Abstract
The field of eDNA is growing exponentially in response to the need for detecting rare and invasive species for management and conservation decisions. Developing technologies and standard protocols within the biosecurity sector must address myriad challenges associated with marine environments, including salinity, temperature, advective and deposition processes, hydrochemistry and pH, and contaminating agents. These approaches must also provide a robust framework that meets the need for biosecurity management decisions regarding threats to human health, environmental resources, and economic interests, especially in areas with limited clean-laboratory resources and experienced personnel. This contribution aims to facilitate dialogue and innovation within this sector by reviewing current approaches for sample collection, post-sampling capture and concentration of eDNA, preservation, and extraction, all through a biosecurity monitoring lens.
1. Introduction
Biological invasions have followed human activities for centuries [1], with cross-regional transfer of non-indigenous species (NIS) having amplified rapidly over the last few decades [2,3]. In the marine realm, this is largely attributed to the massive increase in seaborne trade beginning in the 1950s [4,5], which has served as the major pathway for marine biological invasions [1,6,7]. Continued growth in global maritime traffic and an associated 3- to 20-fold increase in global invasion risk is predicted for the next few decades [8]. Disrupting a potential invasion at the earliest stage of propagation is key, since downstream eradication in highly dynamic marine environments is difficult at best. Although managing the spread of unwanted organisms remains a high priority for regional, national, and international jurisdictions (e.g., Marine Strategy Framework Directive; European Union (EU) Invasive Species Regulation; New Zealand Biosecurity Act), a lack of operational tools and technologies for early detection has been a long-term hurdle [9]. Molecular methods have been used for decades to aid environmental monitoring [10]. The use of tissue or blood samples from an individual to obtain a genetic signal alleviates issues surrounding taxonomic identification, while still relying on visual detection and collection of specimens at the sampled area [11]. More recently, the application of DNA and RNA (collectively termed nucleic acids (NAs)), recovered from environmental samples and referred to as environmental DNA (eDNA) and RNA (eRNA), is increasingly advocated for to be used in biodiversity assessments (e.g., [12,13,14]). The non-invasive manner of sampling and non-reliance on visual observations, combined with the advancements of molecular techniques [15,16], has resulted in a rapid expansion of research to determine if this novel monitoring method meets surveillance and management needs in marine (e.g., [17]), freshwater (e.g., [18]), and terrestrial (e.g., [19]) systems. As with any detection system, balance must be struck between precision, accuracy and sensitivity of methods, logistical requirements, and associated costs. These considerations must further be balanced within the larger framework of resource management decisions [20,21,22].
The sensitivity of NA-based techniques has led to the unprecedented ability to characterize organismal assemblages across multiple trophic levels (e.g., [23,24]), detect and monitor rare species in complex environments (e.g., [25]), expand the known range of organisms (e.g., [26]), and identify potential threats (e.g., [27,28]) that have eluded traditional sampling approaches (e.g., electrofishing, imaging, settlement plates, dive surveys, plankton tows). While these examples demonstrate the immense benefits of using NA-based methodologies for screening environmental samples, several comparison studies have revealed apparent limitations in signal detection [29,30] that can originate from a variety of error sources within the workflow—from sample collection through to processing and bioinformatics (e.g., [31,32,33,34]). These limitations can be particularly problematic in trying to meet the demands of high quality assurance/quality control standards in areas such as biosecurity [18,22,35], involving the detection, monitoring and management of pests, diseases, and nuisance species that can cause problems for humans, animals, plants, or the environment (https://www.marinebiosecurity.org.nz/what-are-marine-pests/; accessed on 13 April 2021).
There is an ever-growing body of research aimed at methodological comparisons of NA-based technologies, starting with sample collection techniques through data analyses pipelines. However, no agreement has been reached on best practices for sample collection and processing (e.g., [36] and references therein). Given the explosive growth in the use of eDNA and the emergence of eRNA [37] in myriad environmental contexts, the establishment of a fully standardized protocol throughout the sample collection and processing pipeline is a challenging task, particularly in light of the reporting of inconclusive or contradictory results [35]. Nevertheless, the development of best practice guidelines for each environment that address biosecurity surveillance needs is desirable, especially for the steps encompassing sampling through to the extraction of NAs. Here, we review the challenges for using NAs within a marine environment, and the current methods for sample collection, post-sampling capture and concentration of eDNA, preservation, and extraction through a biosecurity monitoring lens. We end with an outlook on how emerging methodologies can shape the future of invasive species detection. See Table 1 for common definitions, as used in the context of this manuscript.

Table 1.
Common definitions as used in the context of this manuscript.
2. Applications of Molecular Tools for Marine Biosecurity Surveillance
Targeted and multispecies molecular techniques are increasingly promoted for marine biosecurity applications [18,38,39,40,41], including early (pre- and post-border) detection of unwanted organisms, identification of putative NIS, surveillance of high-priority pest species, determination of the source and pathways of invasion, as well as the genetic structure of founding populations [42]. One advantage of using molecular approaches for surveillance purposes is their sensitivity in detecting and identifying NIS when populations (and therefore concentrations of expelled NAs) are sparse; however, there are numerous considerations to take into account (reviewed extensively in [43]). This aspect, for instance, brings an additional challenge for sampling representativeness, requiring increased field replication [44], sample volumes [45,46], or both, to reduce the risk of false negative results [31,47]. The ability to effectively concentrate NA material over large spatial/temporal scales would provide a valuable trade-off for achieving desirable sensitivity at an economical level, both cost- and timewise (Figure 1).
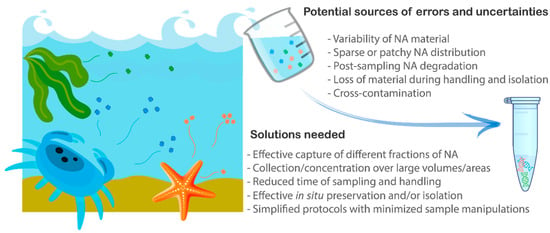
Figure 1.
Potential sources of detection errors (false negatives or false positives) and uncertainties within the primary steps of the molecular surveillance workflow, from sample collection through to nucleic acid (NA) extraction. (Figure created with BioRender.com).
The genetic material most routinely recovered from an environmental sample is DNA [48], while RNA is used to provide additional insights into the ecology of alive and viable organisms within biosecurity contexts [49,50,51,52,53]. The stability of DNA, which can persist for extended periods in the environment, depending on the conditions (days to years [54,55,56]), enables easier handling of samples until extraction. However, the influence of varying abiotic factors on the persistence of eDNA complicates the sampling design and interpretation of results, e.g., in the context of the current presence and affinity of a viable population of target (pest) species [57,58,59]. On the other hand, RNA is a fragile molecule that degrades rapidly (typically hours to days [12]). The fast degradation of RNA allows better approximation of the signals from alive and viable organisms in a sample [49]. The fate and prevalence of particle-bound eRNA is currently not well understood, questioning the applicability of eRNA for routine monitoring purposes [12].
The variable states that NAs are found in the environment, ranging from free-floating molecules to molecules bound to larger complexes (inorganic particles, cell debris, organic floc, tissues, scales), or even intact organisms and their propagules, introduces another level of complexity for effective NA capture from a sample [44]. The decisions to be made at this step (e.g., optimal filter type, pore size, need for pre-filtration or precipitation) are often impeded by poor knowledge of target NA fractions and their variability (spatial and temporal) in the environment. A variety of factors within the marine context can influence this variability and detectability: salinity, temperature, advective and deposition processes, hydrochemistry and pH, and contaminating agents (e.g., [60,61,62,63]). Inherent biological factors, such as cellular degradation (influenced by cell health) can also affect detectability [62]. All of these influences need to be taken into consideration when optimizing protocols around “sampling windows” as part of any sampling approach, from grab samples to passive sampling devices.
Another challenging aspect for biosecurity practitioners and the broader application of molecular techniques is related to the complexity of sample collection and handling protocols [21,22,64]. Thus, the exceptional sensitivity of molecular detection requires an abundance of caution to prevent cross-contamination at every stage of the workflow and avoid false positive detections—the major concern of biosecurity managers hesitant to implement molecular surveillance [18,35]. Furthermore, the nature of the sampled material makes it prone to degradation (e.g., [65]). This can be prevented by minimizing handling time in the field and stabilizing the material as soon as possible after collection, in a cost-efficient and preferably non-toxic way (Figure 1).
In the following sections, we review the current methods available for (i) sample collection, (ii) post-sampling capture and concentration of NAs (including filter types), (iii) preservation, and (iv) extraction and subsequent storage, all within the utility of the marine biosecurity surveillance sector. The major existing gaps are addressed in the context of future perspectives. We do not consider sampling design in this review, as it is case-specific and difficult to generalize. A fit-for-purpose sampling strategy should ultimately be developed for each coastal location, taking into account parameters like harbor size, local hydrodynamic peculiarities (exposure, tidal regime), proximity to introduction pathways, potential sources of inhibitory contaminants, and historical detection of target species.
3. Sample Collection
Testing for the presence of invasive species via NAs within the biosecurity sector ideally requires inexpensive technology that is easy to use across all skill levels, is not prone to cross-contamination, and consistently leads to robust and reliable results. Identifying equipment that meets these criteria will help with its adoption into sustained monitoring frameworks. This rapidly growing field has predominantly incorporated existing methods for sample collection (e.g., [66]); however, there have been calls from management entities for fit-for-purpose technology (e.g., [67]), especially since many traditional methods pose constraints for eDNA sampling [21,43].
Marine biosecurity surveillance targets a whole range of organisms representing different taxonomic groups, functional traits, and lifestyles—from sediment-dwelling or biofouling invertebrates to algae and fish inhabiting the water column. Also, in the case of untargeted surveillance (e.g., passive “screening” for new incursions), the life form, behavior, abundance, and peculiarities of NA distribution of the organisms are generally unknown. Therefore, it is difficult to develop a sampling protocol that is generalized across the multiple habitats in which an incursion may occur. Sampling different environmental matrices, such as seafloor sediments, biofouling of infrastructure, and the water column (which is a rather complex environment in itself), may not always be possible due to logistical constrains (e.g., accessibility of sampling sites in busy ports), health and safety risks (e.g., sample collection by SCUBA divers from open coast hard bottoms), and associated cost and time efforts (Table 2). A few studies have compared the efficiency of targeting different matrices for biodiversity assessment and species detection [66,68,69,70]. Some studies have found important differences between, e.g., sediments and water at different depths [71,72,73]. A comprehensive biosecurity surveillance program should ideally consider collecting representative samples from different habitat types, enabling detection of a wide range of organisms (e.g., [74]). However, in the context of often limited and consistently diminishing monitoring resources, it is important to find a meaningful trade-off between cost-efficiency and the extent of surveillance effort ([75] and references therein). Surface water samples are usually logistically easier to collect and are standardizable at larger scales [76]; thus, they can be considered an efficient alternative for obtaining signals from both planktonic and benthic communities [75,77]. Hereinafter, we focus on methods relevant for waterborne NAs (unless stated otherwise).

Table 2.
Considerations in the nucleic acid (NA) surveillance workflow with regards to choice of biological matrix to sample, as well as possible approaches to collection, concentration, and preservation.
The majority of studies targeting waterborne NAs have employed sampling with bottles, carboys, or buckets (or some combination), and have been amended with a filtration step for sample concentration when needed [58,78]. Plankton net tows have been used for concentrating material from a large volume of water [30,42,53,70,79], an approach that is desirable for detecting low concentrations and overcoming the patchiness of targets (e.g., [80]). However, the configuration of these nets renders them highly prone to cross-contamination across sampling sites/dates, given the propensity of cells and particles to stick in the numerous crevices. Furthermore, the nylon used in constructing plankton nets is often not conducive to repeated, bleach-based sterilization (von Ammon, Pochon, and Zaiko, personal observations). Alternatively, the plastics used for Niskin bottles and van Dorn samplers (both utilized for capturing whole water from discrete depths) are better suited for sterilization via diluted bleach (e.g., [81]) or acid washing (e.g., [82]); however neither of these devices allow for in situ concentration of particles.
