Profile of the Spatial Distribution Patterns of the Human and Bacteriophage Virome in a Wastewater Treatment Plant Located in the South of Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Wastewater Treatment Plant Characterization
2.2. Sample Collection and Nucleic Acid Extraction
2.3. Visualization of Viral Particles by TEM
2.4. Bioinformatics Pipeline
2.5. α-Diversity Indices and Similarity Analyses of Samples
2.6. Differential Abundance of Viral Phylotypes
2.7. Multivariate Redundancy Analysis
3. Results and Discussion
3.1. Quality of the Viral Nucleic Acid Extraction Protocol
3.2. Viral Particle Morphology by TEM
3.3. Viral Phylotypes in the Wastewater Treatment Plant
3.4. Distribution of Viral Phylotypes in the Wastewater Treatment Plant
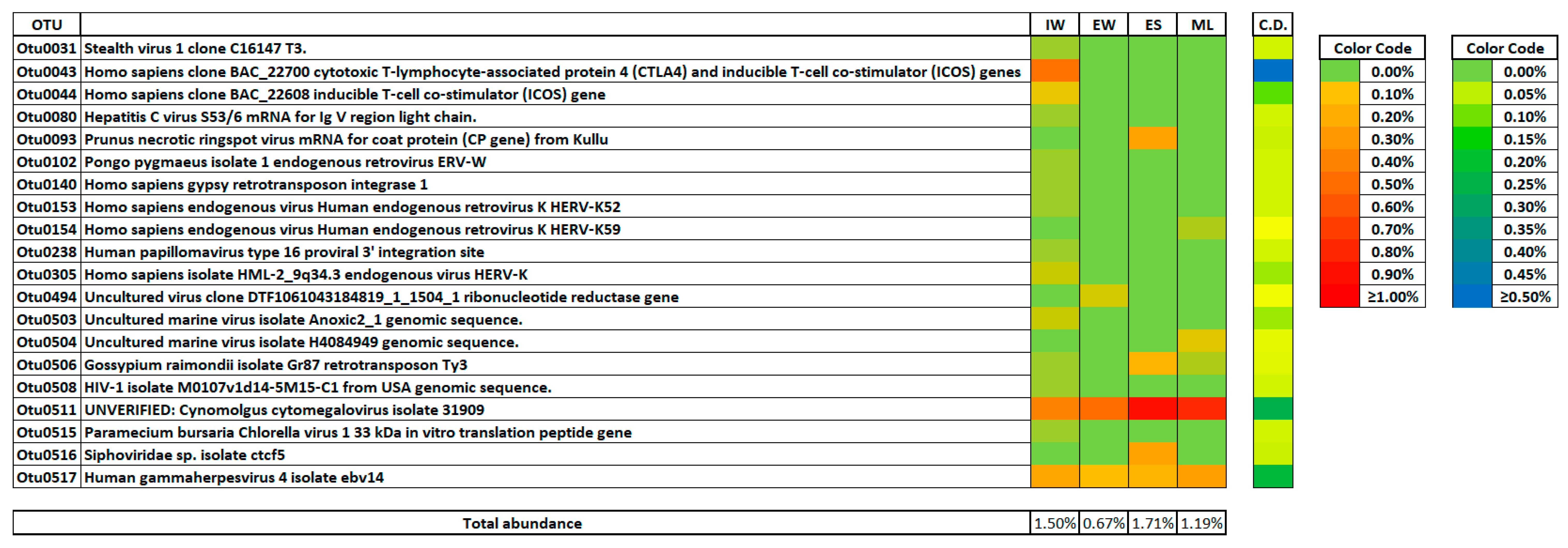
3.5. Sensitive Viral Phylotypes
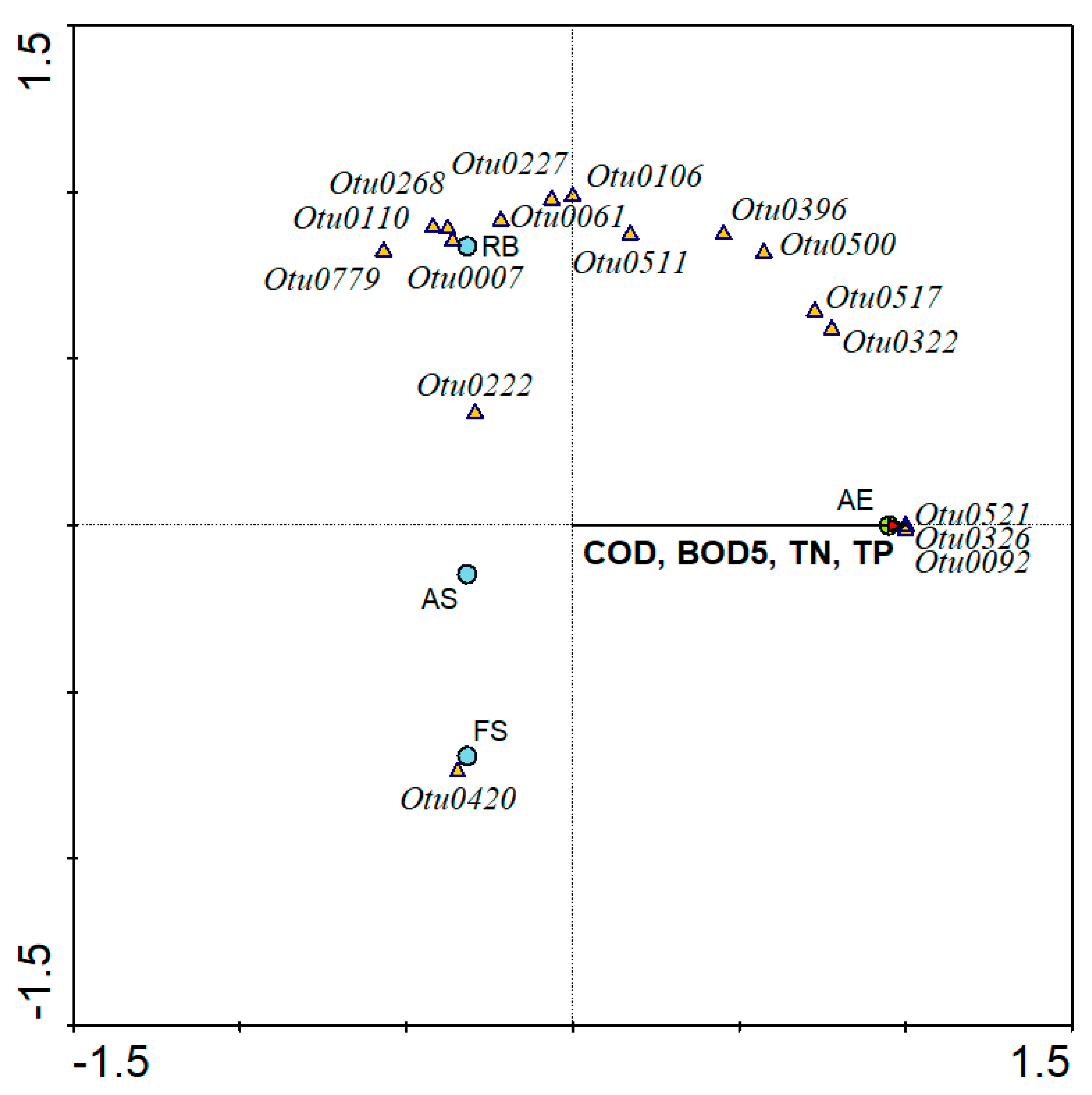
3.6. Linkage between Operational Conditions and Viral Phylotypes
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Kaas, L.; Gourinat, A.C.; Urbès, F.; Langlet, J. A 1-year study on the detection of human enteric viruses in New Caledonia. Food Environ. Virol. 2016, 8, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Li, Q.; Lee, B.E.; Ruecker, N.J.; Neumann, N.F.; Ashbolt, N.J.; Pang, X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018, 147, 73–81. [Google Scholar] [CrossRef]
- O’Brien, E.; Munir, M.; Marsh, T.; Heran, M.; Lesage, G.; Tarabara, V.V.; Xagoraraki, I. Diversity of DNA viruses in effluents of membrane bioreactors in Traverse City, MI (USA) and La Grande Motte (France). Water Res. 2017, 111, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J. Microbial contamination of drinking water and human health from community water systems. Curr. Environ. Health Rep. 2015, 2, 95–106. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Novel Coronavirus (2019-nCoV) Technical Guidance; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Bibby, K. Metagenomic identification of viral pathogens. Trends Biotechnol. 2013, 31, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Sibanda, T.; Gusha, S.S. Inadequately treated wastewater as a source of human enteric viruses in the environment. Int. J. Environ. Res. Public Health 2010, 7, 2620–2637. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Liu, L.; Li, B.; Zhang, T. High-resolution temporal and spatial patterns of Virome in wastewater treatment systems. Environ. Sci. Technol. 2018, 52, 10337–10346. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Saunders, A.M.; Hansen, A.A.; Larsen, P.; Nielsen, J.L. Microbial communities involved in enhanced biological phosphorus removal from wastewater—A model system in environmental biotechnology. Curr. Opin. Biotechnol. 2012, 23, 452–459. [Google Scholar] [CrossRef]
