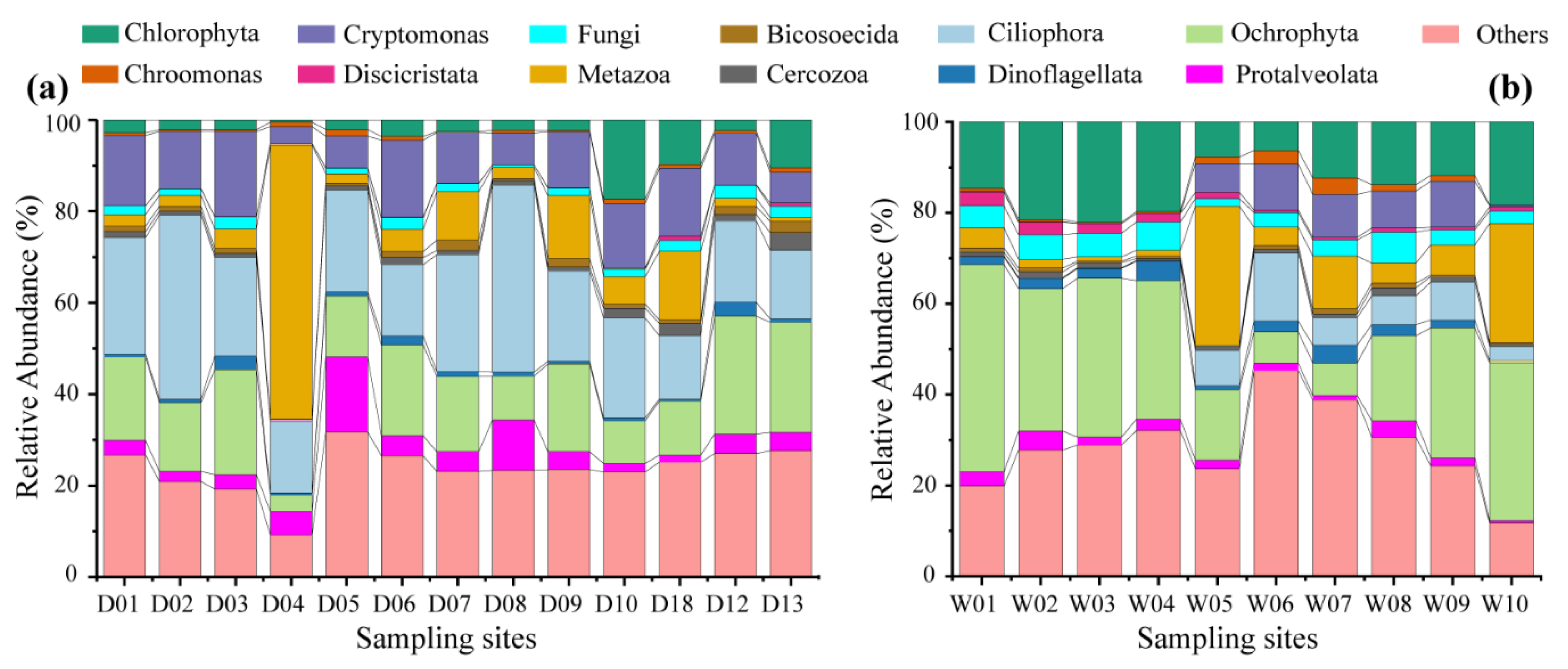
3. Results
3.1. Community Composition and Alpha Diversity
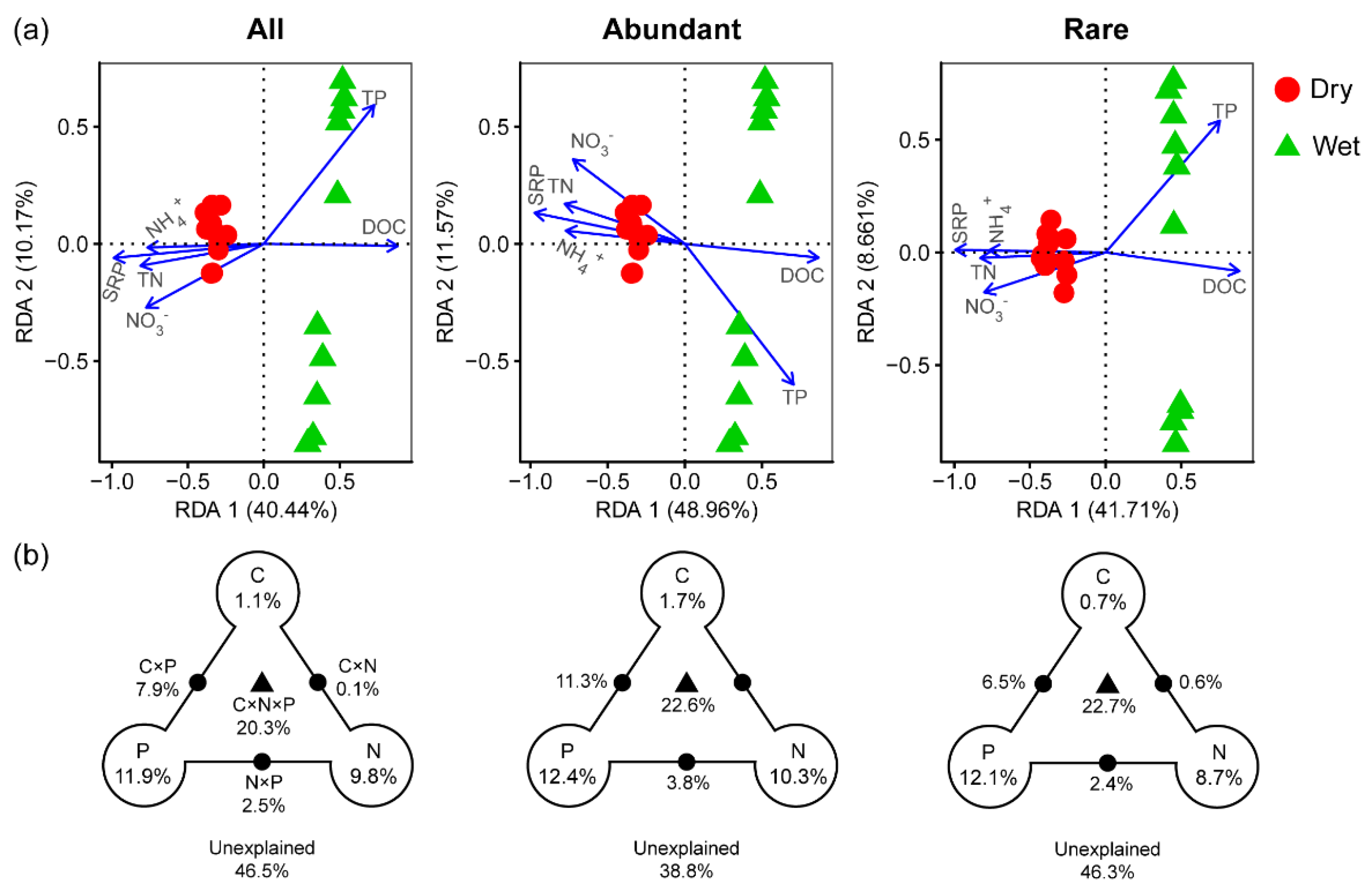
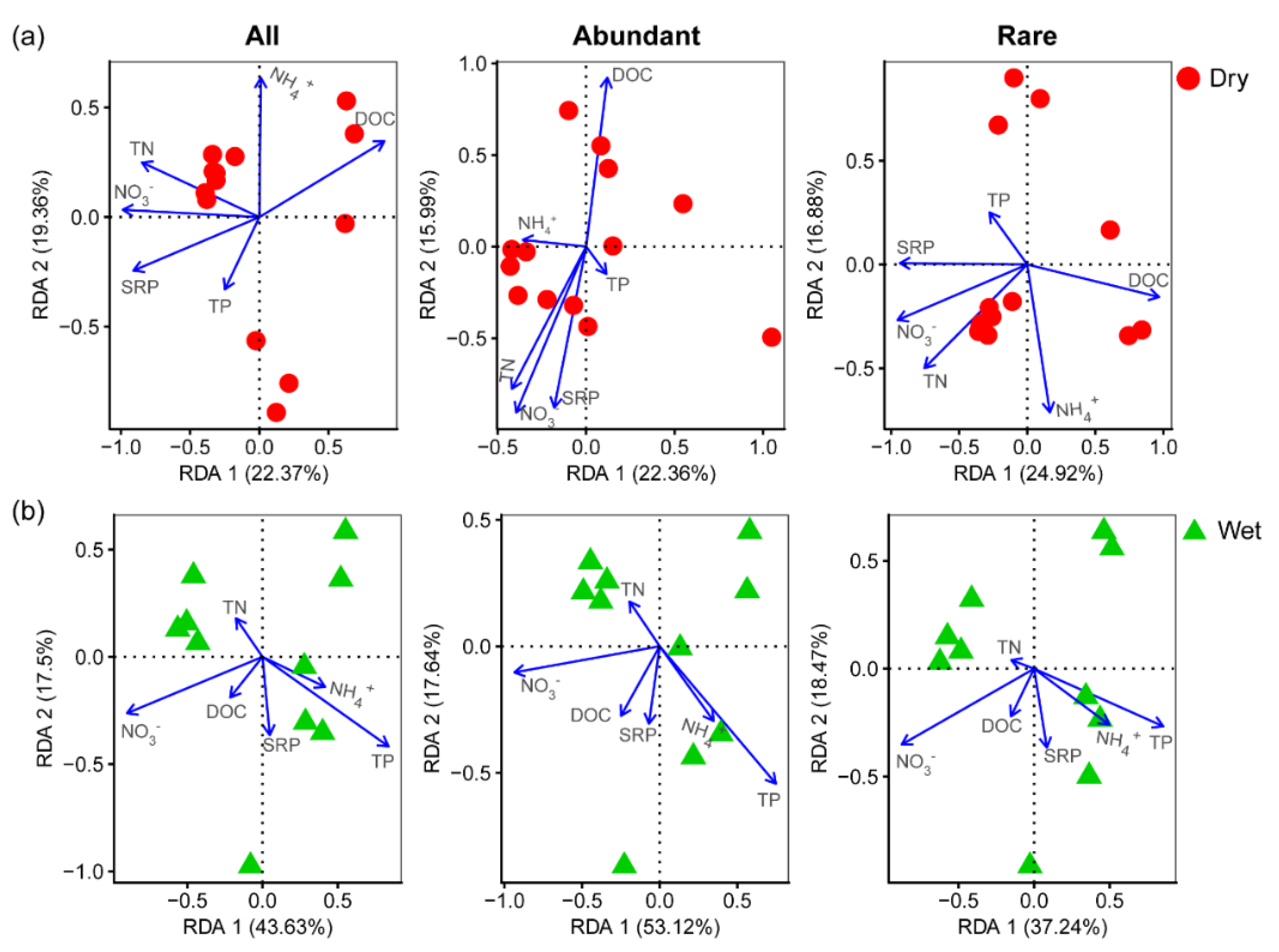
3.2. Relationships between Community and Environmental Variables
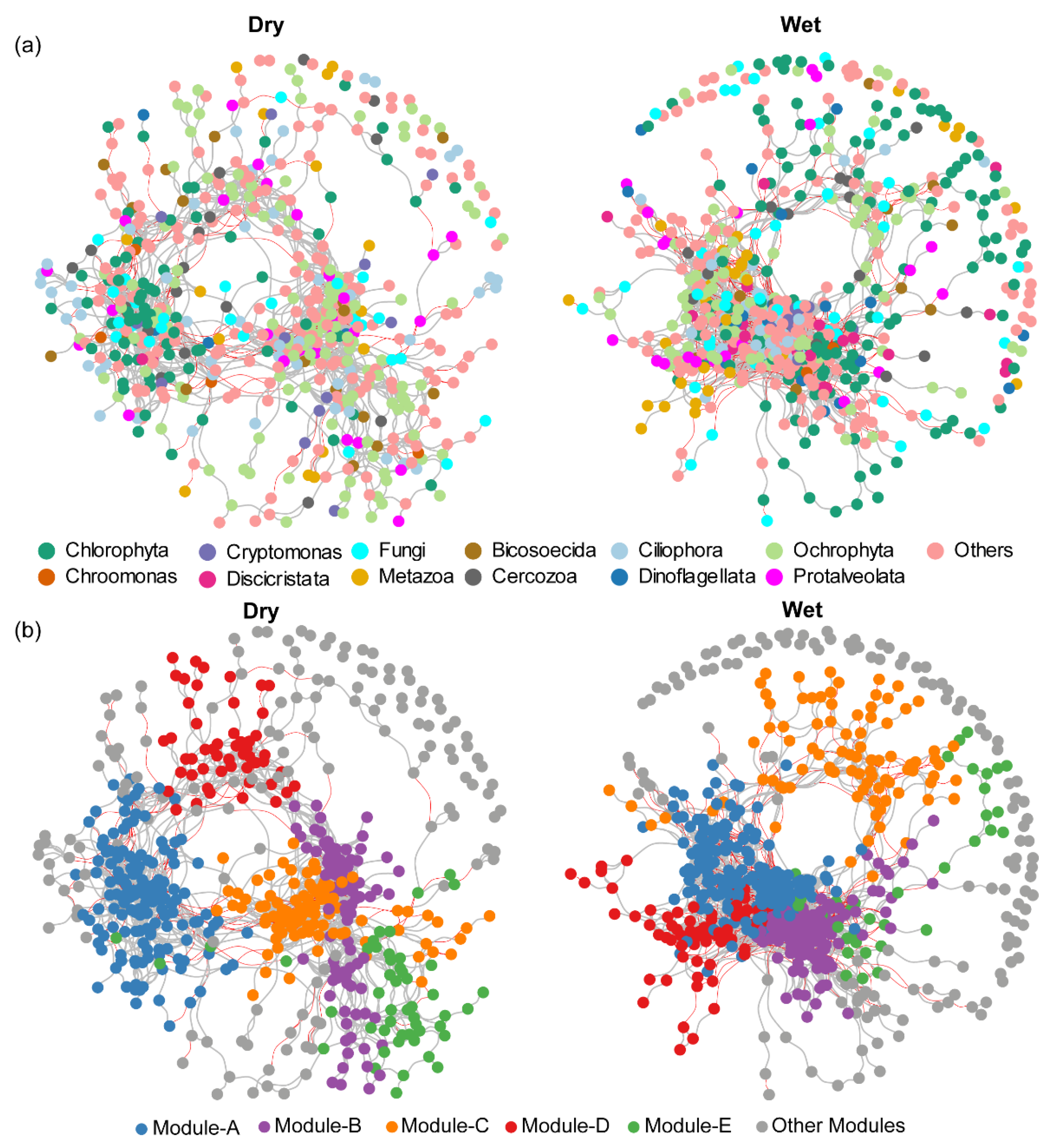
3.3. Co-Occurrence Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gownaris, N.J.; Rountos, K.J.; Kaufman, L.; Kolding, J.; Lwiza, K.M.M.; Pikitch, E.K. Water level fluctuations and the ecosystem functioning of lakes. J. Great Lakes. Res. 2018, 44, 1154–1163. [Google Scholar] [CrossRef]
- Evtimova, V.V.; Donohue, I. Water-level fluctuations regulate the structure and functioning of natural lakes. Freshw. Biol. 2016, 61, 251–264. [Google Scholar] [CrossRef]
- Leira, M.; Cantonati, M. Effects of water-level fluctuations on lakes: An annotated bibliography. Hydrobiologia 2008, 613, 171–184. [Google Scholar] [CrossRef]
- Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Peng, W. Distinct Bacterial Communities in Wet and Dry Seasons during a Seasonal Water Level Fluctuation in the Largest Freshwater Lake (Poyang Lake) in China. Front. Microbiol. 2019, 10, 1167. [Google Scholar] [CrossRef] [PubMed]
- Zohary, T.; Ostrovsky, I. Ecological impacts of excessive water level fluctuations in stratified freshwater lakes. Inland Waters 2011, 1, 47–59. [Google Scholar] [CrossRef]
- Feng, L.; Hu, C.; Chen, X.; Cai, X.; Tian, L.; Gan, W. Assessment of inundation changes of Poyang Lake using MODIS observations between 2000 and 2010. Remote Sens. Environ. 2012, 121, 80–92. [Google Scholar] [CrossRef]
- Haddeland, I.; Heinke, J.; Biemans, H.; Eisner, S.; Florke, M.; Hanasaki, N.; Konzmann, M.; Ludwig, F.; Masaki, Y.; Schewe, J.; et al. Global water resources affected by human interventions and climate change. Proc. Natl. Acad. Sci. USA 2014, 111, 3251–3256. [Google Scholar] [CrossRef]
- Poff, N.L.; Zimmerman, J.K.H. Ecological responses to altered flow regimes: A literature review to inform the science and management of environmental flows. Freshw. Biol. 2010, 55, 194–205. [Google Scholar] [CrossRef]
- Wantzen, K.M.; Rothhaupt, K.-O.; Mörtl, M.; Cantonati, M.; G-Tóth, L.; Fischer, P. Ecological effects of water-level fluctuations in lakes: An urgent issue. Hydrobiologia 2008, 613, 1–4. [Google Scholar] [CrossRef]
