Quantification and Characterization of Antimicrobial Resistance in Greywater Discharged to the Environment
Abstract
1. Introduction
2. Material and Methods
2.1. Study Setting and Sample Collection
2.2. Physiochemical Parameters
2.3. Cultivation and Identification of Antimicrobial-Resistant Bacteria
2.4. Characterization and Antimicrobial Susceptibility Testing of ARB
2.5. DNA Extraction from Untreated GW
2.6. Quantification of ARGs by Quantitative PCR (qPCR)
2.7. Statistics
3. Results
3.1. Physiochemical Parameters and Microbial Counts
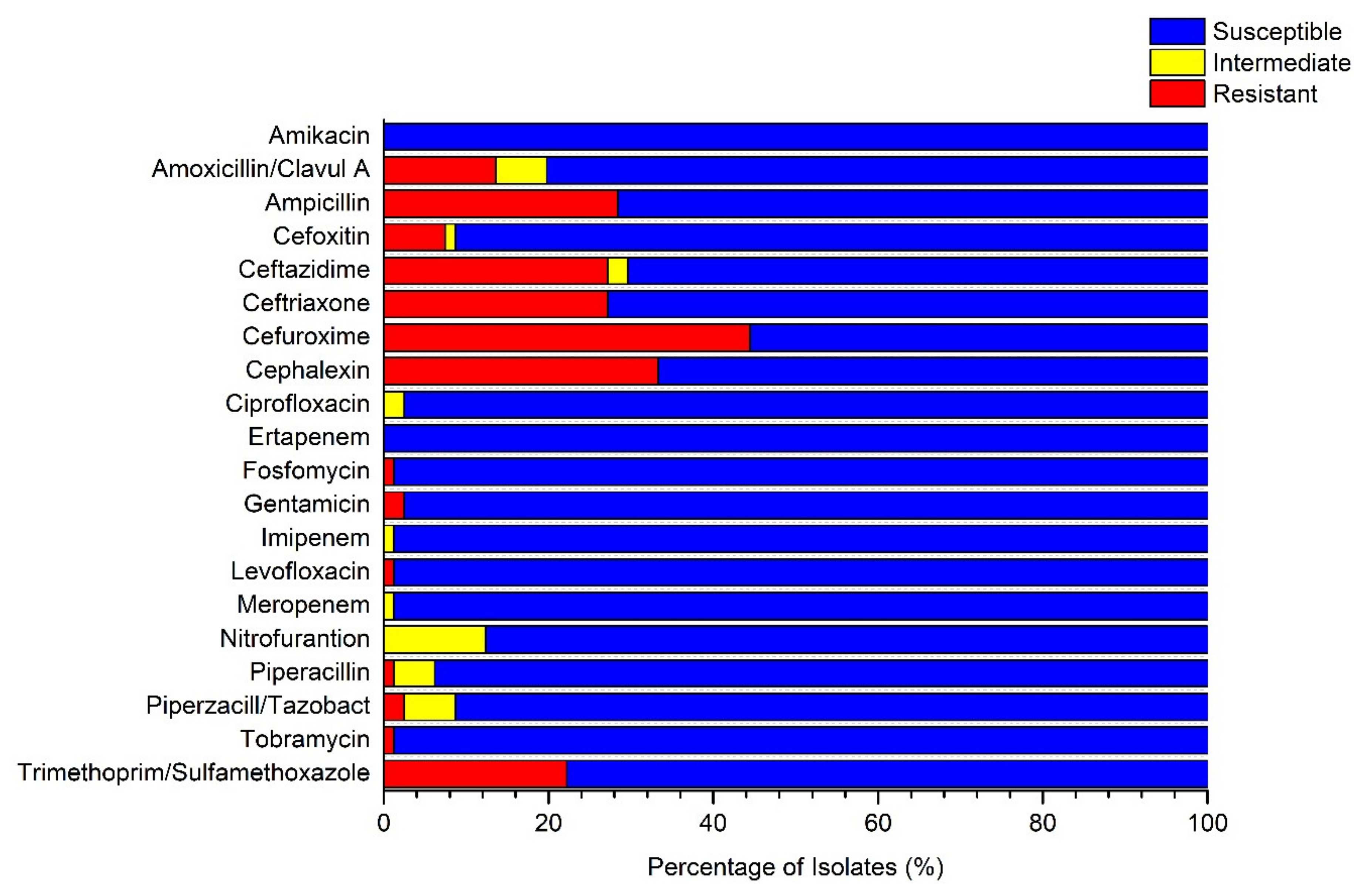
3.2. Presence of Multidrug-Resistant Isolates in Untreated GW Samples
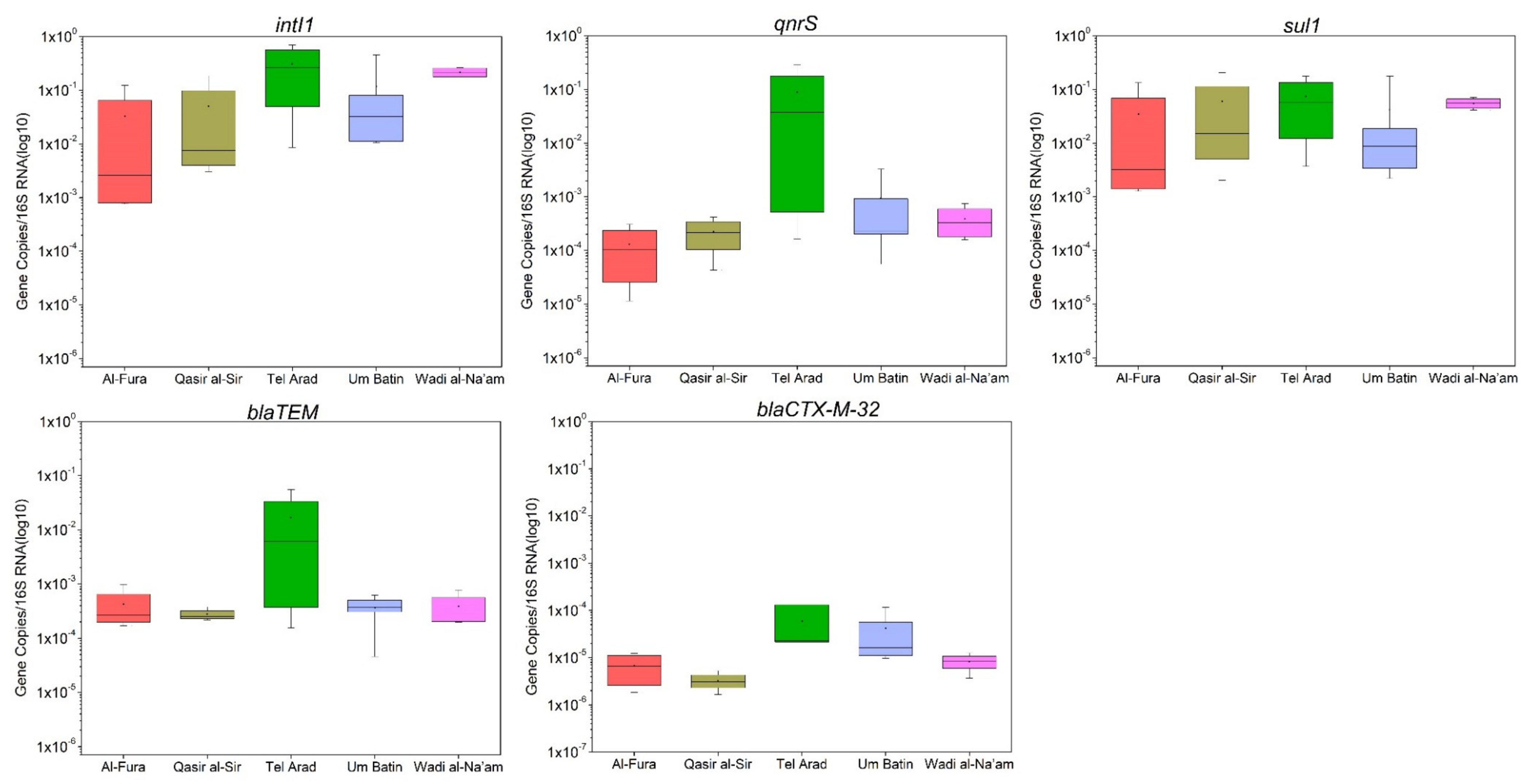
3.3. Quantification of Antimicrobial Resistance Genes in GW
4. Discussion
4.1. Summary of Findings
4.2. ARG Occurrence in Untreated GW
4.3. Presence of MDR ESBL Isolates in GW
4.4. Limitations
4.5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Benami, M.; Gillor, O.; Gross, A. Potential health and environmental risks associated with onsite greywater reuse: A review. Built Environ. 2016, 42, 212–229. [Google Scholar] [CrossRef]
- Casanova, L.M.; Little, V.; Frye, R.J.; Gerba, C.P. A survey of the microbial quality of recycled household graywater. J. Am. Water Resour. Assoc. 2001, 37, 1313–1319. [Google Scholar] [CrossRef]
- Birks, R.; Colbourne, J.; Hobson, R. Microbiological water quality in a large in-building, water recycling facility. Water Sci. Technol. 2004, 50, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Abdouslam, N.; Hawkey, P.M.; Wellington, E.M.H. Incidence of Class 1 Integrons in a Quaternary Ammonium Compound-Polluted Environment. Antimicrob. Agents Chemother. 2005, 49, 1802. [Google Scholar] [CrossRef]
- Kothari, C.; Gaind, R.; Singh, L.C.; Sinha, A.; Kumari, V.; Arya, S.; Chellani, H.; Saxena, S.; Deb, M. Community acquisition of β-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol. 2013, 13, 136. [Google Scholar] [CrossRef]
- Quan, J.; Zhao, D.; Liu, L.; Chen, Y.; Zhou, J.; Jiang, Y.; Du, X.; Zhou, Z.; Akova, M.; Yu, Y. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J. Antimicrob. Chemother. 2016, 72, 273–280. [Google Scholar] [CrossRef]
- Bailie, R.S.; Stevens, M.; McDonald, E.L. The impact of housing improvement and socio-environmental factors on common childhood illnesses: A cohort study in Indigenous Australian communities. J. Epidemiol. Community Health 2012, 66, 821–831. [Google Scholar] [CrossRef]