The use of sterile disposable bottles can be economically feasible depending on sample size; however, this is not an option for bulk water collection devices and downstream filtration apparatus. More commonly, sampling containers and equipment are rigorously sterilized with bleach [83,84,85] or acid [82,86,87] and rinsed several times with deionized water, followed by site-specific water to remove sterilizing agents. False positives can occur with any breach in the protocol (e.g., reduced sterilization times, improper concentrations). Nucleic acid/contaminant removal is of the utmost importance for NA sampling; however, the above measures can be cumbersome in a biosecurity context, where considerable time, effort, and oversight are needed to ensure proper decontamination steps are carried out.
Recent gear adaptations provide promising alternatives for reducing sample handling and associated contamination. Adrian-Kalchhauser and Burkhardt-Holm [64] modified a commercially available telescoping water sampler to combine the capability of discrete depth sampling with the ability to use a new bottle for each deployment. This technique still requires decontamination of the main apparatus between samples. However, the water mass is collected in a separate disposable vessel, and is not subjected to a re-useable chamber (e.g., van Dorn or Niskin bottle). The Smith–Root eDNA backpack sampler [88] is a “fit-for-purpose” system for sampling waterborne eDNA. The unit monitors and regulates the pressure and flow of water across a collection filter. Partially biodegradable filtration units can be used to self-preserve the filter via desiccation, and this has been shown to be comparable to ethanol preservation, even over long storage periods [88]. The use of a negative-pressure inline filtration system means that the water sample can be taken up directly by the intake tubing to the filter, while all pumping occurs downstream of the filtering unit. This approach drastically reduces the chance for cross-contamination between sampling events. Newly configured autonomous platforms are in development and offer great promise towards more comprehensive sampling approaches (Mesobot, [89]; 3G Environmental Sample Processor (ESP), [90]).
One prominent disadvantage of many environmental NA sampling systems and strategies is that they offer only point measures for species detection; thus, there is a move towards systems that might aggregate an eDNA signal over time to provide a window into biosecurity threats across a wider time span. One such device is the continuous, low-level aquatic monitoring (Continuous Low-Level Aquatic Monitoring, C.L.A.M.; https://aqualytical.com/, accessed on 13 April 2021) system that allows for a time-integrated sample (up to 36 h), with the option to tow or allow the device to drift to also capture spatial coverage. The device employs filtration and media sequestering steps for sampling totals and dissolved eDNA. Other lower-tech, cost-effective, and environmentally friendly aggregate strategies are also emerging. One such approach is to deploy passive eDNA samplers that capture eDNA to a physical matrix, such as montmorillonite clay [91], artificial reef structures [92], or settlement plate arrays [24,93]. Such strategies could lead to longer-term, low-cost surveillance of eDNA in many systems, perhaps with a reduction in the need for laborious filtering (see below) and a decrease in the use of disposable plastics. Unfortunately, due to its instability, the collection of eRNA is likely to always require sterile, specialized equipment.
Collectively, these new technologies are addressing many of the criteria needed for sampling waterborne NAs within a biosecurity framework. Easy-to-operate enclosed systems greatly minimize the opportunity for cross-contamination between samples. Furthermore, immediate in situ fixation steps eliminate any lag times to sample preservation (described below). Taken together, these aspects help streamline areas in the pipeline where false positive or false negative information can originate. Gaps in knowledge for these types of collection systems include: (1) robustness and reproducibility of different filter types and fixatives across diverse taxonomic groups and varied aquatic environments (e.g., turbid waters, hydrological influences); (2) utility in saltwater environments; and (3) incorporation of pre-filtration steps to allow for greater volumes to be processed, in order to increase detectability of rare taxa.
4. Post-Sampling Capture and Concentration of eDNA
Most commonly, waterborne NAs from environmental samples are concentrated via filtration. Filter types and pore sizes have been tested for a variety of target organisms (e.g., [82,94,95,96,97,98,99,100]). However, unsurprisingly no “one-size-fits-all” consensus has emerged. Environmental DNA studies have reported using filters ranging in pore size from 0.22 microns [57,85,101] to as large as 20.00 microns [102], although many publications state pore sizes of one micron or less [94,103]. Ideally, the smallest pore size would be used in order to capture all sub-cellular particles. However, capturing adequate material (particle size and abundance) to maximize meaningful molecular signals downstream must be balanced with the time it takes to filter large volumes of water and the increased chance of clogging, which reduces the volumes that can be filtered. Too much material can potentially introduce inhibitory substances (e.g., humic compounds, suspended sediment) or potentially swamp out the molecular signal from rare targets [104,105]. A limited number of studies have shown that larger pore filters can be as informative as smaller pore filters in detecting even rare targets [106,107]. Results from these studies are promising; however, it is unclear how they will translate across a broad range of trophic levels, particle sizes, and environmental variability.
More data are needed on the amount and type of NA material captured and lost when using various pore sizes, to select the optimal combination of time and signal detection efficiency for particular biosecurity applications. As we move towards increasing that body of data, one option is to filter sequentially through multiple pore sizes. This approach avoids the impact of unpredictable episodic disturbances (e.g., influx of particles after a storm surge; highly dynamic marine and estuarine systems; seasonal variability in allochthonous material) on a standardized sampling pipeline that is limited to one pore size. This strategy also increases the opportunity for capturing a broad range of taxa from different NA fractions, and avoids rapid clogging of the small pore size filter (e.g., [108,109]).
Some studies have employed sequential filtration to assess the optimum pore size for capturing eDNA, and in some cases the remaining effluent harbored a significant amount of eDNA. For common carp (Cyprinus carpio), Turner et al. [94] determined that the molecular signal (via qPCR) was most abundant from fractions captured between 1 and 10 microns. However, they also confirmed a large pool of predominately non-target eDNA in their ethanol-precipitated (an alternative to filtration), 0.2 micron filtrate, a fraction that would have remained unexploited in a broad-scale taxa study or monitoring program. Moushomi et al. [110] used qPCR to demonstrate greater copy numbers of nuclear and mitochondrial targets of Daphnia magna (a small, planktonic freshwater crustacean) in eDNA that was ethanol-precipitated and extracted from the 0.2 micron filtrate, versus that extracted from the filter itself. Notably, the eDNA decay rate was lower in the effluent compared to material in the <1.0 to 0.2 micron fraction. These studies suggest that sub-cellular material can be a significant and more stable source of information, and should be explored in future studies.
As an alternative to filtration, Turner et al. [94] and Moushami et al. [110] incorporated ethanol precipitation steps to retrieve eDNA from effluent samples, but this step is limited to small volumes, and can be problematic for seawater samples, due to inherent salt interference (discussed below). However, Sassoubre et al. [86] were able to successfully precipitate eDNA from mesocosm tanks housing two species of marine fishes, and they demonstrated significantly greater genetic copy numbers (via qPCR) for both species in unfiltered tank water (55% and 26%) compared to the cumulative values from three size fractionations (10.0, 1.0, and 0.2 micron filters), plus the remaining effluent. These results warrant additional study to test the reproducibility of this approach for NA-based monitoring.
The majority of studies on environmental NA states and particle sizes to date have been carried out in freshwater environments or have involved a limited number of target species. Extrapolation to a biosecurity framework is complex, given the desire to construct a robust pipeline that maintains an ability to detect newly introduced species, which may include a broad array of taxa that shed particles of different size (e.g., subcellular, colonial), type (e.g., epithelial, slime, waste products), and abundance, depending on organism dimensions, behavior (including seasonality), or physiological state (e.g., egg and veliger release, aging population). In addition, it remains unclear how long the various particle types stay in suspension, which will affect surveillance decisions. Perhaps casting a wider net at this point in time (i.e., analysis of multiple filtered and precipitated fractions) will limit the potential of missing key species, while simultaneously working towards the development of comprehensive databases and new technologies.
In terms of filter type, cellulose-nitrate filters are most commonly used in eDNA studies, and have been shown to consistently perform well for eDNA capture and extraction [84,111]. However, the current body of literature for filter performance does not yet adequately address areas of interest for the biosecurity realm. It should also be taken into consideration that not all filters are available in all desired pore sizes, with some having regionally limited access or imposed additional costs (e.g., the dangerous goods tax on importing cellulose nitrate filters; authors’ personal observation).
To optimize the filtration step for biosecurity applications, available filter types need to be tested for their efficiency in capturing a wide range of NA fractions in waters with varying levels of organic matter and fluctuating water quality parameters (e.g., pH, salinity). Ultimately, we need to balance capture efficiency (range of NA fractions in the presence of organic substances), assess compatibility with eRNA applications, and determine the feasibility of filters in an enclosed engineered system for reduced cross-contamination, thereby diminishing the risks of false positive signals that are highly undesirable and costly for managers and industry.
5. Preservation
Rapid preservation of NAs, particularly eRNA, during field sampling is critical. The method used ideally needs to exhibit robust performance across a variety of sample characteristics, including low-to-high biomass, a broad pH range, and the presence of inhibitors. Among current studies, cold storage is commonly used to minimize any loss of genetic information between field sampling and lab processing [112,113,114,115,116]. However, the use of ice, dry ice, or liquid nitrogen for snap-freezing, or cooling facilities requiring electricity and specialized equipment, all present practical limitations in the field and for transportation. Preservation solutions can be better suited, and preferably should be simple, cost-effective, sterile, and without requirements for special permits (i.e., non-toxic).
A cheap, room-temperature preservation option for whole water is isopropanol [117] or ethanol [97,118,119,120]. Yamanaka et al. [121] found that the preservation of whole water with benzalkonium chloride was successful in recovering 92% of eDNA (up to 8 h). While cheap and effective, these chemicals require the transport of large volumes, which can be a limiting factor when sampling at sea or remote coastal locations. More commonly, samples are preserved after concentration via filtration, using 20% DMSO buffer [122], RNAlater (Qiagen, Germantown, MD, USA), Longmire’s buffer [123], or cetrimonium bromide (CTAB; Sigma-Aldrich, St. Louis, MO, USA) buffer [95]. Ammonium salts, dimethyl sulfoxide (DMSO), and RNAlater showed high precipitation, leading to inhibition of real-time PCR assays (qPCR), while CTAB and Longmire’s successfully preserved filtered eDNA at 20 °C over a two-week period [95]. Salt-saturated DMSO (DESS) buffers containing EDTA have also been suggested as a preservative [124]. Some preservation solutions have been shown to be biased toward taxonomic groups [125] or species [126], while Longmire’s buffer even appeared to enhance eDNA recovery, likely by increasing cell lysis efficiency [127,128]. This could suggest Longmire’s buffer as the best option for sample preservation at ambient temperature conditions.
A few studies indicate that snap-freezing of whole water samples instantly after collection outperforms other preservation methods [125,126]. This is true for one-off analyses, but where samples may be used repeatedly, the issue of freeze–thaw-induced degradation becomes important. In our own experience, even one cycle of freeze–thawing can reduce the eDNA signal. The effect of multiple defrosting episodes on eRNA is largely unknown, but is expected to be substantial as well. Therefore, where possible, it is preferable to avoid freezing samples in the field and instead keep them chilled if they can be delivered to the lab within a reasonably short time (up to ~24 h [119,129]), where they can be immediately processed for filtration and NA extraction (or storage). When specifically targeting eRNA, samples need to be filtered expeditiously and snap-frozen, or stored in expensive RNA-compatible preservation buffers (e.g., RNAlater, LifeGuard, DNA/RNA-Shield, etc.) in the field. Quaternary ammonium salts have been reported to be efficient preservation buffers in some [130,131] but not all [132] RNA studies.
Recent technologies have explored desiccating, self-preserving, and fully encapsulated filter membranes (e.g., Smith-Root self-preserving eDNA Filter Packs), which could achieve higher detection sensitivity on field samples than with buffer preservation [88]. Sterivex and Acrodisc syringe filters work with similar technologies using silica desiccant. Syringes are then chilled at 0 °C and stored at −10 °C or lower until extraction. There is, however, a large knowledge gap in how these approaches perform for eRNA.