- Carducci, A.; Tozzi, E.; Rubulotta, E.; Casini, B.; Cantiani, L.; Rovini, E.; Muscillo, M.; Pacini, R. Assessing airborne biological hazard from urban wastewater treatment. Water Res. 2000, 34, 1173–1178. [Google Scholar] [CrossRef]
- US EPA. Prepublication of the Ground Water Rule Federal Register Notice; US EPA: Washington, DC, USA, 2006.
- Marion, J.W.; Lee, C.; Lee, C.S.; Wang, Q.; Lemeshow, S.; Buckley, T.J.; Saif, L.J.; Lee, J. Integrating bacterial and viral water quality assessment to predict swimming-associated illness at a freshwater beach: A cohort study. PLoS ONE 2014, 11, e112029. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Solis, J.J.; David, Z.; Ramon, C.; Lalucat, J. Comparative reductions of bacterial indicators, bacteriophage-infecting enteric bacteria and enteroviruses in wastewater tertiary treatments by lagooning and UV-radiation. Water Sci. Technol. 2008, 58, 2223–2233. [Google Scholar] [CrossRef] [PubMed]
- Simmons, F.J.; Xagoraraki, I. Release of infectious human enteric viruses by full-scale wastewater utilities. Water Res. 2011, 45, 3590–3598. [Google Scholar] [CrossRef]
- Carducci, A.; Morici, P.; Pizzi, F.; Battistini, R.; Rovini, E.; Verani, M. Study of the viral removal efficiency in a urban wastewater treatment plant. Water Sci. Technol. 2008, 58, 893–897. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.; Wichelns, D.; Raschid-Sally, L.; Minhas, P.S.; Drechsel, P.; Bahri, A.; Ensink, J.H. Agricultural use of marginal-quality water—Opportunities and challenges. In Water for Food, Water for Life. A Comprehensive Assessment of Water Management in Agriculture; IWMI & Earthscan: London, UK; Ottawa, ON, Canada, 2017; No. 612-2016-40578. [Google Scholar]
- Molle, B.; Brelle, F.; Bessy, J.; Gatel, D. Which water quality for which uses? Overcoming over-zealous use of the precautionary principle to reclaim wastewater for appropriate irrigation uses. Irrig. Drain. 2012, 61, 87–94. [Google Scholar] [CrossRef]
- Drechsel, P.; Evans, A.E.V. Wastewater reuse in irrigated agriculture. Irrig. Drain. Syst. 2010, 24, 1–152. [Google Scholar] [CrossRef]