- Treister-Goltzman, Y.; Peleg, R. What is Known About Health and Morbidity in the Pediatric Population of Muslim Bedouins in Southern Israel: A Descriptive Review of the Literature from the Past Two Decades. J. Immigr. Minority Health 2015, 17, 940–946. [Google Scholar] [CrossRef]
- Ezery, Z. Inadequate Wastewater Infrastructure’s Effect on the Environmental Health of the Bedouin Population in the Negev. Master’s Thesis, Ben-Gurion University of the Negev, Be’er Sheva, Israel, 2016. [Google Scholar]
- Natalya, B.; Fraser, D.; Givon-Lavi, N.; Dagan, R. A Decade (1989–1998) of Pediatric Invasive Pneumococcal Disease in 2 Populations Residing in 1 Geographic Location: Implications for Vaccine Choice. Clin. Infect. Dis. 2001, 33, 421–427. [Google Scholar] [CrossRef]
- Dagan, R.; Landau, D.; Haikin, H.; Tal, A. Hospitalization of Jewish and Bedouin infants in southern Israel for bronchiolitis caused by respiratory syncytial virus. Pediatr. Infect. Dis. J. 1993, 12, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Adler, A.; Givon-Lavi, N.; Moses, A.E.; Block, C.; Dagan, R. Carriage of Community-Associated Methicillin-Resistant Staphylococcus aureus in a Cohort of Infants in Southern Israel: Risk Factors and Molecular Features. J. Clin. Microbiol. 2010, 48, 531. [Google Scholar] [CrossRef] [PubMed]
- Elnasasra, A.; Alnsasra, H.; Smolyakov, R.; Riesenberg, K.; Nesher, L. Ethnic Diversity and Increasing Resistance Patterns of Hospitalized Community-Acquired Urinary Tract Infections in Southern Israel: A Prospective Study. Isr. Med. Assoc. J. 2017, 19, 538–542. [Google Scholar] [PubMed]
- Gatica, J.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in the soil microbiome. Environ. Sci. Pollut. Res. 2013, 20, 3529–3538. [Google Scholar] [CrossRef] [PubMed]
- Troiano, E.; Beneduce, L.; Gross, A.; Ronen, Z. Antibiotic-Resistant Bacteria in Greywater and Greywater-Irrigated Soils. Front. Microbiol. 2018, 9, 2666. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.-C.; Michael, I.; Fatta-Kassinos, D. Urban Wastewater Treatment Plants as Hotspots for Antibiotic Resistant Bacteria and Genes Spread into the Environment: A Review. Sci. Environ. 2013, 447C, 345–360. [Google Scholar] [CrossRef]
- APHA. WPCF (2005) Standard Methods for the Examination of Water and Wastewater; APHA: Washington, DC, USA, 2005. [Google Scholar]