6. Extraction of Nucleic Acids
Extraction of high-quality NAs from environmental samples is paramount for successful, molecular-based marine biosecurity surveillance. A plethora of NA extraction methods have been developed since the inception of DNA isolation in 1869 [133]. The two fundamental steps for extracting NAs are (1) cell lysis and extraction of intracellular NAs into aqueous solution—this is done either enzymatically by incubation with hydrolyzing enzymes (e.g., proteinase K; Sigma-Aldrich, St. Louis, MO, USA [134,135]); mechanically through bead-beating, freeze-thawing or grinding [136,137]; or chemically, using detergents (e.g., sodium dodecyl sulfate (SDS; Sigma-Aldrich, St. Louis, MO, USA), guanidinium thiocyanate (Sigma-Aldrich, St. Louis, MO, USA), or CTAB (Sigma-Aldrich, St. Louis, MO, USA)) that solubilize cell membrane components [138,139,140]—and (2) purification and isolation of NAs from the aqueous phase. This is done via washing with detergent and/or organic solutions (e.g., phenol–chloroform–isoamyl (PCI) [141]), precipitation with isopropanol, ethanol, or polyethylene glycol [142,143], as well as filtration through gels, silica columns, magnetic beads, or ion-exchange resins [142,143].
While traditional lab-based NA isolation protocols using SDS, CTAB, or PCI are the least expensive, and have been successfully applied for monitoring aquatic biodiversity from both pelagic and benthic aquatic environments [94,138,144,145,146], they are time-consuming and include the use of toxic chemicals that restrict their application to specialized personnel and labs only [36]. Additionally, traditional lab-based protocols are often modified by laboratories to improve the characterization of either specific groups of organisms or sample types, and therefore no single such protocol will ever fit all the situations required by marine biosecurity surveillance programs. Commercial kits are now most commonly used in eDNA studies [36,124]. They present significant advantages over traditional lab-based protocols, including ready-to-use QC/QAed reagents, standardized extraction procedures, and PCR inhibitor removal solutions, with an absence of dangerous chemicals and relatively low cost and time per sample. In the biosecurity context, standardization is critical [35], and therefore the use of NA extraction kits is highly desirable, as they ensure an appreciable degree of consistency, which can be optimized through the automation of extraction using robotics [147]. Despite these advantages, no universal extraction kit that performs best for all sample types and research goals has been developed; there exist numerous DNA or RNA kits designed for specific applications, but a comparatively much smaller selection of kits enabling the co-extraction of both DNA and RNA from environmental samples [145].
Lear et al. [148] undertook a comprehensive data mining of 584 research articles, focusing on “extracellular DNA”, and recommended the use of specific kits for DNA extraction of organisms in environmental samples, as follows: (1) Qiagen DNeasy PowerSoil and/or PowerMax kits from soil, sediment, feces and leaf litter; and (2) Qiagen DNeasy Blood and Tissue kits and/or DNeasy PowerWater kits for extraction of DNA from water and ice. Other researchers have also compared a number of commercial kits, with the leading contenders being Qiagen DNeasy Blood and Tissue and the PowerWater kits (see [149] for review). Hinlo et al. [150] found the DNeasy kit gave better DNA yields, however PowerWater has an additional step to remove downstream enzyme inhibitors. This means that even though the nucleic acid yield may be higher for the Qiagen DNeasy Blood and Tissue kit, for samples high in naturally-occurring PCR inhibitors, PowerWater may give greater sensitivity and accuracy. Therefore, judicious selection of a commercial kit best suited to the environmental sample can significantly improve outcomes. Comparable estimates of biodiversity have been described from these two kits; however, representation should be investigated prior to committing to a strategy [29,82].
A significant challenge for environmental NA studies arises from the co-purification of naturally occurring enzyme inhibitors. The most well-described of these are humic, tannic, and fluvic acids [151,152,153,154]. At high concentrations, these inhibit polymerases, preventing downstream detection of target sequences and potentially interfering with metabarcoding studies. Two approaches are generally employed to prevent inhibitory molecules from biasing environmental nucleic acid surveys: using nucleic acid extraction systems that remove inhibitors (e.g., PowerWater or other inhibitor removal kits, [150,155]), or the use of downstream polymerase enzymes that are resistant to inhibition. A new technique built around hydroxyl-coated magnetic beads offers the promise of rapid, inhibitor-free, high-yield nucleic acid isolation from environmental samples. In a pioneering study undertaken by Yuan et al. [156], this method increased yield from approximately 10% for traditional methods to over 90% from wastewater treatment plant-activated sludge when magnetic beads were used. Yuan et al. [156] was able to fine-tune the eDNA fraction captured using magnetic beads by combining with sample filtration to examine free DNA, intracellular DNA, or DNA absorbed to extracellular particles. Sanches and Schrier [157] combined glass-fiber filters and magnetic beads to capture eDNA from estuarine samples that they argue contain some of the highest levels of inhibitory molecules for any environment. However, with only these studies to date, further work on volume limits, ratio of magnetic beads to sample, and nucleic acid purity all require further investigation.
For marine biosecurity applications, a fundamental problem with available kits, apart from surety of supply, relates to the actual amount of biofouling or sediment material that can be processed, ranging from 0.25 g (DNeasy PowerSoil; Qiagen, Germantown, MD, USA) to 10 g (DNeasy PowerMax; Qiagen, Germantown, MD, USA), with the latter being approximately four times more expensive than the former. Therefore, biosecurity researchers often face the difficult choice of either increasing the replication of homogenized (e.g., freeze-dried or bead-beaten) samples; extracting DNA from only 0.2 g, using the cheaper kit unless kits can be modified (e.g., [83]); or processing fewer samples while maximizing the recovery of rare species from larger amounts of starting material, using the more expensive kit. A combination of approaches may be employed, which increases the difficulty of performing statistical analyses.
Another significant problem for long-term biosecurity surveys is the proprietary nature of reagent recipes, as well as the systematic fine-tuning of available commercial kits that offer no guarantee for extended consistency in protocols and results. The recent discontinuation of the former DNeasy PowerSoil kit (now PowerSoil Pro; Qiagen, Germantown, MD, USA) is a good example [147]. Nevertheless, while non-commercial protocols can be easier to maintain through time, and provide higher amounts of extracted material, they are much more likely to introduce contamination in the workflow and generally yield much lower NA quality compared with commercial kits, which are able to more effectively remove PCR inhibitors. Therefore, one could argue that quality is more important than quantity for minimizing false positive/negative results, and that the use of commercial kits with appropriate replication and sample isolation workflow is the best approach for routine marine biosecurity surveillance efforts. Integration of an internal positive control (IPC) into any extraction protocol (to assess extraction efficiency and impacts from inhibitors) needs to be considered as part of a routine pipeline to ensure a robust downstream dataset (e.g., [158,159]).
Methods for isolating eRNA are broadly similar to those targeting eDNA; however, the generally acknowledged (though recently challenged) view of increased eRNA susceptibility to degradation makes this area more challenging [12,50,160]. eRNA recovery rates have been reported to range from 70% to as little as 5% of the original concentration [161,162]. This is reportedly due to the rapid degradation that can occur during standard enzymatic treatment with DNAse (to avoid cross-contamination from carry-over genomic DNA) and the reverse transcription process (used to generate cDNA), both of which involve long exposures to the elevated temperatures needed for enzyme activation/de-activation. Without actively inhibiting or excluding environmental RNAses through using standard RNA handling precautions, reliable and reproducible capture of eRNA will be difficult and inconsistent.
In the current era of rapidly evolving molecular and analytical technologies, it is particularly important to build robust and quality-assured baselines (e.g., in the form of eDNA/eRNA sample and data archives) for ensuring continuity and intercalibration of ever-emerging tools [163]. Therefore, it is important to ensure that extracted NAs remain stable, and there is no or minimal degradation over time. Post-extraction stabilization of NAs can be achieved through deep-freezing or appropriate buffering. Ideally, curated cryopreservation archives should be established for long-term sample storage and back-ups. Moreover, access to such archives would also allow retrospective refinement of biodiversity information if required for conservation or biosecurity purposes.
7. Outlook
Several factors that can introduce variation and uncertainties when sampling NA within a marine biosecurity context still need to be adequately addressed [47,163], in order to ensure that the produced data meet the requirements for precision and accuracy, and are balanced with logistical and economical constraints. First, the effects of salinity, advective and deposition processes, peculiar hydrochemistry, and contaminating agents may compromise the capture of NAs. Second, unique workflow requirements are needed for optimized eRNA-based surveillance, which has recently been advocated as a promising alternative for biodiversity science [50], and more specifically, for biosecurity surveillance [12,49,69]. Third, there is a need for simplistic, yet robust and sensitive, non-toxic collection procedures (e.g., minimal sample volumes, enclosed filtration and subsequent elution and extraction of material, passive sampling options) that do not compromise sample integrity (e.g., through degradation or contamination) in the hands of non-laboratory personnel. In a marine biosecurity context, optimized fit-for-purpose and time-efficient technologies for sampling and purifying NAs from environmental samples would transform our ability to undertake cost-effective surveillance, aid in early detection of potential threats, and revolutionize biosecurity management potential (Figure 2). Refinement and validation of NA-based methods to fit these criteria will enhance molecular surveillance by biosecurity practitioners and citizen science programs.
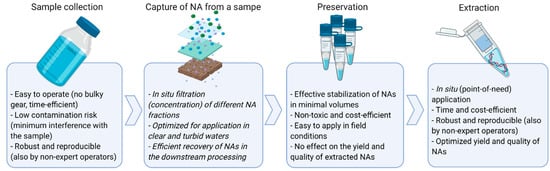
Figure 2.
Desirable features of a methodological workflow around eDNA/eRNA sample collection through to nucleic acid extraction for robust implementation in routine biosecurity surveillance (Figure created with BioRender.com).
In order to address the outstanding challenges of molecular (NA-based) biosecurity surveillance, we should proactively explore and embrace methodological advances in environmental genomics and those technologies emerging in adjacent fields (e.g., chemical, biotechnological, or medical applications). For example, nanofibers and nanoparticles used for immobilizing and the delivery of DNA [164,165,166] can be explored for applicability in eDNA/eRNA sampling devices. Selective binding and stabilization of waterborne NAs would be beneficial for both snapshot and time-integrated sample collection, with the latter of particular interest for biosecurity applications, as it would allow for increased detection probabilities of weak molecular signals from new incursions in highly dynamic coastal environments. The concept of time-integrated (or “passive”) sample collection is well-exploited for cost-effective monitoring of nutrients and contaminants in water [167,168]. However, such technology has not yet been extended to NA capture. There is a range of common compounds with promising properties for NA adsorption from ambient water, including clay minerals, silica, and alumina [91,169,170,171,172], as well as less conventional substrates that show potential for NA binding, e.g., functionalized graphene, gold nanoparticles, polyamidoamines [165,173,174]. The use of marine invertebrates, such as sponges [175], as natural samplers and bioaccumulators, is also an area of promise for ongoing, low-cost, surveillance, and is worthy of being explored further.
Recent advances in the field of microfluidics has resulted in the development of several approaches for separating, sorting, concentrating, and characterizing microparticles [176]. Non-membrane-based approaches are finding application in biomedical, industrial, and environmental fields, and allow for the separation of particles with particular characteristics [177,178,179]. These approaches use the motion of suspended particles in fluids to concentrate and separate them out based on size, shape, and density (inertial microfluidic devices), allowing for much larger volumes of liquids to be processed, but require information about target particle characteristics (e.g., size, shape, density). For most marine pests, the size/density characteristics of the suspended material harboring NAs remains unclear and needs to be better characterized. Nevertheless, these new technologies offer the potential for greatly increasing water sample volumes and NA capture, thereby significantly increasing our chances of detecting low-abundance species.
We also see value in developing sequence-specific hybridization of NAs, based on substrates enriched with custom synthesized biomolecules (e.g., peptide nucleic acids) to bind with high specificity to NA strands originating from a target species (e.g., NIS or pathogen). Peptide nucleic acids are synthetic polypeptide backbones with nucleic acid bases attached as side chains, that can form Watson–Crick pairings with complementary DNA. The artificial, synthetic, polyamide backbone increases resistance to degradation by nuclease and protease enzymes in the environment [180]. Protein−nucleic acid interactions are an important field of modern molecular biology, and have been extensively explored for developing DNA machineries and biomedical applications [181,182]. However, there are significant hurdles to overcome to identify materials suitable for highly dynamic biological and physico-chemical aquatic environments. There is a requirement for high specificity for target organism(s), and resilience to changes in environmental conditions and non-organic contamination, as well as the potential to recover genetic material for downstream analyses (NA extraction and quantification).