- Nuanualsuwan, S.; Thongtha, P.; Kamolsiripichaiporn, S.; Subharat, S. UV inactivation and model of UV inactivation of foot-and-mouth disease viruses in suspension. Int. J. Food Microbiol. 2008, 127, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Puchol, S.; Rusiñol, M.; Fernández-Cassi, X.; Timoneda, N.; Itarte, M.; Andrés, C.; Anton, C.; Abril, J.F.; Girones, R.; Bofill-Mas, S. Characterisation of the sewage virome: Comparison of NGS tools and occurrence of significant pathogens. Sci. Total Environ. 2020, 713, 136604. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Farkas, K.; Harrison, C.; Jones, D.L.; Allison, H.E.; McCarthy, A.J. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. mSystems 2018, 3. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Timoneda, N.; Martínez-Puchol, S.; Rusinol, M.; Rodriguez-Manzano, J.; Figuerola, N.; Bofill-Mas, S.; Abril, J.F.; Girones, R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2018, 618, 870–880. [Google Scholar] [CrossRef]
- Maza-Márquez, P.; Castellano-Hinojosa, A.; González-Martínez, A.; Juárez-Jiménez, B.; González-López, J.; Rodelas, B. Abundance of total and metabolically active Candidatus Microthrix and fungal populations in three full-scale wastewater treatment plants. Chemosphere 2019, 232, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Aw, T.G.; Howe, A.; Rose, J.B. Metagenomic approaches for direct and cell culture evaluation of the virological quality of wastewater. J. Virol. Methods 2014, 210, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Semenkovich, N.P.; Whiteson, K.; Rohwer, F.; Gordon, J.I. Going viral: Next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 2012, 10, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Marston, D.A.; McElhinney, L.M.; Ellis, R.J.; Horton, D.L.; Wise, E.L.; Leech, S.L.; David, D.; de Lamballerie, X.; Fooks, A.R. Nextion sequencing of viral RNA genomes. BMC Genom. 2013, 14, 444. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Nilsson, C.; Lim, Y.W.; Ruan, Y.; Breitbart, M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009, 11, 2806–2820. [Google Scholar] [CrossRef] [PubMed]
- Hayat, M.A.; Miller, S.E. Negative Staining; McGraw-Hill: New York, NY, USA, 1990. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Altschul, S.F. BLAST algorithm. ELS 2001. [Google Scholar] [CrossRef]
- Zhao, G.; Wu, G.; Lim, E.S.; Droit, L.; Krishnamurthy, S.; Barouch, D.H.; Virgin, H.W.; Wang, D. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology 2017, 503, 21–30. [Google Scholar] [CrossRef]