- Gross, A.; Sklarz, M.Y.; Yakirevich, A.; Soares, M.I.M. Small scale recirculating vertical flow constructed wetland (RVFCW) for the treatment and reuse of wastewater. Water Sci. Technol. 2008, 58, 487–494. [Google Scholar] [CrossRef]
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of multi-resistant Gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Faron, M.L.; Buchan, B.W.; Hyke, J.; Madisen, N.; Lillie, J.L.; Granato, P.A.; Wilson, D.A.; Procop, G.W.; Novak-Weekley, S.; Marlowe, E.; et al. Multicenter Evaluation of the Bruker MALDI Biotyper CA System for the Identification of Clinical Aerobic Gram-Negative Bacterial Isolates. PLoS ONE 2015, 10, e0141350. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicorbial Susceptibility Testing, 29th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Habib, J.; Kittinger, C.; Luxner, J.; Zarfel, G. Multiresistant Bacteria Isolated from Activated Sludge in Austria. Int. J. Environ. Res. Public Health 2018, 15, 479. [Google Scholar] [CrossRef]
- Jiang, X.; Cui, X.; Xu, H.; Liu, W.; Tao, F.; Shao, T.; Pan, X.; Zheng, B. Whole Genome Sequencing of Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli Isolated From a Wastewater Treatment Plant in China. Front. Microbiol. 2019, 10, 1797. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.; Cacace, D.; Kampouris, I.; Guilloteau, H.; Jäger, T.; Marano, R.B.M.; Karaolia, P.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; et al. Inter-laboratory calibration of quantitative analyses of antibiotic resistance genes. J. Environ. Chem. Eng. 2020, 8, 102214. [Google Scholar] [CrossRef]
- Auerbach, E.; Seyfried, E.; McMahon, K. Tetracycline Resistance Genes in Activated Sludge Wastewater Treatment Plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Lucassen, R.; Rehberg, L.; Heyden, M.; Bockmühl, D. Strong correlation of total phenotypic resistance of samples from household environments and the prevalence of class 1 integrons suggests for the use of the relative prevalence of intI1 as a screening tool for multi-resistance. PLoS ONE 2019, 14, e0218277. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Robleto, E.; Dumont, T.; Levy, S.B. The Frequency of Antibiotic-Resistant Bacteria in Homes Differing in Their Use of Surface Antibacterial Agents. Curr. Microbiol. 2012, 65, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Oteng-Peprah, M.; Acheampong, M.A.; deVries, N.K. Greywater Characteristics, Treatment Systems, Reuse Strategies and User Perception—A Review. Water Air Soil Pollut. 2018, 229, 255. [Google Scholar] [CrossRef] [PubMed]