Targeted eDNA capture can be achieved via DNA enrichment techniques, using methods recently developed to remove non-target DNA from a DNA mixture in order to increase the probability of detecting the target [183]. DNA complementary to the targets is synthesized with biotin attached, and then bound to streptavidin-labelled magnetic beads that can be held by magnets. The DNA capture beads are commonly known as “baits”. Baits were recently used to isolate low concentrations of ancient Pleistocene human DNA from soil samples collected from cave floors [184], and DNA target enrichment has been used to increase the DNA barcoding regions for the metagenomic characterization of aquatic invertebrate communities and remove non-target organisms, such as bacteria [183].
The performance of these approaches for environmental applications, however, should be thoroughly examined before they can be considered for biosecurity surveillance. For example, it is important to assess their specificity for different fractions of environmental NAs (versus binding non-target compounds); sensitivity or ability to absorb NAs at low concentrations; cumulative capacity—that is, the ability to accumulate NAs over a sustained period of time; saturation and magnitude of NA signal/integrity loss over time; and compatibility with common NA isolation procedures and analytical workflows. It is also crucial to carefully evaluate the cost-efficiency of these techniques, as it might determine the attractiveness of their uptake for routine biosecurity applications.
As biosecurity depends on finding rare NA signals, and those from biologically viable taxa, there is a strong need for more improvements in the eRNA sampling pipeline. To that end, challenges unique to eRNA need to be taken into consideration throughout the myriad steps that cross-cut aspects of collection, concentration, and preservation. The increase in time and financial resources need to be weighed against the benefits of the information that can be gleaned from eRNA. Furthermore, it would be advantageous for eRNA-specific characteristics to be tested and included in the design of deployable systems that are easy to use (few manipulations) by all skill levels, yet remain reliable and not conducive to contamination or impacts from environmental variables. With increasing efficiency in laboratory processing, eRNA detection limits can improve and are promising to deliver sensitive information about the living fraction of the sampled biomass.
Recent technological advances now mean that the gap between sample collection and laboratory-based diagnostics is narrowing, with many assays now looking to circumvent or simplify the extraction step. Such advances are often driven by the development of new in situ diagnostic approaches that offer rapid, user-friendly, sensitive, and cost-effective field-ready tools (point-of-care or point-of-need tests). Among the most promising approaches are loop-mediated isothermal amplification (LAMP [185]) and recombinase polymerase amplification (RPA), both of which involve the rapid amplification of DNA/RNA using an isothermal amplification reaction with similar sensitivities and specificities to laboratory-based PCR assays [186,187], as well as the CRISPR/Cas13-based SHERLOCK system [188,189]. These approaches do not require thermal cycling and operate at much lower temperatures, with the end-point analysis of amplified products in the field possible using lateral flow (LF) strips, similar to those used in pregnancy test kits [190]. These assays have already been developed and applied to a number of pests, diseases, and ecological monitoring [187,188,191,192], but have not yet been widely applied in the marine biosecurity context. The assays are particularly useful for rapid field-based diagnostics, since once established, the tests themselves require minimal technical skills and laboratory infrastructure. However, extraction of high-quality nucleic acids is still needed, and represents a bottleneck in the widespread adoption of RPA and LAMP for point-of-care tests [193]. Several new, simple, and rapid extraction methods have been developed [194,195,196], but have not been widely tested for environmental applications.
The emergence of simple, low-cost biosecurity tools also opens up the prospect of engaging the wider community and citizen science groups to undertake monitoring and surveillance [9]. A time when NA-based surveillance data might be collected by school children, community groups, and local agencies directly using devices that produce geospatially tagged genotype or sequence data in the field is becoming a reality. Currently, devices such as the Nanopore Flongle [197,198] provide such capability, and this will only become more portable and integrated with our mobile devices in the future. As data acquisition becomes more routine, the emergent challenges will fall to issues of data veracity, storage, and integrity, all towards building databases that will serve biosurveillance interests in the future. As has happened with the acquisition and storage of image data, cloud-based repositories for these emerging perspectives on our natural world, akin to tools like iNaturalist (https://www.inaturalist.org/; accessed on 16 April 2021) or Find-A-Pest (http://www.findapest.nz/; accessed on 16 April 2021), will be vital.
8. Concluding Remarks
There is much promise emerging around the targeting and capture of NAs in biosecurity monitoring. Technological advances make the capture, extraction, and detection of even early incursions increasingly realistic at time scales where management actions can make a difference to the establishment and spread of NIS. However, standardization and QA/QC protocols for eDNA/eRNA applications are crucial for the biosecurity sector. Data on species presence/absence, which is not properly caveated with an understanding of the limitations of the technology, particularly around its accuracy and precision, may trigger unwarranted management responses [35], cause widespread reluctance of the uptake of the technology by stakeholders, or contribute to the ongoing propagation of unwanted pests.
The nature of the type of data generated is inherently complicated, which means the research community must work closely with biosecurity surveillance managers to ensure protocol steps are feasible, and that data-associated caveats are clearly understood and communicated. This robust approach is particularly important in New Zealand, where the current biosecurity strategy calls for building a team of 5 million (New Zealand’s current population) to aid in protecting the country from the spread of unwanted organisms (https://www.mpi.govt.nz/protection-and-response/biosecurity/biosecurity-2025/biosecurity-2025/; accessed on 16 April 2021). In support of these efforts, we envision a (semi)autonomous pipeline that adheres to and expands on minimum reporting guidelines and sampling conduct recommendations, as outlined in Goldberg et al. [129], while overcoming the variability and compromises inherent to environments with a scarcity of clean laboratory resources and experienced personnel. Coupled with systematic efforts and flexibility, a transparent level of standardization between researchers and end-users can be achieved that meets the over-arching goal of protecting human health, economic interests, and environmental resources within the biosecurity sector.
Author Contributions
Conceptualization, H.A.B. and A.Z.; funding acquisition, A.Z.; content curation, H.A.B. and A.Z.; writing, H.A.B., A.Z., J.-A.L.S., N.G, X.P., C.D.H.S., U.v.A. and G.-J.J.; review, H.A.B., A.Z., N.G., X.P., C.D.H.S., U.v.A. and G.-J.J.; editing, H.A.B., A.Z., J.-A.L.S., N.G., X.P., C.D.H.S., U.v.A. and G.-J.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the New Zealand Ministry of Business, Innovation and Employment funding (CAWX1904—a toolbox to underpin and enable tomorrow’s marine biosecurity system).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
Special thanks to Margaret Ryan (Anatomy Department, Otago University) for preliminary data and suggested literature. We also thank two anonymous reviewers for their suggestions that ultimately strengthened the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ojaveer, H.; Olenin, S.; Narščius, A.; Florin, A.-B.; Ezhova, E.; Gollasch, S.; Jensen, K.R.; Lehtiniemi, M.; Minchin, D.; Normant-Saremba, M.; et al. Dynamics of Biological Invasions and Pathways over Time: A Case Study of a Temperate Coastal Sea. Biol. Invasions 2017, 19, 799–813. [Google Scholar] [CrossRef]
- Cohen, A.N.; Carlton, J.T. Accelerating Invasion Rate in a Highly Invaded Estuary. Science 1998, 279, 555–558. [Google Scholar] [CrossRef]
- Seebens, H.; Blackburn, T.M.; Dyer, E.E.; Genovesi, P.; Hulme, P.E.; Jeschke, J.M.; Pagad, S.; Pyšek, P.; Winter, M.; Arianoutsou, M.; et al. No Saturation in the Accumulation of Alien Species Worldwide. Nat. Commun. 2017, 8, 14435. [Google Scholar] [CrossRef]
- Seebens, H.; Schwartz, N.; Schupp, P.J.; Blasius, B. Predicting the Spread of Marine Species Introduced by Global Shipping. Proc. Natl. Acad. Sci. USA 2016, 113, 5646–5651. [Google Scholar] [CrossRef] [PubMed]
- Ojaveer, H.; Galil, B.S.; Carlton, J.T.; Alleway, H.; Goulletquer, P.; Lehtiniemi, M.; Marchini, A.; Miller, W.; Occhipinti-Ambrogi, A.; Peharda, M.; et al. Historical Baselines in Marine Bioinvasions: Implications for Policy and Management. PLos ONE 2018, 13, e0202383. [Google Scholar] [CrossRef]
- Wonham, M.; Walton, W.; Ruiz, G.; Frese, A.; Galil, B. Going to the Source: Role of the Invasion Pathway in Determining Potential Invaders. Mar. Ecol. Prog. Ser. 2001, 215, 1–12. [Google Scholar] [CrossRef]
- Leppäkoski, E.; Gollasch, S.; Olenin, S. Invasive Aquatic Species of Europe. Distribution, Impacts and Management; Springer: Dordrecht, The Netherlands, 2002; ISBN 978-94-015-9956-6. [Google Scholar]
- Sardain, A.; Sardain, E.; Leung, B. Global Forecasts of Shipping Traffic and Biological Invasions to 2050. Nat. Sustain. 2019, 2, 274–282. [Google Scholar] [CrossRef]
- Larson, E.R.; Graham, B.M.; Achury, R.; Coon, J.J.; Daniels, M.K.; Gambrell, D.K.; Jonasen, K.L.; King, G.D.; LaRacuente, N.; Perrin-Stowe, T.I.; et al. From EDNA to Citizen Science: Emerging Tools for the Early Detection of Invasive Species. Front. Ecol. Environ. 2020, 18, 194–202. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Singer, G.A.C.; Hebert, P.D.N.; Hickey, D.A. DNA Barcoding: How It Complements Taxonomy, Molecular Phylogenetics and Population Genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef]
- Wood, S.A.; Biessy, L.; Latchford, J.L.; Zaiko, A.; von Ammon, U.; Audrezet, F.; Cristescu, M.E.; Pochon, X. Release and Degradation of Environmental DNA and RNA in a Marine System. Sci. Total Environ. 2020, 704, 135314. [Google Scholar] [CrossRef]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A New Paradigm in Ecosystem Assessment Made Possible by next-Generation DNA Sequencing: NEWS AND VIEWS: OPINION. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The Future of Biotic Indices in the Ecogenomic Era: Integrating (e)DNA Metabarcoding in Biological Assessment of Aquatic Ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef]
- Ansorge, W.J. Next-Generation DNA Sequencing Techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Liu, S.; Yang, Q.; Su, X.; Zhou, L.; Tang, M.; Fu, R.; Li, J.; Huang, Q. Ultra-Deep Sequencing Enables High-Fidelity Recovery of Biodiversity for Bulk Arthropod Samples without PCR Amplification. GigaSci 2013, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Keeley, N.; Wood, S.A.; Pochon, X. Development and Preliminary Validation of a Multi-Trophic Metabarcoding Biotic Index for Monitoring Benthic Organic Enrichment. Ecol. Indic. 2018, 85, 1044–1057. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Nelson, N.M.; Jerde, C.L.; Luikart, G. Are Environmental DNA Methods Ready for Aquatic Invasive Species Management? Trends Ecol. Evol. 2020, 35, 668–678. [Google Scholar] [CrossRef]