- Goodacre, N.; Aljanahi, A.; Nandakumar, S.; Mikailov, M.; Khan, A.S. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Bian, G.; Gloor, G.B.; Gong, A.; Jia, C.; Zhang, W.; Hu, J.; Burton, J.P. The Gut Microbiota of Healthy Aged Chinese Is Similar to That of the Healthy Young. mSphere 2018, 2. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.-C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Muñoz-Palazon, B.; Rodriguez-Sanchez, A.; Hurtado-Martinez, M.; Santana, F.; Gonzalez-Lopez, J.; Mack, L.; Gonzalez-Martinez, A. Polar Arctic Circle biomass enhances performance and stability of aerobic granular sludge systems operated under different temperatures. Bioresour. Technol. 2020, 300, 122650. [Google Scholar] [CrossRef] [PubMed]
- Rosani, U.; Gerdol, M. A bioinformatics approach reveals seven nearly-complete RNA-virus genomes in bivalve RNA-seq data. Virus Res. 2017, 239, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, R.; Fane, A.G. The roles of bacteriophages in membrane-based water and wastewater treatment processes: A review. Water Res. 2017, 110, 120–132. [Google Scholar] [CrossRef]
- Tanji, Y.; Mizoguchi, K.; Yoichi, M.; Morita, M.; Kijima, N.; Kator, H.; Unno, H. Seasonal change and fate of coliphages infected to Escherichia coli O157: H7 in a wastewater treatment plant. Water Res. 2003, 37, 1136–1142. [Google Scholar] [CrossRef]
- Zhang, Y.; Hunt, H.K.; Hu, Z. Application of bacteriophages to selectively remove Pseudomonas aeruginosa in water and wastewater filtration systems. Water Res. 2013, 47, 4507–4518. [Google Scholar] [CrossRef]
- Kitajima, M.; Iker, B.C.; Pepper, I.L.; Gerba, C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes—Identification of potential viral indicators. Sci. Total Environ. 2014, 488, 290–296. [Google Scholar] [CrossRef]
- Hata, A.; Kitajima, M.; Katayama, H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J. Appl. Microbiol. 2013, 114, 545–554. [Google Scholar] [CrossRef]
- Ateba, C.N.; Akindolire, M.A. Isolation and characterisation of bacteriophages with lytic activity against virulent Escherichia coli O157: H7: Potential bio-control agents. Preprints 2019, 2019010132. [Google Scholar] [CrossRef]
- Hewitt, J.; Leonard, M.; Greening, G.E.; Lewis, G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011, 45, 6267–6276. [Google Scholar] [CrossRef]
- Gulino, K.; Rahman, J.; Badri, M.; Morton, J.; Bonneau, R.; Ghedin, E. Initial Mapping of the New York City Wastewater Virome. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.; Newman, C.; Knowles, M.; Bolton, F.J.; Hollyoak, V.; Richards, S.; Daley, P.; Counter, D.; Smith, H.R.; Keppie, N.E. Coli O157 phage type 21/28 outbreak in North Cumbria associated with pasteurized milk. Epidemiol. Infect. 