- Loiselle, S.A.; Bracchini, L.; Cózar, A.; Dattilo, A.M.; Rossi, C. Extensive spatial analysis of the light environment in a subtropical shallow lake, Laguna Iberá, Argentina. Hydrobiologia 2005, 534, 181–191. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Yao, J. Investigation of Residence and Travel Times in a Large Floodplain Lake with Complex Lake-River Interactions: Poyang Lake (China). Water 2015, 7, 1991–2012. [Google Scholar] [CrossRef]
- Hideo, O.; Shuichi, E.; Toshiyuki, I.; Yasuaki, O.; Shinji, T. Seasonal Changes in Water Quality as Affected by Water Level Fluctuations in Lake Tonle Sap, Cambodia. Geogr. Rev. Jpn. Ser. B 2017, 90, 53–65. [Google Scholar] [CrossRef][Green Version]
- Dinka, M.; Agoston-Szabo, E.; Berczik, A.; Kutrucz, G. Influence of water level fluctuation on the spatial dynamic of the water chemistry at Lake Ferto/Neusiedler See. Limnological 2004, 34, 48–56. [Google Scholar] [CrossRef]
- Coops, H.; Beklioglu, M.; Crisman, T.L. The role of water-level fluctuations in shallow lake ecosystems—Workshop conclusions. Hydrobiologia 2003, 506, 23–27. [Google Scholar] [CrossRef]
- Norris, R.H.; Barbour, M.T. Bioassessment of Aquatic Ecosystems. In Encyclopedia of Inland Waters; Elsevier: Oxford, UK, 2009; pp. 21–28. [Google Scholar]
- Ren, Z.; Qu, X.; Peng, W.; Yu, Y.; Zhang, M. Nutrients Drive the Structures of Bacterial Communities in Sediments and Surface Waters in the River-Lake System of Poyang Lake. Water 2019, 11, 930. [Google Scholar] [CrossRef]
- Soldatova, E.; Guseva, N.; Sun, Z.; Bychinsky, V.; Boeckx, P.; Gao, B. Sources and behaviour of nitrogen compounds in the shallow groundwater of agricultural areas (Poyang Lake basin, China). J. Contam. Hydrol. 2017, 202, 59–69. [Google Scholar] [CrossRef]
- Baumgärtner, D.; Mörtl, M.; Rothhaupt, K.-O. Effects of water-depth and water-level fluctuations on the macroinvertebrate community structure in the littoral zone of Lake Constance. Hydrobiologia 2008, 613, 97–107. [Google Scholar] [CrossRef]
- Brauns, M.; Garcia, X.-F.; Pusch, M.T. Potential effects of water-level fluctuations on littoral invertebrates in lowland lakes. Hydrobiologia 2008, 613, 5–12. [Google Scholar] [CrossRef]
- Sutela, T.; Vehanen, T. Effects of water-level regulation on the nearshore fish community in boreal lakes. Hydrobiologia 2008, 613, 13–20. [Google Scholar] [CrossRef]
- White, M.S.; Xenopoulos, M.A.; Hogsden, K.; Metcalfe, R.A.; Dillon, P.J. Natural lake level fluctuation and associated concordance with water quality and aquatic communities within small lakes of the Laurentian Great Lakes region. Hydrobiologia 2008, 613, 21–31. [Google Scholar] [CrossRef]