- van der Heyde, M.; Bunce, M.; Wardell-Johnson, G.; Fernandes, K.; White, N.E.; Nevill, P. Testing Multiple Substrates for Terrestrial Biodiversity Monitoring Using Environmental DNA Metabarcoding. Mol. Ecol. Resour 2020, 20, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Vander Zanden, M.J.; Hansen, G.J.A.; Higgins, S.N.; Kornis, M.S. A Pound of Prevention, plus a Pound of Cure: Early Detection and Eradication of Invasive Species in the Laurentian Great Lakes. J. Great Lakes Res. 2010, 36, 199–205. [Google Scholar] [CrossRef]
- Trebitz, A.S.; Hoffman, J.C.; Darling, J.A.; Pilgrim, E.M.; Kelly, J.R.; Brown, E.A.; Chadderton, W.L.; Egan, S.P.; Grey, E.K.; Hashsham, S.A.; et al. Early Detection Monitoring for Aquatic Non-Indigenous Species: Optimizing Surveillance, Incorporating Advanced Technologies, and Identifying Research Needs. J. Environ. Manag. 2017, 202, 299–310. [Google Scholar] [CrossRef]
- Lyal, C.H.C.; Miller, S.E. Capacity of United States Federal Government and Its Partners to Rapidly and Accurately Report the Identity (Taxonomy) of Non-Native Organisms Intercepted in Early Detection Programs. Biol. Invasions 2020, 22, 101–127. [Google Scholar] [CrossRef]
- Chain, F.J.J.; Brown, E.A.; MacIsaac, H.J.; Cristescu, M.E. Metabarcoding Reveals Strong Spatial Structure and Temporal Turnover of Zooplankton Communities among Marine and Freshwater Ports. Divers. Distrib. 2016, 22, 493–504. [Google Scholar] [CrossRef]
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Tait, L.; Lavery, S.; Inglis, G.; Pochon, X. The Impact of Artificial Surfaces on Marine Bacterial and Eukaryotic Biofouling Assemblages: A High-Throughput Sequencing Analysis. Mar. Environ. Res. 2018, 133, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Brys, R.; Halfmaerten, D.; Neyrinck, S.; Mauvisseau, Q.; Auwerx, J.; Sweet, M.; Mergeay, J. Reliable EDNA Detection and Quantification of the European Weather Loach ( Misgurnus Fossilis ). J. Fish. Biol. 2020, jfb.14315. [Google Scholar] [CrossRef]
- Witzel, N.A.; Taheri, A.; Miller, B.T.; Hardman, R.H.; Withers, D.I.; Spear, S.F.; Sutton, W.B. Validation of an Environmental DNA Protocol to Detect a Stream-breeding Amphibian, the Streamside Salamander (Ambystoma Barbouri). Environ. DNA 2020, 2, 554–564. [Google Scholar] [CrossRef]
- Kim, Y.; Aw, T.G.; Teal, T.K.; Rose, J.B. Metagenomic Investigation of Viral Communities in Ballast Water. Environ. Sci. Technol. 2015, 49, 8396–8407. [Google Scholar] [CrossRef]
- Darling, J.A.; Martinson, J.; Gong, Y.; Okum, S.; Pilgrim, E.; Lohan, K.M.P.; Carney, K.J.; Ruiz, G.M. Ballast Water Exchange and Invasion Risk Posed by Intracoastal Vessel Traffic: An Evaluation Using High Throughput Sequencing. Environ. Sci. Technol. 2018, 52, 9926–9936. [Google Scholar] [CrossRef]
- Deiner, K.; Walser, J.-C.; Mächler, E.; Altermatt, F. Choice of Capture and Extraction Methods Affect Detection of Freshwater Biodiversity from Environmental DNA. Biol. Conserv. 2015, 183, 53–63. [Google Scholar] [CrossRef]
- Wood, S.A.; Pochon, X.; Ming, W.; von Ammon, U.; Woods, C.; Carter, M.; Smith, M.; Inglis, G.; Zaiko, A. Considerations for Incorporating Real-Time PCR Assays into Routine Marine Biosecurity Surveillance Programmes: A Case Study Targeting the Mediterranean Fanworm ( Sabella Spallanzanii ) and Club Tunicate ( Styela Clava ). Genome 2019, 62, 137–146. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From Molecules to Management: Adopting DNA-Based Methods for Monitoring Biological Invasions in Aquatic Environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.C.; Coghlan, M.L.; Bunce, M. From Benchtop to Desktop: Important Considerations When Designing Amplicon Sequencing Workflows. PLos ONE 2015, 10, e0124671. [Google Scholar] [CrossRef] [PubMed]
- Furlan, E.M.; Davis, J.; Duncan, R.P. Identifying Error and Accurately Interpreting Environmental DNA Metabarcoding Results: A Case Study to Detect Vertebrates at Arid Zone Waterholes. Mol. Ecol. Resour. 2020, 20, 1259–1276. [Google Scholar] [CrossRef]
- Thalinger, B.; Deiner, K.; Harper, L.R.; Rees, H.C.; Blackman, R.C.; Sint, D.; Traugott, M.; Goldberg, C.S.; Bruce, K. A Validation Scale to Determine the Readiness of Environmental DNA Assays for Routine Species Monitoring; Molecular Biology: Hoboken, NJ, USA, 2020. [Google Scholar]
- Darling, J.A.; Pochon, X.; Abbott, C.L.; Inglis, G.J.; Zaiko, A. The Risks of Using Molecular Biodiversity Data for Incidental Detection of Species of Concern. Divers. Distrib. 2020, 26, 1116–1121. [Google Scholar] [CrossRef]
- Kumar, G.; Eble, J.E.; Gaither, M.R. A Practical Guide to Sample Preservation and Pre-PCR Processing of Aquatic Environmental DNA. Mol. Ecol. Resour. 2020, 20, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tsuri, K.; Ikeda, S.; Hirohara, T.; Shimada, Y.; Minamoto, T.; Yamanaka, H. Messenger RNA Typing of Environmental RNA (ERNA): A Case Study on Zebrafish Tank Water with Perspectives for the Future Development of ERNA Analysis on Aquatic Vertebrates. Environ. DNA 2021, 3, 14–21. [Google Scholar] [CrossRef]
- Bott, N.J.; Ophel-Keller, K.M.; Sierp, M.T.; Herdina; Rowling, K.P.; McKay, A.C.; Loo, M.G.K.; Tanner, J.E.; Deveney, M.R. Toward Routine, DNA-Based Detection Methods for Marine Pests. Biotechnol. Adv. 2010, 28, 706–714. [Google Scholar] [CrossRef]
- Wood, S.; Smith, K.; Banks, J.; Tremblay, L.; Rhodes, L.; Mountfort, D.; Cary, S.; Pochon, X. Molecular Genetic Tools for Environmental Monitoring of New Zealand’s Aquatic Habitats, Past, Present and the Future. New Zealand J. Mar. Freshw. Res. 2013, 47, 90–119. [Google Scholar] [CrossRef]
- Rius, M.; Bourne, S.; Hornsby, H.G.; Chapman, M.A. Applications of Next-Generation Sequencing to the Study of Biological Invasions. Curr. Zool. 2015, 61, 488–504. [Google Scholar] [CrossRef]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and Limitations of Environmental DNA/RNA Tools for Marine Biosecurity: Management and Surveillance of Non-Indigenous Species. Front. Mar. Sci. 2018, 5, 322. [Google Scholar] [CrossRef]
- Richardson, M.F.; Sherman, C.D.H.; Lee, R.S.; Bott, N.J.; Hirst, A.J. Multiple Dispersal Vectors Drive Range Expansion in an Invasive Marine Species. Mol. Ecol. 2016, 25, 5001–5014. [Google Scholar] [CrossRef] [PubMed]
- Beng, K.C.; Corlett, R.T. Applications of Environmental DNA (EDNA) in Ecology and Conservation: Opportunities, Challenges and Prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Furlan, E.M.; Gleeson, D. Improving Reliability in Environmental DNA Detection Surveys through Enhanced Quality Control. Mar. Freshw. Res. 2017, 68, 388. [Google Scholar] [CrossRef]
- Foote, A.D.; Thomsen, P.F.; Sveegaard, S.; Wahlberg, M.; Kielgast, J.; Kyhn, L.A.; Salling, A.B.; Galatius, A.; Orlando, L.; Gilbert, M.T.P. Investigating the Potential Use of Environmental DNA (EDNA) for Genetic Monitoring of Marine Mammals. PLos ONE 2012, 7, e41781. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-Generation Monitoring of Aquatic Biodiversity Using Environmental DNA Metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Mathieu, C.; Hermans, S.M.; Lear, G.; Buckley, T.R.; Lee, K.C.; Buckley, H.L. A Systematic Review of Sources of Variability and Uncertainty in EDNA Data for Environmental Monitoring. Front. Ecol. Evol. 2020, 8, 135. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA: ENVIRONMENTAL DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; Zaiko, A.; Fletcher, L.M.; Laroche, O.; Wood, S.A. Wanted Dead or Alive? Using Metabarcoding of Environmental DNA and RNA to Distinguish Living Assemblages for Biosecurity Applications. PLos ONE 2017, 12, e0187636. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, M.E. Can Environmental RNA Revolutionize Biodiversity Science? Trends Ecol. Evol. 2019, 34, 694–697. [Google Scholar] [CrossRef]
- Kitahashi, T.; Sugime, S.; Inomata, K.; Nishijima, M.; Kato, S.; Yamamoto, H. Meiofaunal Diversity at a Seamount in the Pacific Ocean: A Comprehensive Study Using Environmental DNA and RNA. Deep Sea Res. Part. I: Oceanogr. Res. Pap. 2020, 160, 103253. [Google Scholar] [CrossRef]
- Brandt, M.I.; Trouche, B.; Henry, N.; Liautard-Haag, C.; Maignien, L.; de Vargas, C.; Wincker, P.; Poulain, J.; Zeppilli, D.; Arnaud-Haond, S. An Assessment of Environmental Metabarcoding Protocols Aiming at Favoring Contemporary Biodiversity in Inventories of Deep-Sea Communities. Front. Mar. Sci. 2020, 7, 234. [Google Scholar] [CrossRef]
- Zaiko, A.; Wood, S.A.; Pochon, X.; Biessy, L.; Laroche, O.; Croot, P.; Garcia-Vazquez, E. Elucidating Biodiversity Shifts in Ballast Water Tanks during a Cross-Latitudinal Transfer: Complementary Insights from Molecular Analyses. Environ. Sci. Technol. 2020, 54, 8443–8454. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Danovaro, R. Extracellular DNA Plays a Key Role in Deep-Sea Ecosystem Functioning. Science 2005, 309, 2179. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Beolchini, F.; Dell’Anno, A. Damage and Degradation Rates of Extracellular DNA in Marine Sediments: Implications for the Preservation of Gene Sequences. Mol. Ecol. 2008, 17, 3939–3951. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Poulenard, J.; Sabatier, P.; Messager, E.; Gielly, L.; Leloup, A.; Etienne, D.; Bakke, J.; Malet, E.; Fanget, B.; et al. DNA from Lake Sediments Reveals Long-Term Ecosystem Changes after a Biological Invasion. Sci. Adv. 2018, 4, eaar4292. [Google Scholar] [CrossRef]
- Andruszkiewicz, E.A.; Sassoubre, L.M.; Boehm, A.B. Persistence of Marine Fish Environmental DNA and the Influence of Sunlight. PLos ONE 2017, 12, e0185043. [Google Scholar] [CrossRef] [PubMed]
- Pinfield, R.; Dillane, E.; Runge, A.K.W.; Evans, A.; Mirimin, L.; Niemann, J.; Reed, T.E.; Reid, D.G.; Rogan, E.; Samarra, F.I.P.; et al. False-negative Detections from Environmental DNA Collected in the Presence of Large Numbers of Killer Whales ( Orcinus Orca ). Environ. DNA 2019, 1, 316–328. [Google Scholar] [CrossRef]
- Murakami, H.; Yoon, S.; Kasai, A.; Minamoto, T.; Yamamoto, S.; Sakata, M.K.; Horiuchi, T.; Sawada, H.; Kondoh, M.; Yamashita, Y.; et al. Dispersion and Degradation of Environmental DNA from Caged Fish in a Marine Environment. Fish. Sci. 2019, 85, 327–337. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence EDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying Effects of UV-B, Temperature, and PH on EDNA Degradation in Aquatic Microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the Fate of EDNA in the Environment and Implications for Studying Biodiversity. Proc. R. Soc. B. 2019, 286, 20191409. [Google Scholar] [CrossRef] [PubMed]
- Jo, T.; Murakami, H.; Yamamoto, S.; Masuda, R.; Minamoto, T. Effect of Water Temperature and Fish Biomass on Environmental DNA Shedding, Degradation, and Size Distribution. Ecol. Evol 2019, 9, 1135–1146. [Google Scholar] [CrossRef]
- Adrian-Kalchhauser, I.; Burkhardt-Holm, P. An EDNA Assay to Monitor a Globally Invasive Fish Species from Flowing Freshwater. PLos ONE 2016, 11, e0147558. [Google Scholar] [CrossRef]
- Kasai, A.; Takada, S.; Yamazaki, A.; Masuda, R.; Yamanaka, H. The Effect of Temperature on Environmental DNA Degradation of Japanese Eel. Fish. Sci. 2020, 86, 465–471. [Google Scholar] [CrossRef]
- Rey, A.; Basurko, O.C.; Rodriguez-Ezpeleta, N. Considerations for Metabarcoding-based Port Biological Baseline Surveys Aimed at Marine Nonindigenous Species Monitoring and Risk Assessments. Ecol. Evol. 2020, 10, 2452–2465. [Google Scholar] [CrossRef] [PubMed]
- Bouchet, P.; Phillips, C.; Huang, Z.; Meeuwig, J.; Foster, S.; Przeslawski, R. Comparative Assessment of Pelagic Sampling Methods Used in Marine Monitoring. Report to the National Environmental Science Programme; Marine Biodiversity Hub, 2018; p. 149. Available online: https://www.researchgate.net/profile/Phil-Bouchet/publication/327237111_Comparative_assessment_of_pelagic_sampling_methods_used_in_marine_monitoring/links/5b836fce4585151fd134f890/Comparative-assessment-of-pelagic-sampling-methods-used-in-marine-monitoring.pdf (accessed on 16 April 2021).