2002, 129, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Gabisoniya, T.G.; Loladze, M.Z.; Nadiradze, M.M.; Chakhunashvili, N.K.; Alibegashvili, M.G.; Tamarashvili, N.G.; Pushkina, V.A. Effects of bacteriophages on biofilm formation by strains of Pseudomonas aeruginosa. Appl. Biochem. Microbiol. 2016, 52, 293–297. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z. Combined treatment of Pseudomonas aeruginosa biofilms with bacteriophages and chlorine. Biotechnol. Bioeng. 2013, 110, 286–295. [Google Scholar] [CrossRef] [PubMed]
- De Smet, J.; Hendrix, H.; Blasdel, B.G.; Danis-Wlodarczyk, K.; Lavigne, R. Pseudomonas predators: Understanding and exploiting phage–host interactions. Nat. Rev. Microbiol. 2017, 15, 517. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Munir, M.; Marsh, T.; Xagoraraki, I. Metagenomic insights into microbial diversity and resistance to antibiotics in wastewater treatment plants. In Environmental Engineering—Doctor of Philosophy; Michigan State University: Lansing, MI, USA, 2014; p. 10. [Google Scholar]
- Shapiro, O.H.; Kushmaro, A. Bacteriophage ecology in environmental biotechnology processes. Curr. Opin. Biotech. 2011, 22, 449–455. [Google Scholar] [CrossRef]
- Calero-Cáceres, W.; Muniesa, M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- Wong, M.V.; Hashsham, S.A.; Gulari, E.; Rouillard, J.M.; Aw, T.G.; Rose, J.B. Detection and characterization of human pathogenic viruses circulating in community wastewater using multi target microarrays and polymerase chain reaction. J. Water Health 2013, 11, 659–670. [Google Scholar] [CrossRef]
- Alhamlan, F.S.; Ederer, M.M.; Brown, C.J.; Coats, E.R.; Crawford, R.L. Metagenomics-based analysis of viral communities in dairy lagoon wastewater. J. Microbiol. Meth. 2013, 92, 183–188. [Google Scholar] [CrossRef]
- Foster, A.L.; Gaisa, M.M.; Hijdra, R.M.; Turner, S.S.; Morey, T.J.; Jacobson, K.B.; Fierer, D.S. Shedding of hepatitis C virus into the rectum of HIV-infected men who have sex with men. Clin. Infect. Dis. 2016, 64, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Withey, S.; Cartmell, E.; Avery, L.M.; Stephenson, T. Bacteriophages—Potential for application in wastewater treatment processes. Sci. Total Environ. 2005, 339, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gerba, C.P.; Betancourt, W.Q.; Kitajima, M.; Rock, C.M. Reducing uncertainty in estimating virus reduction by advanced water treatment processes. Water Res. 2018, 133, 282–288. [Google Scholar] [CrossRef] [PubMed]





| Parameter | Value |
|---|---|
| Bioreactor volume (m3) | 3754 |
| Equivalent inhabitants | 223,000 |
| Flow rate (m3/month) | 552,295 ± 9480 |
| HRT (h) | 4.9 ± 0.1 |
| SRT (day) | 4.0 ± 0.2 |