- Schaechter, M. Eukaryotic Microbes; Academic Press and Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Zubkov, M.V.; Tarran, G.A. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature 2008, 455, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Fabian, J.; Zlatanovic, S.; Mutz, M.; Premke, K. Fungal-bacterial dynamics and their contribution to terrigenous carbon turnover in relation to organic matter quality. ISME J. 2017, 11, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Khomich, M.; Davey, M.L.; Kauserud, H.; Rasconi, S.; Andersen, T. Fungal communities in Scandinavian lakes along a longitudinal gradient. Fungal Ecol. 2017, 27, 36–46. [Google Scholar] [CrossRef]
- Mohamed, D.J.; Martiny, J.B. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 2011, 5, 379–388. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, Z.; Chen, C.; Jia, Z. Soil microbial community structure and diversity are largely influenced by soil pH and nutrient quality in 78-year-old tree plantations. Biogeosciences 2017, 14, 2101–2111. [Google Scholar] [CrossRef]
- Capo, E.; Debroas, D.; Arnaud, F.; Perga, M.E.; Chardon, C.; Domaizon, I. Tracking a century of changes in microbial eukaryotic diversity in lakes driven by nutrient enrichment and climate warming. Environ. Microbiol. 2017, 19, 2873–2892. [Google Scholar] [CrossRef]
- Logares, R.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.M.; Decelle, J.; Dolan, J.R.; Dunthorn, M.; et al. Patterns of rare and abundant marine microbial eukaryotes. Curr. Biol. 2014, 24, 813–821. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, H.; Yang, J.R.; Liu, M.; Huang, B.; Yang, J. Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 2018, 12, 2263–2277. [Google Scholar] [CrossRef]
- Pedrós-Alió, C. The rare bacterial biosphere. Ann. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef]
- Lepère, C.; Boucher, D.; Jardillier, L.; Domaizon, I.; Debroas, D. Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin). Appl. Environ. Microbiol. 2006, 72, 2971–2981. [Google Scholar] [CrossRef]
- Caron, D.A.; Countway, P.D. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat. Microb. Ecol. 2009, 57, 227–238. [Google Scholar] [CrossRef]
- Debroas, D.; Hugoni, M.; Domaizon, I. Evidence for an active rare biosphere within freshwater protists community. Mol. Ecol. 2015, 24, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.J.; Yu, L.; Heidelberg, J.F.; Kirchman, D.L. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. USA 2011, 108, 12776–12781. [Google Scholar] [CrossRef]