- Koziol, A.; Stat, M.; Simpson, T.; Jarman, S.; DiBattista, J.D.; Harvey, E.S.; Marnane, M.; McDonald, J.; Bunce, M. Environmental DNA Metabarcoding Studies Are Critically Affected by Substrate Selection. Mol. Ecol. Resour. 2019, 19, 366–376. [Google Scholar] [CrossRef]
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Lavery, S.D.; Inglis, G.J.; Pochon, X. Linking Environmental DNA and RNA for Improved Detection of the Marine Invasive Fanworm Sabella Spallanzanii. Front. Mar. Sci. 2019, 6, 621. [Google Scholar] [CrossRef]
- Schabacker, J.C.; Amish, S.J.; Ellis, B.K.; Gardner, B.; Miller, D.L.; Rutledge, E.A.; Sepulveda, A.J.; Luikart, G. Increased EDNA Detection Sensitivity Using a Novel High-volume Water Sampling Method. Environ. DNA 2020, 2, 244–251. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Howland, K.; Normandeau, E.; Grey, E.K.; Archambault, P.; Deiner, K.; Lodge, D.M.; Hernandez, C.; Leduc, N.; Bernatchez, L. eDNA Metabarcoding as a New Surveillance Approach for Coastal Arctic Biodiversity. Ecol. Evol. 2018, 8, 7763–7777. [Google Scholar] [CrossRef] [PubMed]
- Holman, L.E.; de Bruyn, M.; Creer, S.; Carvalho, G.; Robidart, J.; Rius, M. Detection of Introduced and Resident Marine Species Using Environmental DNA Metabarcoding of Sediment and Water. Sci. Rep. 2019, 9, 11559. [Google Scholar] [CrossRef] [PubMed]
- Antich, A.; Palacín, C.; Cebrian, E.; Golo, R.; Wangensteen, O.S.; Turon, X. Marine Biomonitoring with EDNA: Can Metabarcoding of Water Samples Cut It as a Tool for Surveying Benthic Communities? Mol. Ecol. 2020, mec.15641. [Google Scholar] [CrossRef]
- Outinen, O.; Forsström, T.; Yli-Rosti, J.; Vesakoski, O.; Lehtiniemi, M. Monitoring of Sessile and Mobile Epifauna—Considerations for Non-Indigenous Species. Mar. Pollut. Bull. 2019, 141, 332–342. [Google Scholar] [CrossRef]
- Pearman, J.K.; Ammon, U.; Laroche, O.; Zaiko, A.; Wood, S.A.; Zubia, M.; Planes, S.; Pochon, X. Metabarcoding as a Tool to Enhance Marine Surveillance of Nonindigenous Species in Tropical Harbors: A Case Study in Tahiti. Environ. DNA 2021, 3, 173–189. [Google Scholar] [CrossRef]
- Grey, E.K.; Bernatchez, L.; Cassey, P.; Deiner, K.; Deveney, M.; Howland, K.L.; Lacoursière-Roussel, A.; Leong, S.C.Y.; Li, Y.; Olds, B.; et al. Effects of Sampling Effort on Biodiversity Patterns Estimated from Environmental DNA Metabarcoding Surveys. Sci. Rep. 2018, 8, 8843. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, Á.; Muxika, I.; Rodríguez-Ezpeleta, N. Adapting Metabarcoding-Based Benthic Biomonitoring into Routine Marine Ecological Status Assessment Networks. Ecol. Indic. 2018, 95, 194–202. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Laramie, M.B.; MacCoy, D.; Maclean, S. Integration of EDNA-Based Biological Monitoring within the U.S. Geological Survey’s National Streamgage Network. J. Am. Water Resour Assoc. 2019, 55, 1505–1518. [Google Scholar] [CrossRef]
- von Ammon, U.; Jeffs, A.; Zaiko, A.; van der Rees, A.; Goodwin, D.; Beckley, L.E.; Malpot, E.; Pochon, X. A Portable Cruising Speed Net: Expanding Global Collection of Sea Surface Plankton Data. Frontiers in Marine Science. in press.
- Bessey, C.; Jarman, S.N.; Berry, O.; Olsen, Y.S.; Bunce, M.; Simpson, T.; Power, M.; McLaughlin, J.; Edgar, G.J.; Keesing, J. Maximizing Fish Detection with EDNA Metabarcoding. Environ. DNA 2020, 2, 493–504. [Google Scholar] [CrossRef]
- Carim, K.J.; Christianson, K.R.; McKelvey, K.M.; Pate, W.M.; Silver, D.B.; Johnson, B.M.; Galloway, B.T.; Young, M.K.; Schwartz, M.K. Environmental DNA Marker Development with Sparse Biological Information: A Case Study on Opossum Shrimp (Mysis Diluviana). PLos ONE 2016, 11, e0161664. [Google Scholar] [CrossRef]
- Djurhuus, A.; Port, J.; Closek, C.J.; Yamahara, K.M.; Romero-Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B.; et al. Evaluation of Filtration and DNA Extraction Methods for Environmental DNA Biodiversity Assessments across Multiple Trophic Levels. Front. Mar. Sci. 2017, 4, 314. [Google Scholar] [CrossRef]
- Spens, J.; Evans, A.R.; Halfmaerten, D.; Knudsen, S.W.; Sengupta, M.E.; Mak, S.S.T.; Sigsgaard, E.E.; Hellström, M. Comparison of Capture and Storage Methods for Aqueous Macrobial eDNA Using an Optimized Extraction Protocol: Advantage of Enclosed Filter. Methods Ecol. Evol. 2017, 8, 635–645. [Google Scholar] [CrossRef]
- Jeunen, G.; Knapp, M.; Spencer, H.G.; Taylor, H.R.; Lamare, M.D.; Stat, M.; Bunce, M.; Gemmell, N.J. Species-level Biodiversity Assessment Using Marine Environmental DNA Metabarcoding Requires Protocol Optimization and Standardization. Ecol. Evol. 2019, 9, 1323–1335. [Google Scholar] [CrossRef]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of Environmental DNA in Marine Systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef]
- Sassoubre, L.M.; Yamahara, K.M.; Gardner, L.D.; Block, B.A.; Boehm, A.B. Quantification of Environmental DNA (EDNA) Shedding and Decay Rates for Three Marine Fish. Environ. Sci. Technol. 2016, 50, 10456–10464. [Google Scholar] [CrossRef] [PubMed]
- Salter, I. Seasonal Variability in the Persistence of Dissolved Environmental DNA (EDNA) in a Marine System: The Role of Microbial Nutrient Limitation. PLos ONE 2018, 13, e0192409. [Google Scholar] [CrossRef]
- Thomas, A.C.; Nguyen, P.L.; Howard, J.; Goldberg, C.S. A Self-preserving, Partially Biodegradable EDNA Filter. Methods Ecol. Evol. 2019, 10, 1136–1141. [Google Scholar] [CrossRef]
- Yoerger, D.R.; Curran, M.; Fujii, J.; German, C.R.; Gomez-Ibanez, D.; Govindarajan, A.F.; Howland, J.C.; Llopiz, J.K.; Wiebe, P.H.; Hobson, B.W.; et al. Mesobot: An Autonomous Underwater Vehicle for Tracking and Sampling Midwater Targets. In Proceedings of the 2018 IEEE/OES Autonomous Underwater Vehicle Workshop (AUV), Porto, Portugal, 6–9 November 2018; pp. 1–7. [Google Scholar]
- Yamahara, K.M.; Preston, C.M.; Birch, J.; Walz, K.; Marin, R.; Jensen, S.; Pargett, D.; Roman, B.; Ussler, W.; Zhang, Y.; et al. In Situ Autonomous Acquisition and Preservation of Marine Environmental DNA Using an Autonomous Underwater Vehicle. Front. Mar. Sci. 2019, 6, 373. [Google Scholar] [CrossRef]
- Kirtane, A.; Atkinson, J.D.; Sassoubre, L. Design and Validation of Passive Environmental DNA Samplers Using Granular Activated Carbon and Montmorillonite Clay. Environ. Sci. Technol. 2020, 54, 11961–11970. [Google Scholar] [CrossRef] [PubMed]
- Leray, M.; Knowlton, N. DNA Barcoding and Metabarcoding of Standardized Samples Reveal Patterns of Marine Benthic Diversity. Proc. Natl. Acad. Sci. USA 2015, 112, 2076–2081. [Google Scholar] [CrossRef]
- Zaiko, A.; Schimanski, K.; Pochon, X.; Hopkins, G.A.; Goldstien, S.; Floerl, O.; Wood, S.A. Metabarcoding Improves Detection of Eukaryotes from Early Biofouling Communities: Implications for Pest Monitoring and Pathway Management. Biofouling 2016, 32, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.R.; Barnes, M.A.; Xu, C.C.Y.; Jones, S.E.; Jerde, C.L.; Lodge, D.M. Particle Size Distribution and Optimal Capture of Aqueous Macrobial eDNA. Methods Ecol. Evol. 2014, 5, 676–684. [Google Scholar] [CrossRef]
- Renshaw, M.A.; Olds, B.P.; Jerde, C.L.; McVeigh, M.M.; Lodge, D.M. The Room Temperature Preservation of Filtered Environmental DNA Samples and Assimilation into a Phenol–Chloroform–Isoamyl Alcohol DNA Extraction. Mol. Ecol. Resour 2015, 15, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of Temperature and Trophic State on Degradation of Environmental DNA in Lake Water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Minamoto, T.; Fukuda, M.; Katsuhara, K.R.; Fujiwara, A.; Hidaka, S.; Yamamoto, S.; Takahashi, K.; Masuda, R. Environmental DNA Reflects Spatial and Temporal Jellyfish Distribution. PLos ONE 2017, 12, e0173073. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Lopez, J.; Bourne, S.; Holman, L.; Seymour, M.; Grey, E.K.; Lacoursière, A.; Li, Y.; Renshaw, M.A.; Pfrender, M.E.; et al. Optimising the Detection of Marine Taxonomic Richness Using Environmental DNA Metabarcoding: The Effects of Filter Material, Pore Size and Extraction Method. MBMG 2018, 2, e28963. [Google Scholar] [CrossRef]
- Capo, E.; Spong, G.; Königsson, H.; Byström, P. Effects of Filtration Methods and Water Volume on the Quantification of Brown Trout ( Salmo Trutta ) and Arctic Char ( Salvelinus Alpinus ) EDNA Concentrations via Droplet Digital PCR. Environ. DNA 2020, 2, 152–160. [Google Scholar] [CrossRef]
- Liu, Y.; Wikfors, G.H.; Rose, J.M.; McBride, R.S.; Milke, L.M.; Mercaldo-Allen, R. Application of Environmental DNA Metabarcoding to Spatiotemporal Finfish Community Assessment in a Temperate Embayment. Front. Mar. Sci. 2019, 6, 674. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using Environmental DNA to Census Marine Fishes in a Large Mesocosm. PLos ONE 2014, 9, e86175. [Google Scholar] [CrossRef]
- Egan, S.P.; Barnes, M.A.; Hwang, C.-T.; Mahon, A.R.; Feder, J.L.; Ruggiero, S.T.; Tanner, C.E.; Lodge, D.M. Rapid Invasive Species Detection by Combining Environmental DNA with Light Transmission Spectroscopy: EDNA-LTS Species Detection. Conserv. Lett. 2013, 6, 402–409. [Google Scholar] [CrossRef]
- Barnes, M.A.; Chadderton, W.L.; Jerde, C.L.; Mahon, A.R.; Turner, C.R.; Lodge, D.M. Environmental Conditions Influence EDNA Particle Size Distribution in Aquatic Systems. Environ. DNA 2020, edn3.160. [Google Scholar] [CrossRef]
- Jane, S.F.; Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Schwartz, M.K.; Lowe, W.H.; Letcher, B.H.; Whiteley, A.R. Distance, Flow and PCR Inhibition: EDNA Dynamics in Two Headwater Streams. Mol. Ecol. Resour. 2015, 15, 216–227. [Google Scholar] [CrossRef]
- Hunter, M.E.; Ferrante, J.A.; Meigs-Friend, G.; Ulmer, A. Improving EDNA Yield and Inhibitor Reduction through Increased Water Volumes and Multi-Filter Isolation Techniques. Sci. Rep. 2019, 9, 5259. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Schabacker, J.; Smith, S.; Al-Chokhachy, R.; Luikart, G.; Amish, S.J. Improved Detection of Rare, Endangered and Invasive Trout in Using a New Large-volume Sampling Method for EDNA Capture. Environ. DNA 2019, 1, 227–237. [Google Scholar] [CrossRef]