| TSS (g/L) | 5.05 ± 0.57 |
| Maximum–minimum temperature (°C) | 16.5 ± 3.7 to 2.8 ± 3.4 |
| Average temperature (°C) | 9.3 ± 2.9 |
| Relative humidity (%) | 74.8 ± 10.0 |
| Wind speed (m/s) | 1.1 ± 0.6 |
| Average daily solar radiation (Mj/m2) | 12.5 ± 6.6 |
| Precipitation (mm/day) | 1.0 ± 3.4 |
| COD | BOD5 | TN | TP | pH | |
|---|---|---|---|---|---|
| Influent | 653.8 ± 27.7 mg O2/L | 374.4 ± 23.5 mg O2/L | 97.4 ± 0.2 mg/L | 11.5 ± 2.3 mg/L | 7.7 ± 2 |
| Effluent | 70.0 ± 2.4 mg O2/L | 12.2 ± 1.3 mg O2/L | 52.3 ± 8.3 mg/L | 1.0 ± 0.2 mg/L | 7.7 ± 2 |
| Removal | 92.0 ± 0.6% | 97.4 ± 0.2% | 43.4 ± 9.9 mg/L | 90.8 ± 2.6 mg/L |
| Sample | DNA | RNA | cDNA | DNA + cDNA | Abs260/Abs280 |
|---|---|---|---|---|---|
| Influent water (IW) | 61 | 40 | 1310 | 71 | 1.98 |
| Mixed liquor (MX) | 56 | 50 | 1612 | 61 | 1.91 |
| Excess sludge (ES) | 57 | 70 | 1413 | 65 | 1.84 |
| Effluent water (EW) | 108 | 52 | 1372 | 112 | 1.96 |
| Samples | Chao-1 | Shannon–Wiener | Simpson |
|---|---|---|---|
| Influent water (IW) | 1566.6 | 2.7 | 0.4 |
| Effluent water (EW) | 412.3 | 1.1 | 0.7 |
| Excess sludge (ES) | 333.8 | 1.2 | 0.6 |
| Mixed liquor (ML) | 441.4 | 1.2 | 0.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Fontana, C.; Rodriguez-Sanchez, A.; Muñoz-Palazon, B.; Gonzalez-Martinez, A.; Vela-Cano, M.; Gonzalez-Lopez, J. Profile of the Spatial Distribution Patterns of the Human and Bacteriophage Virome in a Wastewater Treatment Plant Located in the South of Spain. Water 2020, 12, 2316. https://doi.org/10.3390/w12082316
García-Fontana C, Rodriguez-Sanchez A, Muñoz-Palazon B, Gonzalez-Martinez A, Vela-Cano M, Gonzalez-Lopez J. Profile of the Spatial Distribution Patterns of the Human and Bacteriophage Virome in a Wastewater Treatment Plant Located in the South of Spain. Water. 2020; 12(8):2316. https://doi.org/10.3390/w12082316
Chicago/Turabian StyleGarcía-Fontana, Cristina, Alejandro Rodriguez-Sanchez, Barbara Muñoz-Palazon, Alejandro Gonzalez-Martinez, Maria Vela-Cano, and Jesus Gonzalez-Lopez. 2020. "Profile of the Spatial Distribution Patterns of the Human and Bacteriophage Virome in a Wastewater Treatment Plant Located in the South of Spain" Water 12, no. 8: 2316. https://doi.org/10.3390/w12082316
APA StyleGarcía-Fontana, C., Rodriguez-Sanchez, A., Muñoz-Palazon, B., Gonzalez-Martinez, A., Vela-Cano, M., & Gonzalez-Lopez, J. (2020). Profile of the Spatial Distribution Patterns of the Human and Bacteriophage Virome in a Wastewater Treatment Plant Located in the South of Spain. Water, 12(8), 2316. https://doi.org/10.3390/w12082316