- Pedrós-Alió, C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006, 14, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Hugoni, M.; Taib, N.; Debroas, D.; Domaizon, I.; Jouan Dufournel, I.; Bronner, G.; Salter, I.; Agogue, H.; Mary, I.; Galand, P.E. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Natl. Acad. Sci. USA 2013, 110, 6004–6009. [Google Scholar] [CrossRef]
- Liu, L.; Yang, J.; Lv, H.; Yu, X.; Wilkinson, D.M.; Yang, J. Phytoplankton Communities Exhibit a Stronger Response to Environmental Changes than Bacterioplankton in Three Subtropical Reservoirs. Environ. Sci. Technol. 2015, 49, 10850–10858. [Google Scholar] [CrossRef]
- Logares, R.; Mangot, J.F.; Massana, R. Rarity in aquatic microbes: Placing protists on the map. Res. Microbiol. 2015, 166, 831–841. [Google Scholar] [CrossRef]
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef]
- Raven, J.A.; Giordano, M. Acquisition and metabolism of carbon in the Ochrophyta other than diatoms. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373. [Google Scholar] [CrossRef]
- Tas, B.; Gonulol, A. An ecologic and taxonomic study on phytoplankton of a shallow lake, Turkey. J. Environ. Biol. 2007, 28, 439–445. [Google Scholar] [PubMed]
- Chen, M.; Chen, F.; Zhao, B.; Wu, Q.L.; Kong, F. Seasonal variation of microbial eukaryotic community composition in the large, shallow, subtropical Taihu Lake, China. Aquat. Ecol. 2010, 44, 1–12. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, S.; Zhang, W.; Feng, M.; Yu, R. A rule of hydrological regulating on nutritional status of Poyang Lake, since the operation of the Three Gorges Dam. Ecol. Indic. 2019, 104, 535–542. [Google Scholar] [CrossRef]
- Tzoraki, O.; Nikolaidis, N.P.; Amaxidis, Y.; Skoulikidis, N.T. In-stream biogeochemical processes of a temporary river. Environ. Sci. Technol. 2007, 41, 1225–1231. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, X.; Tang, C.; Xu, H.; Jiang, X.; Zhang, Y. Influence of long-term inundation and nutrient addition on denitrification in sandy wetland sediments from Poyang Lake, a large shallow subtropical lake in China. Environ. Pollut. 2016, 219, 440–449. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Zhang, Y.; Peng, W. Microbial community structure and functional properties in permanently and seasonally flooded areas in Poyang Lake. Sci. Rep. 2020, 10, 4819. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Y.; Huang, B.; Wei, Z.; Miao, A.; Yang, L. Nitrogen and phosphorus limitation of phytoplankton growth in different areas of Lake Taihu, China. J. Freshw. Ecol. 2014, 30, 113–127. [Google Scholar] [CrossRef]