- Ghaitidak, D.M.; Yadav, K.D. Characteristics and treatment of greywater—A review. Environ. Sci. Pollut. Res. 2013, 20, 2795–2809. [Google Scholar] [CrossRef]
- Bakare, B.F.; Mtsweni, S.; Rathilal, S. Characteristics of greywater from different sources within households in a community in Durban, South Africa. J. Water Reuse Desalin. 2016, 7, 520–528. [Google Scholar] [CrossRef]
- Noutsopoulos, C.; Andreadakis, A.; Kouris, N.; Charchousi, D.; Mendrinou, P.; Galani, A.; Mantziaras, I.; Koumaki, E. Greywater characterization and loadings – Physicochemical treatment to promote onsite reuse. J. Environ. Manag. 2018, 216, 337–346. [Google Scholar] [CrossRef]
- De Gisi, S.; Casella, P.; Notarnicola, M.; Farina, R. Grey water in buildings: A mini-review of guidelines, technologies and case studies. Civ. Eng. Environ. Syst. 2016, 33, 35–54. [Google Scholar] [CrossRef]
- Ottoson, J.; Stenström, T.A. Faecal contamination of greywater and associated microbial risks. Water Res. 2003, 37, 645–655. [Google Scholar] [CrossRef]
- Winward, G.P.; Avery, L.M.; Frazer-Williams, R.; Pidou, M.; Jeffrey, P.; Stephenson, T.; Jefferson, B. A study of the microbial quality of grey water and an evaluation of treatment technologies for reuse. Ecol. Eng. 2008, 32, 187–197. [Google Scholar] [CrossRef]
- Ronen, Z.; Guerrero, A.; Gross, A. Greywater disinfection with the environmentally friendly Hydrogen Peroxide Plus (HPP). Chemosphere 2010, 78, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Craddock, H.A.; Rjoub, Y.; Rosen, D.; Greif, J.; Lipchin, C.; Mongodin, E.F.; Sapkota, A.R. Antibiotic-resistant Escherichia coli and Klebsiella sp. in greywater reuse systems and pond water used for agricultural irrigation in the West Bank, Palestinian Territories. Environ. Res. 2020, in press. [Google Scholar]
- Boyjoo, Y.; Pareek, V.K.; Ang, M. A review of greywater characteristics and treatment processes. Water Sci. Technol. 2013, 67, 1403–1424. [Google Scholar] [CrossRef] [PubMed]
- Cogan, T.A.; Bloomfield, S.F.; Humphrey, T.J. The effectiveness of hygiene procedures for prevention of cross-contamination from chicken carcases in the domestic kitchen. Lett. Appl. Microbiol. 1999, 29, 354–358. [Google Scholar] [CrossRef]