- Wittwer, C.; Nowak, C.; Strand, D.A.; Vrålstad, T.; Thines, M.; Stoll, S. Comparison of Two Water Sampling Approaches for EDNA-Based Crayfish Plague Detection. Limnologica 2018, 70, 1–9. [Google Scholar] [CrossRef]
- Not, F.; del Campo, J.; Balagué, V.; de Vargas, C.; Massana, R. New Insights into the Diversity of Marine Picoeukaryotes. PLos ONE 2009, 4, e7143. [Google Scholar] [CrossRef]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahe, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic Plankton Diversity in the Sunlit Ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Moushomi, R.; Wilgar, G.; Carvalho, G.; Creer, S.; Seymour, M. Environmental DNA Size Sorting and Degradation Experiment Indicates the State of Daphnia Magna Mitochondrial and Nuclear EDNA Is Subcellular. Sci. Rep. 2019, 9, 12500. [Google Scholar] [CrossRef]
- Majaneva, M.; Diserud, O.H.; Eagle, S.H.C.; Boström, E.; Hajibabaei, M.; Ekrem, T. Environmental DNA Filtration Techniques Affect Recovered Biodiversity. Sci. Rep. 2018, 8, 4682. [Google Scholar] [CrossRef] [PubMed]
- Mahon, A.R.; Jerde, C.L.; Galaska, M.; Bergner, J.L.; Chadderton, W.L.; Lodge, D.M.; Hunter, M.E.; Nico, L.G. Validation of EDNA Surveillance Sensitivity for Detection of Asian Carps in Controlled and Field Experiments. PLos ONE 2013, 8, e58316. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Doi, H. Using Environmental DNA to Estimate the Distribution of an Invasive Fish Species in Ponds. PLos ONE 2013, 8, e56584. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-Unseen” Detection of Rare Aquatic Species Using Environmental DNA: EDNA Surveillance of Rare Aquatic Species. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Wilcox, T.M.; Carim, K.J.; Young, M.K.; McKelvey, K.S.; Franklin, T.W.; Schwartz, M.K. Comment: The Importance of Sound Methodology in Environmental DNA Sampling. North. Am. J. Fish. Manag. 2018, 38, 592–596. [Google Scholar] [CrossRef]
- Sales, N.G.; Wangensteen, O.S.; Carvalho, D.C.; Mariani, S. Influence of Preservation Methods, Sample Medium and Sampling Time on EDNA Recovery in a Neotropical River. Environ. DNA 2019, 1, 119–130. [Google Scholar] [CrossRef]
- Doi, H.; Uchii, K.; Matsuhashi, S.; Takahara, T.; Yamanaka, H.; Minamoto, T. Isopropanol Precipitation Method for Collecting Fish Environmental DNA. Limnol. Oceanogr. Methods 2017, 15, 212–218. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a New Method for Early Detection of New Zealand Mudsnails ( Potamopyrgus Antipodarum ). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating Occupancy and Abundance of Stream Amphibians Using Environmental DNA from Filtered Water Samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Spall, J.L.; Shokralla, S.; van Konynenburg, S. Assessing Biodiversity of a Freshwater Benthic Macroinvertebrate Community through Non-Destructive Environmental Barcoding of DNA from Preservative Ethanol. BMC Ecol. 2012, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, H.; Minamoto, T.; Matsuura, J.; Sakurai, S.; Tsuji, S.; Motozawa, H.; Hongo, M.; Sogo, Y.; Kakimi, N.; Teramura, I.; et al. A Simple Method for Preserving Environmental DNA in Water Samples at Ambient Temperature by Addition of Cationic Surfactant. Limnology 2017, 18, 233–241. [Google Scholar] [CrossRef]
- Seutin, G.; White, B.N.; Boag, P.T. Preservation of Avian Blood and Tissue Samples for DNA Analyses. Can. J. Zool. 1991, 69, 82–90. [Google Scholar] [CrossRef]
- Longmire, J.; Maltbie, M.; Baker, R. Use of ‘Lysis Buffer’ in DNA Isolation and Its Implications for Museum Collections. Mus. Tex. Tech. Univ. 1997, 163, 1–3. [Google Scholar]
- van der Loos, L.M.; Nijland, R. Biases in Bulk: DNA Metabarcoding of Marine Communities and the Methodology Involved. Mol. Ecol. 2020, mec.15592. [Google Scholar] [CrossRef]
- Mäki, A.; Salmi, P.; Mikkonen, A.; Kremp, A.; Tiirola, M. Sample Preservation, DNA or RNA Extraction and Data Analysis for High-Throughput Phytoplankton Community Sequencing. Front. Microbiol. 2017, 8, 1848. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.A.; Pratte, Z.A.; Kellogg, C.A. Comparison of DNA Preservation Methods for Environmental Bacterial Community Samples. FEMS MicroBiol. Ecol. 2013, 83, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Wegleitner, B.J.; Jerde, C.L.; Tucker, A.; Chadderton, W.L.; Mahon, A.R. Long Duration, Room Temperature Preservation of Filtered EDNA Samples. Conserv. Genet. Resour. 2015, 7, 789–791. [Google Scholar] [CrossRef]
- Williams, K.E.; Huyvaert, K.P.; Piaggio, A.J. No Filters, No Fridges: A Method for Preservation of Water Samples for EDNA Analysis. BMC Res. Notes 2016, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S.; et al. Critical Considerations for the Application of Environmental DNA Methods to Detect Aquatic Species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Bachoon, D.S.; Chen, F.; Hodson, R.E. RNA Recovery and Detection of MRNA by RT-PCR from Preserved Prokaryotic Samples. Fems Microbiol. Lett. 2001, 201, 127–132. [Google Scholar] [CrossRef]
- van Eijsden, R.G.E.; Stassen, C.; Daenen, L.; Van Mulders, S.E.; Bapat, P.M.; Siewers, V.; Goossens, K.V.Y.; Nielsen, J.; Delvaux, F.R.; Van Hummelen, P.; et al. A Universal Fixation Method Based on Quaternary Ammonium Salts (RNAlater) for Omics-Technologies: Saccharomyces Cerevisiae as a Case Study. Biotechnol. Lett. 2013, 35, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, A.J.; Kurhela, E.; Aho, T.; Oittinen, T.; Tiirola, M. Storage of Environmental Samples for Guaranteeing Nucleic Acid Yields for Molecular Microbiological Studies. Appl. MicroBiol. Biotechnol. 2010, 88, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Dahm, R. Discovering DNA: Friedrich Miescher and the Early Years of Nucleic Acid Research. Hum. Genet. 2008, 122, 565–581. [Google Scholar] [CrossRef]
- Goldenberger, D.; Perschil, I.; Ritzler, M.; Altwegg, M. A Simple “Universal” DNA Extraction Procedure Using SDS and Proteinase K Is Compatible with Direct PCR Amplification. Genome Res. 1995, 4, 368–370. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA Recovery from Soils of Diverse Composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Ogram, A.; Sayler, G.S.; Barkay, T. The Extraction and Purification of Microbial DNA from Sediments. J. Microbiol. Methods 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Tsai, Y.L.; Olson, B.H. Rapid Method for Direct Extraction of DNA from Soil and Sediments. Appl. Environ. Microbiol. 1991, 57, 1070–1074. [Google Scholar] [CrossRef]
- Natarajan, V.P.; Zhang, X.; Morono, Y.; Inagaki, F.; Wang, F. A Modified SDS-Based DNA Extraction Method for High Quality Environmental DNA from Seafloor Environments. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Tkach, V.; Pawlowski, J.W. A New Method of DNA Extraction from the Ethanol-Fixed Parasitic Worms. Acta Parasitol. 1999, 44, 147–148. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant. Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Purification of Nucleic Acids by Extraction with Phenol: Chloroform. Cold Spring Harb. Protoc. 2006. [Google Scholar] [CrossRef]
- Porteous, L.A.; Seidler, R.J.; Watrud, L.S. An Improved Method for Purifying DNA from Soil for Polymerase Chain Reaction Amplification and Molecular Ecology Applications. Mol. Ecol. 1997, 6, 787–791. [Google Scholar] [CrossRef]
- Manoli, F.; Dalas, E. Spontaneous Precipitation of Calcium Carbonate in the Presence of Ethanol, Isopropanol and Diethylene Glycol. J. Cryst. Growth 2000, 218, 359–364. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport Distance of Invertebrate Environmental DNA in a Natural River. PLos ONE 2014, 9, e88786. [Google Scholar] [CrossRef]
- Lever, M.A.; Torti, A.; Eickenbusch, P.; Michaud, A.B.; Šantl-Temkiv, T.; Jørgensen, B.B. A Modular Method for the Extraction of DNA and RNA, and the Separation of DNA Pools from Diverse Environmental Sample Types. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Geerts, A.N.; Boets, P.; Van den Heede, S.; Goethals, P.; Van der heyden, C. A Search for Standardized Protocols to Detect Alien Invasive Crayfish Based on Environmental DNA (EDNA): A Lab and Field Evaluation. Ecol. Indic. 2018, 84, 564–572. [Google Scholar] [CrossRef]
- Pearman, J.K.; Keeley, N.B.; Wood, S.A.; Laroche, O.; Zaiko, A.; Thomson-Laing, G.; Biessy, L.; Atalah, J.; Pochon, X. Comparing Sediment DNA Extraction Methods for Assessing Organic Enrichment Associated with Marine Aquaculture. PeerJ 2020, 8, e10231. [Google Scholar] [CrossRef] [PubMed]
- Lear, G.; Dickie, I.; Banks, J.; Boyer, S.; Buckley, H.; Buckley, T.; Cruickshank, R.; Dopheide, A.; Handley, K.; Hermans, S.; et al. Methods for the Extraction, Storage, Amplification and Sequencing of DNA from Environmental Samples. NZ 2018, 42, 1A–50A. [Google Scholar] [CrossRef]
- Tsuji, S.; Takahara, T.; Doi, H.; Shibata, N.; Yamanaka, H. The Detection of Aquatic Macroorganisms Using Environmental DNA Analysis—A Review of Methods for Collection, Extraction, and Detection. Environ. DNA 2019, 1, 99–108. [Google Scholar] [CrossRef]
- Hinlo, R.; Gleeson, D.; Lintermans, M.; Furlan, E. Methods to Maximise Recovery of Environmental DNA from Water Samples. PLos ONE 2017, 12, e0179251. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Jansson, L.; Nilsson, E.; Noppa, L.; Forsman, M.; Rådström, P.; Hedman, J. Humic Substances Cause Fluorescence Inhibition in Real-Time Polymerase Chain Reaction. Anal. Biochem. 2015, 487, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in QPCR, DPCR and MPS—Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Uchii, K.; Doi, H.; Okahashi, T.; Katano, I.; Yamanaka, H.; Sakata, M.K.; Minamoto, T. Comparison of Inhibition Resistance among PCR Reagents for Detection and Quantification of Environmental DNA. Environ. DNA 2019, 1, 359–367. [Google Scholar] [CrossRef]
- Lance, R.F.; Guan, X. Variation in Inhibitor Effects on QPCR Assays and Implications for EDNA Surveys. Can. J. Fish. Aquat. Sci. 2020, 77, 23–33. [Google Scholar] [CrossRef]