- Fand, C.; Lai, Z.; Yang, J.; Xu, Y. Study on the Nonuniform Spatial Distribution of Water Level in Poyang Lake Based on ASAR Images and DEM. Procedia Environ. Sci. 2011, 10, 2540–2546. [Google Scholar]
- Zhao, J.; Li, J.; Yan, H.; Zheng, L.; Dai, Z. Analysis on the Water Exchange between the Main Stream of the Yangtze River and the Poyang Lake. Procedia Environ. Sci. 2011, 10, 2256–2264. [Google Scholar] [CrossRef]
- Hui, F.; Xu, B.; Huang, H.; Yu, Q.; Gong, P. Modelling spatial-temporal change of Poyang Lake using multitemporal Landsat imagery. Int. J. Remote Sens. 2008, 29, 5767–5784. [Google Scholar] [CrossRef]
- Wang, L.; Liang, T. Distribution characteristics of phosphorus in the sediments and overlying water of Poyang lake. PLoS ONE 2015, 10, e0125859. [Google Scholar] [CrossRef] [PubMed]
- Shankman, D.; Keim, B.D.; Song, J. Flood frequency in China’s Poyang Lake region: Trends and teleconnections. Int. J. Climatol. 2006, 26, 1255–1266. [Google Scholar] [CrossRef]
- Shankman, D.; Liang, Q. Landscape Changes and Increasing Flood Frequencyin China’s Poyang Lake Region. Prof. Geogr. 2003, 55, 434–445. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Hawkins, J.A.; Jones, S.K., Jr.; Finkelstein, I.J.; Press, W.H. Indel-correcting DNA barcodes for high-throughput sequencing. Proc. Natl. Acad. Sci. USA 2018, 115, E6217–E6226. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Chen, W.; Pan, Y.; Yu, L.; Yang, J.; Zhang, W. Patterns and Processes in Marine Microeukaryotic Community Biogeography from Xiamen Coastal Waters and Intertidal Sediments, Southeast China. Front. Microbiol. 2017, 8, 1912. [Google Scholar] [CrossRef]
- Manthey, M.; Fridley, J.D. Beta Diversity Metrics and the Estimation of Niche Width via Species Co-Occurrence Data: Reply to Zeleny. J. Ecol. 2009, 97, 18–22. [Google Scholar] [CrossRef]
- Levins, R. Evolution in Changing Environments; Princeton University Press: Princeton, NJ, USA, 1968. [Google Scholar]
- Göthe, E.; Baattrup-Pedersen, A.; Wiberg-Larsen, P.; Graeber, D.; Kristensen, E.A.; Friberg, N. Environmental and spatial controls of taxonomic versus trait composition of stream biota. Freshwater. Biol. 2017, 62, 397–413. [Google Scholar] [CrossRef]
- Liu, L.; Chen, H.; Liu, M.; Yang, J.R.; Xiao, P.; Wilkinson, D.M.; Yang, J. Response of the eukaryotic plankton community to the cyanobacterial biomass cycle over 6 years in two subtropical reservoirs. ISME. J. 2019, 13, 2196–2208. [Google Scholar] [CrossRef] [PubMed]