- Maimon, A.; Tal, A.; Friedler, E.; Gross, A. Safe on-site reuse of greywater for irrigation—A critical review of current guidelines. Environ. Sci. Technol. 2010, 44, 3213–3220. [Google Scholar] [CrossRef]
- Maimon, A.; Friedler, E.; Gross, A. Parameters affecting greywater quality and its safety for reuse. Sci. Total Environ. 2014, 487, 20–25. [Google Scholar] [CrossRef]
- Leonard, M.; Gilpin, B.; Robson, B.; Wall, K. Field study of the composition of greywater and comparison of microbiological indicators of water quality in on-site systems. Environ. Monit. Assess. 2016, 188, 475. [Google Scholar] [CrossRef]
- Marano, R.B.M.; Zolti, A.; Jurkevitch, E.; Cytryn, E. Antibiotic resistance and class 1 integron gene dynamics along effluent, reclaimed wastewater irrigated soil, crop continua: Elucidating potential risks and ecological constraints. Water Res. 2019, 164, 114906. [Google Scholar] [CrossRef]
- Fluit, A.C.; Schmitz, F.J. Class 1 Integrons, Gene Cassettes, Mobility, and Epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 761–770. [Google Scholar] [CrossRef]
- Gillings, M. DNA as a Pollutant: The Clinical Class 1 Integron. Curr. Pollut. Rep. 2018, 4, 1–7. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.-C. Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 2012, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Carattoli, A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef]
- Friedman, C.R.; Whitney, C.G. It’s Time for a Change in Practice: Reducing Antibiotic Use Can Alter Antibiotic Resistance. J. Infect. Dis. 2008, 197, 1082–1083. [Google Scholar] [CrossRef]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. B Biol. Sci. 2018, 285, 20180332. [Google Scholar] [CrossRef]
| Parameter | Al-Fura | Qasr al-Sir | Tel Arad | Um Batin | Wadi al-Na’am | All Villages | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | |
| pH | 3.80–6.58 | 4.81 ± 1.22 | 4.7–7.3 | 5.6 ± 1.1 | 5.8–9.5 | 7.8 ± 1.5 | 4.9–7.4 | 6.1 ± 1.25 | 5.5–6.7 | 6 ± 0.5 | 3.80–9.59 | 6.09 ± 1.46 |
| EC (mS/cm) | 1.02–10.17 | 3.98 ± 4.2 | 0.4–0.9 | 0.6 ± 0.1 | 0.7–4.4 | 2.3 ± 1.6 | 1.5–3.7 | 2.3 ± 1.05 | 0.7–0.9 | 0.8 ± 0.1 | 0.13–10.17 | 2.04 ± 2.19 |
| TSS (mg/L) | 231.6–2853 | 1156.5 ± 1164 | 22–575 | 260 ± 250.1 | 76–13620 | 3487.5 ± 6755 | 76–338 | 207 ± 185.2 | 21–493 | 212.4 ± 208.6 | 21–13620 | 1160 ± 3177 |