- McKee, A.M.; Spear, S.F.; Pierson, T.W. The Effect of Dilution and the Use of a Post-Extraction Nucleic Acid Purification Column on the Accuracy, Precision, and Inhibition of Environmental DNA Samples. Biol. Conserv. 2015, 183, 70–76. [Google Scholar] [CrossRef]
- Yuan, Q.-B.; Huang, Y.-M.; Wu, W.-B.; Zuo, P.; Hu, N.; Zhou, Y.-Z.; Alvarez, P.J.J. Redistribution of Intracellular and Extracellular Free & Adsorbed Antibiotic Resistance Genes through a Wastewater Treatment Plant by an Enhanced Extracellular DNA Extraction Method with Magnetic Beads. Environ. Int. 2019, 131, 104986. [Google Scholar] [CrossRef]
- Sanches, T.M.; Schreier, A.D. Optimizing an EDNA Protocol for Estuarine Environments: Balancing Sensitivity, Cost and Time. PLos ONE 2020, 15, e0233522. [Google Scholar] [CrossRef]
- Green, H.C.; Field, K.G. Sensitive Detection of Sample Interference in Environmental QPCR. Water Res. 2012, 46, 3251–3260. [Google Scholar] [CrossRef]
- Turner, C.R.; Uy, K.L.; Everhart, R.C. Fish Environmental DNA Is More Concentrated in Aquatic Sediments than Surface Water. Biol. Conserv. 2015, 183, 93–102. [Google Scholar] [CrossRef]
- Marshall, N.T.; Vanderploeg, H.A.; Chaganti, S.R. Environmental (e)RNA Advances the Reliability of EDNA by Predicting Its Age. Sci. Rep. 2021, 11, 2769. [Google Scholar] [CrossRef]
- Munk, B.; Lebuhn, M. Process Diagnosis Using Methanogenic Archaea in Maize-Fed, Trace Element Depleted Fermenters. Anaerobe 2014, 29, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Lebuhn, M.; Derenkó, J.; Rademacher, A.; Helbig, S.; Munk, B.; Pechtl, A.; Stolze, Y.; Prowe, S.; Schwarz, W.; Schlüter, A.; et al. DNA and RNA Extraction and Quantitative Real-Time PCR-Based Assays for Biogas Biocenoses in an Interlaboratory Comparison. Bioengineering 2016, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Leese, F.; Bouchez, A.; Abarenkov, K.; Altermatt, F.; Borja, Á.; Bruce, K.; Ekrem, T.; Čiampor, F.; Čiamporová-Zaťovičová, Z.; Costa, F.O.; et al. Why We Need Sustainable Networks Bridging Countries, Disciplines, Cultures and Generations for Aquatic Biomonitoring 2.0: A Perspective Derived From the DNAqua-Net COST Action. In Advances in Ecological Research; Elsevier: Amsterdam, The Netherlands, 2018; Volume 58, pp. 63–99. ISBN 978-0-12-813949-3. [Google Scholar]
- Peckys, D.B.; Melechko, A.V.; Simpson, M.L.; McKnight, T.E. Immobilization and Release Strategies for DNA Delivery Using Carbon Nanofiber Arrays and Self-Assembled Monolayers. Nanotechnology 2009, 20, 145304. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Adsorption of DNA onto Gold Nanoparticles and Graphene Oxide: Surface Science and Applications. Phys. Chem. Chem. Phys. 2012, 14, 10485. [Google Scholar] [CrossRef]
- Matthew, R.A.; Gopi, M.M.; Menon, P.; Jayakumar, R.; Vijayachandran, L.S. Synthesis of Electrospun Silica Nanofibers for Protein/DNA Binding. Mater. Lett. 2016, 184, 5–8. [Google Scholar] [CrossRef]
- Wood, S.A.; Holland, P.T.; MacKenzie, L. Development of Solid Phase Adsorption Toxin Tracking (SPATT) for Monitoring Anatoxin-a and Homoanatoxin-a in River Water. Chemosphere 2011, 82, 888–894. [Google Scholar] [CrossRef]
- Hale, S.E.; Oen, A.M.P.; Cornelissen, G.; Jonker, M.T.O.; Waarum, I.-K.; Eek, E. The Role of Passive Sampling in Monitoring the Environmental Impacts of Produced Water Discharges from the Norwegian Oil and Gas Industry. Mar. Pollut. Bull. 2016, 111, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gallori, E.; Bazzicalupo, M.; Dal Canto, L.; Fani, R.; Nannipieri, P.; Vettori, C.; Stotzky, G. Transformation of Bacillus Subtilis by DNA Bound on Clay in Non-Sterile Soil. FEMS Microbiol. Ecol. 1994, 15, 119–126. [Google Scholar] [CrossRef]
- Vandeventer, P.E.; Lin, J.S.; Zwang, T.J.; Nadim, A.; Johal, M.S.; Niemz, A. Multiphasic DNA Adsorption to Silica Surfaces under Varying Buffer, PH, and Ionic Strength Conditions. J. Phys. Chem. B 2012, 116, 5661–5670. [Google Scholar] [CrossRef]
- Lozano, H.J.; Busto, N.; Lari, M.; Leal, J.M.; García, B. Binding of Aluminium/Cacodylate Complexes with DNA and RNA. Experimental and “ in Silico ”Study. New J. Chem. 2018, 42, 8137–8144. [Google Scholar] [CrossRef]
- Sheng, X.; Qin, C.; Yang, B.; Hu, X.; Liu, C.; Waigi, M.G.; Li, X.; Ling, W. Metal Cation Saturation on Montmorillonites Facilitates the Adsorption of DNA via Cation Bridging. Chemosphere 2019, 235, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Bhattacharjee, N.; Searson, P.C. Electrochemically Programmed Release of Biomolecules and Nanoparticles. Nano Lett. 2006, 6, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Lomas, H.; Canton, I.; MacNeil, S.; Du, J.; Armes, S.P.; Ryan, A.J.; Lewis, A.L.; Battaglia, G. Biomimetic PH Sensitive Polymersomes for Efficient DNA Encapsulation and Delivery. Adv. Mater. 2007, 19, 4238–4243. [Google Scholar] [CrossRef]
- Mariani, S.; Baillie, C.; Colosimo, G.; Riesgo, A. Sponges as Natural Environmental DNA Samplers. Curr. Biol. 2019, 29, R401–R402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Mukherjee, P.; Gao, H.; Luan, Q.; Papautsky, I. Label-Free Microfluidic Sorting of Microparticles. APL Bioeng. 2019, 3, 041504. [Google Scholar] [CrossRef]
- Watanabe, M.; Kenmotsu, H.; Ko, R.; Wakuda, K.; Ono, A.; Imai, H.; Taira, T.; Naito, T.; Murakami, H.; Abe, M.; et al. Isolation and Molecular Analysis of Circulating Tumor Cells from Lung Cancer Patients Using a Microfluidic Chip Type Cell Sorter. Cancer Sci. 2018, 109, 2539–2548. [Google Scholar] [CrossRef]
- Shakeel Syed, M.; Rafeie, M.; Henderson, R.; Vandamme, D.; Asadnia, M.; Ebrahimi Warkiani, M. A 3D-Printed Mini-Hydrocyclone for High Throughput Particle Separation: Application to Primary Harvesting of Microalgae. Lab. Chip 2017, 17, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Bai, X.; Wu, C.; Li, W.; Liu, K.; Kiani, A. Recent Advances in the Beneficiation of Ultrafine Coal Particles. Fuel Process. Technol. 2018, 178, 104–125. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M. An Introduction to Peptide Nucleic Acid. Curr Issues Mol. Biol. 1999, 1, 89–104. [Google Scholar]
- Metaferia, B.; Wei, J.S.; Song, Y.K.; Evangelista, J.; Aschenbach, K.; Johansson, P.; Wen, X.; Chen, Q.; Lee, A.; Hempel, H.; et al. Development of Peptide Nucleic Acid Probes for Detection of the HER2 Oncogene. PLos ONE 2013, 8, e58870. [Google Scholar] [CrossRef]
- Tutkus, M.; Rakickas, T.; Kopūstas, A.; Ivanovaitė, Š.; Venckus, O.; Navikas, V.; Zaremba, M.; Manakova, E.; Valiokas, R. Fixed DNA Molecule Arrays for High-Throughput Single DNA–Protein Interaction Studies. Langmuir 2019, 35, 5921–5930. [Google Scholar] [CrossRef]
- Dowle, E.; Pochon, X.; Keeley, N.; Wood, S.A. Assessing the Effects of Salmon Farming Seabed Enrichment Using Bacterial Community Diversity and High-Throughput Sequencing. FEMS Microbiol. Ecol. 2015, 91, fiv089. [Google Scholar] [CrossRef]
- Slon, V.; Hopfe, C.; Weiß, C.L.; Mafessoni, F.; de la Rasilla, M.; Lalueza-Fox, C.; Rosas, A.; Soressi, M.; Knul, M.V.; Miller, R.; et al. Neandertal and Denisovan DNA from Pleistocene Sediments. Science 2017, 356, 605–608. [Google Scholar] [CrossRef]
- Notomi, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Macdonald, J. Recombinase Polymerase Amplification: Emergence as a Critical Molecular Technology for Rapid, Low-Resource Diagnostics. Expert Rev. Mol. Diagn. 2015, 15, 1475–1489. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic Acid Detection with CRISPR Nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral Flow (Immuno)Assay: Its Strengths, Weaknesses, Opportunities and Threats. A Literature Survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef]
- Williams, M.; O’Grady, J.; Ball, B.; Carlsson, J.; Eyto, E.; McGinnity, P.; Jennings, E.; Regan, F.; Parle-McDermott, A. The Application of CRISPR-Cas for Single Species Identification from Environmental DNA. Mol. Ecol. Resour. 2019, 19, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, V.B.; Khallaf, B.; Surapaneni, A.; Crosbie, N.D.; Soni, S.K.; Ball, A.S. Detection of Helminth Ova in Wastewater Using Recombinase Polymerase Amplification Coupled to Lateral Flow Strips. Water 2020, 12, 691. [Google Scholar] [CrossRef]
- Hansen, S.; Abd El Wahed, A. Point-Of-Care or Point-Of-Need Diagnostic Tests: Time to Change Outbreak Investigation and Pathogen Detection. Trop. Med. 2020, 5, 151. [Google Scholar] [CrossRef]
- Chowdhury, R.; Ghosh, P.; Khan, M.d.A.A.; Hossain, F.; Faisal, K.; Nath, R.; Baker, J.; Wahed, A.A.E.; Maruf, S.; Nath, P.; et al. Evaluation of Rapid Extraction Methods Coupled with a Recombinase Polymerase Amplification Assay for Point-of-Need Diagnosis of Post-Kala-Azar Dermal Leishmaniasis. Trop. Med. 2020, 5, 95. [Google Scholar] [CrossRef]
- Mondal, D.; Ghosh, P.; Khan, M.A.A.; Hossain, F.; Böhlken-Fascher, S.; Matlashewski, G.; Kroeger, A.; Olliaro, P.; Abd El Wahed, A. Mobile Suitcase Laboratory for Rapid Detection of Leishmania Donovani Using Recombinase Polymerase Amplification Assay. Parasites Vectors 2016, 9, 281. [Google Scholar] [CrossRef]
- Faye, O.; Faye, O.; Soropogui, B.; Patel, P.; El Wahed, A.A.; Loucoubar, C.; Fall, G.; Kiory, D.; Magassouba, N.; Keita, S.; et al. Development and Deployment of a Rapid Recombinase Polymerase Amplification Ebola Virus Detection Assay in Guinea in 2015. Euro Surveill. 2015, 20, 30053. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, N.; Bokma, J.; Haesebrouck, F.; Nauwynck, H.; Boyen, F.; Pardon, B.; Theuns, S. High Quality Genome Assemblies of Mycoplasma Bovis Using a Taxon-Specific Bonito Basecaller for MinION and Flongle Long-Read Nanopore Sequencing. BMC Bioinform. 2020, 21, 517. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.S.; Au, C.H.; Lam, H.Y.; Wang, C.L.N.; Ho, D.N.-Y.; Lam, Y.M.; Chu, D.K.W.; Poon, L.L.M.; Chan, T.L.; Zee, J.S.-T.; et al. Evaluation on the Use of Nanopore Sequencing for Direct Characterization of Coronaviruses from Respiratory Specimens, and a Study on Emerging Missense Mutations in Partial RdRP Gene of SARS-CoV-2. Virol. J. 2020, 17, 183. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).