- de Menezes, A.B.; Prendergast-Miller, M.T.; Richardson, A.E.; Toscas, P.; Farrell, M.; Macdonald, L.M.; Wark, T.; Thrall, P.H. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ. Microbiol. 2015, 17, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Csardi, M.G. Package ’igraph’, Version 0.6.6.; The Network Analysis Package; CZI: Redwood City, CA, USA, 2013. [Google Scholar]
- Qu, X.; Peng, W.; Liu, Y.; Zhang, M.; Ren, Z.; Wu, N.; Liu, X. Networks and ordination analyses reveal the stream community structures of fish, macroinvertebrate and benthic algae, and their responses to nutrient enrichment. Ecol. Indic. 2019, 101, 501–511. [Google Scholar] [CrossRef]
- Jacob, R.; Harikrishnan, K.P.; Misra, R.; Ambika, G. Measure for degree heterogeneity in complex networks and its application to recurrence network analysis. R. Soc. Open Sci. 2017, 4. [Google Scholar] [CrossRef]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Guimerà, R.; Amaral, L.A.N. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef]
- Riis, T.; Hawes, I. Relationships between water level fluctuations and vegetation diversity in shallow water of New Zealand lakes. Aquat. Bot. 2002, 74, 133–148. [Google Scholar] [CrossRef]
- Turner, M.A.; Huebert, D.B.; Findlay, D.L.; Hendzel, L.L.; Jansen, W.A.; Bodaly, R.A.; Armstrong, L.M.; Kasian, S.E.M. Divergent impacts of experimental lake-level drawdown on planktonic and benthic plant communities in a boreal forest lake. Can. J. Fish. Aquat. Sci. 2005, 62, 991–1003. [Google Scholar] [CrossRef]
- Judd, K.E.; Crump, B.C.; Kling, G.W. Variation in dissolved organic matter controls bacterial production and community composition. Ecology 2006, 87, 2068–2079. [Google Scholar] [CrossRef]
- Peter, H.; Ylla, I.; Gudasz, C.; Romani, A.M.; Sabater, S.; Tranvik, L.J. Multifunctionality and diversity in bacterial biofilms. PLoS ONE 2011, 6, e23225. [Google Scholar] [CrossRef]
- Li, B.; Yang, G.; Wan, R.; Zhang, Y.; Dai, X.; Chen, Y. Spatiotemporal Variability in the Water Quality of Poyang Lake and Its Associated Responses to Hydrological Conditions. Water 2016, 8, 296. [Google Scholar] [CrossRef]
- Yao, X.; Wang, S.; Ni, Z.; Jiao, L. The response of water quality variation in Poyang Lake (Jiangxi, People’s Republic of China) to hydrological changes using historical data and DOM fluorescence. Environ. Sci. Pollut. Res. Int. 2015, 22, 3032–3042. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhu, G.; Li, W.; Zhang, Y.; Zhao, L.; Gu, Z. Estimation of the algal-available phosphorus pool in sediments of a large, shallow eutrophic lake (Taihu, China) using profiled SMT fractional analysis. Environ. Pollut. 2013, 173, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Teubner, K.; Chen, Y. Water quality characteristics of Poyang Lake, China, in response to changes in the water level. Hydrol. Res. 2016, 47, 238–248. [Google Scholar] [CrossRef]