| BOD5 (mg/L) | 448.25–572.5 | 492.6 ± 54.8 | 57.5–503 | 280.9 ± 185.0 | 6.7–532.2 | 312.5 ± 247.6 | 101.3–881.8 | 570.2 ± 313.1 | 287.2–491.7 | 378.2 ± 85.0 | 6.75–881.83 | 414.7 ± 220.62 |
| TOC (mg/L) | 543.14–6047 | 2645 ± 2460 | 72.8–407.4 | 245 ± 137.9 | 3.3–833 | 412.1 ± 411.8 | 48.5–3450 | 1300 ± 1304 | 108.5–827 | 323 ± 338.0 | 3.34–6047 | 1000 ± 1460 |
| TN (mg/L) | 21.36–142.81 | 54.52 ± 58.94 | 15.6–29.1 | 21.7 ± 5.5 | 1.3–94.5 | 32.8 ± 42.9 | 8.9–424.8 | 114.9 ± 175.8 | 1.0–7.7 | 4.6 ± 3.3 | 1.05–424.84 | 49.06 ± 93.16 |
| FC (CFU/100 mL) | 1 × 106–1.26 × 109 | 3.8 × 108 ± 5.94 × 108 | 4.7 × 106 –1.1 × 108 | 3.6 × 107 ± 4.5 × 107 | 0–2.2 × 108 | 5.5 × 107 ± 1.1 × 108 | 1.1 × 106–6.0 × 107 | 1.8 × 107 ± 2.5 × 107 | 2.3 × 108–1.5 × 109 | 1.1 × 109 ± 5.8 × 108 | 0–1.5 × 109 | 2.61 × 108 ± 4.94 × 108 |
| ESBL (CFU/100 mL) | 6.90 × 105–9× 106 | 4.87 × 106 ± 4 × 106 | 5.7 × 105–1.0 × 107 | 3.7 × 106 ± 3.34 × 106 | 1.0 × 105–1.0 × 107 | 4.1 × 106 ± 5.7 × 106 | 4.3 × 105–2.8 × 106 | 1.6 × 106 ± 1.7 × 106 | 1.2 × 106–2.5 × 107 | 7.3 × 106 ± 1.7 × 106 | 1.05 × 105–2.56 × 107 | 4.5 × 106 ± 5.89 × 106 |
| E. coli (CFU/100 mL) | 0–4 × 106 | 1.23 × 106 ± 1.88 × 106 | 0–5.5 × 106 | 1.3 × 106 ± 2.1 × 106 | 0–1.9 × 108 | 4.7 × 107 ± 9.4 × 107 | 6 × 106–9.5× 106 | 7.7 × 106 ± 2.4 × 106 | 6.5 × 107–4 × 108 | 1.7 × 10 8± 1.5 × 108 | 0–4 × 108 | 4.15 × 107 ± 9.47 × 107 |
| Variables | TSS (mg/L) | BOD5 (mg/L) | TOC (mg/L) | TN (mg/L) | FC (CFU/100 mL) | ESBL (CFU/100 mL) | 16S | intI1 | sul1 | qnrS | blaTEM | blaCTX-M-32 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSS (mg/L) | 1 | |||||||||||
| BOD5 (mg/L) | 0.385 | 1 | ||||||||||
| TOC (mg/L) | 0.442 | 0.870 | 1 | |||||||||
| TN (mg/L) | 0.356 | 0.412 | 0.515 | 1 | ||||||||
| FC (CFU/100 mL) | 0.108 | 0.491 | 0.261 | −0.032 | 1 | |||||||
| ESBL (CFU/100 mL) | −0.103 | 0.058 | −0.035 | −0.099 | 0.589 | 1 | ||||||
| 16S | 0.183 | 0.297 | 0.350 | 0.017 | 0.672 | 0.359 | 1 | |||||
| intI1 | −0.182 | 0.056 | −0.309 | −0.308 | 0.307 | −0.011 | −0.206 | 1 | ||||
| sul1 | −0.218 | −0.010 | −0.327 | −0.281 | 0.436 | 0.179 | 0.035 | 0.911 | 1 | |||
| qnrS | 0.221 | 0.183 | 0.008 | 0.035 | 0.192 | 0.198 | −0.102 | 0.574 | 0.465 | 1 | ||
| blaTEM | 0.370 | 0.210 | 0.089 | 0.086 | 0.375 | 0.355 | 0.178 | 0.389 | 0.405 | 0.838 | 1 | |