- Toming, K.; Tuvikene, L.; Vilbaste, S.; Agasild, H.; Viik, M.; Kisand, A.; Feldmann, T.; Martma, T.; Jones, R.I.; Nõges, T. Contributions of autochthonous and allochthonous sources to dissolved organic matter in a large, shallow, eutrophic lake with a highly calcareous catchment. Limnol. Oceanogr. 2013, 58, 1259–1270. [Google Scholar] [CrossRef]
- Duan, S.; Kaushal, S.S.; Groffman, P.M.; Band, L.E.; Belt, K.T. Phosphorus export across an urban to rural gradient in the Chesapeake Bay watershed. J. Geophys. Res. Biogeosciences 2012, 117, 117. [Google Scholar] [CrossRef]
- Kronvang, B.; Laubel, A.; Grant, R. Suspended sediment and particulate phosphorus transport and delivery pathways in an arable catchment, Gelaek Stream, Denmark. Hydrol. Process. 1997, 11, 627–642. [Google Scholar] [CrossRef]
- Massey, H.F.; Jackson, M.L. Selective Erosion of Soil Fertility Constituents. Soil. Sci. Soc. Am. J. 1952, 16, 353–356. [Google Scholar] [CrossRef]
- River, M.; Richardson, C.J. Particle size distribution predicts particulate phosphorus removal. Ambio 2018, 47, 124–133. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Microbial community structure and its functional implications. Nature 2009, 459, 193–199. [Google Scholar] [CrossRef] [PubMed]
- McCann, K.S. The diversity-stability debate. Nature 2000, 405, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.M.; Pimm, S.L.; Sole, R.V. Ecological networks and their fragility. Nature 2006, 442, 259–264. [Google Scholar] [CrossRef]
- Saavedra, S.; Stouffer, D.B.; Uzzi, B.; Bascompte, J. Strong Contributors to Network Persistence are the Most Vulnerable to Extinction. Nature 2011, 478, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Baah-Acheamfour, M.; Carlyle, C.N.; Bissett, A.; Richardson, A.E.; Siddique, T.; Bork, E.W.; Chang, S.X. Determinants of bacterial communities in Canadian agroforestry systems. Environ. Microbiol. 2016, 18, 1805–1816. [Google Scholar] [CrossRef]
- Gophen, M. The Impact of Nitrogen and Phosphorus Dynamics on the Kinneret Phytoplankton: II: Chlorophyta, Cyanophyta, Diatoms and Peridinium. Open J. Mod. Hydrol. 2017, 7, 298–313. [Google Scholar] [CrossRef][Green Version]
- Beaver, J.R.; Crisman, T.L.; Bienert, R.W. Distribution of planktonic ciliates in highly coloured subtropical lakes: Comparison with clearwater ciliate communities and the contribution of myxotrophic taxa to total autotrophic biomass. Freshw. Biol. 1988, 20, 51–60. [Google Scholar] [CrossRef]
- Ramos Rodríguez, E.; Pérez-Martínez, C.; Conde-Porcuna, J.M. Strict stoichiometric homeostasis of Cryptomonas pyrenoidifera (Cryptophyceae) in relation to N:P supply ratios. J. Limnol. 2017, 76, 182–189. [Google Scholar] [CrossRef]
- Liu, Y.; Qu, X.; Elser, J.J.; Peng, W.; Zhang, M.; Ren, Z.; Zhang, H.; Zhang, Y.; Yang, H. Impact of Nutrient and Stoichiometry Gradients on Microbial Assemblages in Erhai Lake and Its Input Streams. Water 2019, 11, 1711. [Google Scholar] [CrossRef]

| Topological Parameters | Dry | Wet | ||
|---|---|---|---|---|
| Real | Random | Real | Random | |
| Number of Nodes | 574 | 574 | 682 | 682 |
| Number of Edges | 2263 | 2263 | 5151 | 5151 |
| Negative Edges | 252 (11.1%) | 252 | 2003 (38.9%) | 2003 |
| Connectance | 0.014 | 0.014 | 0.022 | 0.022 |
| Average Degree | 7.885 | 7.885 | 15.106 | 15.106 |
| Average Path Length | 5.525 a | 3.306 ± 0.005 * | 4.508 b | 2.694 ± 0.001 * |
| Diameter | 17.000 a | 5.997 ± 0.263 * | 16.000 b | 4.015 ± 0.122 * |
| Clustering Coefficient | 0.476 a | 0.014 ± 0.002 * | 0.449 b | 0.022 ± 0.001 * |
| Centralization Degree | 0.056 a | 0.017 ± 0.002 * | 0.101 b | 0.020 ± 0.003 * |
| Centralization Betweenness | 0.097 a | 0.013 ± 0.003 * | 0.040 b | 0.006 ± 0.001 * |
| Modularity | 0.653 a | 0.323 ± 0.004 * | 0.437 b | 0.220 ± 0.003 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Peng, W. Seasonal Water Level Fluctuation and Concomitant Change of Nutrients Shift Microeukaryotic Communities in a Shallow Lake. Water 2020, 12, 2317. https://doi.org/10.3390/w12092317
Liu Y, Ren Z, Qu X, Zhang M, Yu Y, Peng W. Seasonal Water Level Fluctuation and Concomitant Change of Nutrients Shift Microeukaryotic Communities in a Shallow Lake. Water. 2020; 12(9):2317. https://doi.org/10.3390/w12092317
Chicago/Turabian StyleLiu, Yang, Ze Ren, Xiaodong Qu, Min Zhang, Yang Yu, and Wenqi Peng. 2020. "Seasonal Water Level Fluctuation and Concomitant Change of Nutrients Shift Microeukaryotic Communities in a Shallow Lake" Water 12, no. 9: 2317. https://doi.org/10.3390/w12092317
APA StyleLiu, Y., Ren, Z., Qu, X., Zhang, M., Yu, Y., & Peng, W. (2020). Seasonal Water Level Fluctuation and Concomitant Change of Nutrients Shift Microeukaryotic Communities in a Shallow Lake. Water, 12(9), 2317. https://doi.org/10.3390/w12092317