| blaCTX-M-32 | 0.427 | 0.222 | 0.149 | 0.049 | 0.346 | 0.014 | 0.384 | 0.166 | 0.075 | 0.437 | 0.541 | 1 |
| Identity of Bacterial Isolate | Source | Village | No. of Isolates |
|---|---|---|---|
| Enterobacter cloacae | Kitchen | Al-Fura | 2 |
| Klebsiella oxytoca | Kitchen | Qasr al-Sir | 3 |
| Klebsiella pneumoniae | Kitchen | Tel Arad | 2 |
| Klebsiella pneumoniae | Mixed GW | Tel Arad | 2 |
| Escherichia coli | Kitchen | Um Batin | 3 |
| Klebsiella pneumoniae | Kitchen | Wadi al-Na’am | 2 |
| Escherichia coli | Kitchen | Wadi al-Na’am | 1 |
| Location | Organism | Amoxicillin/Clavulanic A | Ampicillin | Ceftazidime | Ceftriaxone | Cefuroxime | Cephalexin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (S/I/) | MIC (µg/m) | (S/I/) | MIC (µg/m) | (S/I/) | MIC (µg/m) | (S/I/) | MIC (µg/m) | (S/I/R) | MIC (µg/m) | (S/I/R) | MIC (µg/m) | ||
| Al-Fura | E. cloacae | R | ≥32 | R | ≥32 | R | ≥64 | R | 16 | R | ≥64 | R | ≥64 |
| E. cloacae | R | ≥32 | R | ≥32 | R | ≥64 | R | 16 | R | ≥64 | R | ≥64 | |
| Qasr al -Sir | K. oxytoca | S | ≤2 | R | ≥32 | R* | <=1 | R | 16 | R | ≥64 | R | ≥64 |
| K. oxytoca | S | ≤2 | R | ≥32 | R* | ≤1 | R | 16 | R | ≥64 | R | ≥64 | |
| K. oxytoca | S | ≤2 | R | ≥32 | R* | ≤1 | R | 16 | R | ≥64 | R | ≥64 | |
| Tel Arad | K. pneumoniae | I | 16 | R | ≥32 | R | 8 | R | ≥64 | R | ≥64 | R | ≥64 |
| K. pneumoniae | S | 4 | R | ≥32 | R | 4 | S | ≤1.0 | R | ≥64 | R | ≥64 | |
| K. pneumoniae | S | 8 | R | ≥32 | R* | ≤1 | R | 4 | R | 16 | R | 16 | |
| K. pneumoniae | I | 16 | R | ≥32 | R | 8 | R | ≥64 | R | ≥64 | R | ≥64 | |
| Um Batin | E. coli | S | 4 | R | ≥32 | R | 4 | R | ≥64 | R | ≥64 | R | ≥64 |
| E. coli | S | 4 | R | ≥32 | R | 16 | R | ≥64 | R | ≥64 | R | ≥64 | |
| E. coli | S | 4 | R | ≥32 | R | 16 | R | ≥64 | R | ≥64 | R | ≥64 | |
| Wadi aln’am | E. coli | S | 4 | R | ≥32 | R | 16 | R | ≥64 | R | 16 | R | ≥64 |
| K. pneumoniae | S | 4 | R | ≥32 | R | 4 | R | 16 | R | ≥64 | R | ≥64 | |
| K. pneumoniae | I | 16 | R | ≥32 | R | 8 | R | ≥64 | R | ≥64 | R | ≥64 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porob, S.; Craddock, H.A.; Motro, Y.; Sagi, O.; Gdalevich, M.; Ezery, Z.; Davidovitch, N.; Ronen, Z.; Moran-Gilad, J. Quantification and Characterization of Antimicrobial Resistance in Greywater Discharged to the Environment. Water 2020, 12, 1460. https://doi.org/10.3390/w12051460
Porob S, Craddock HA, Motro Y, Sagi O, Gdalevich M, Ezery Z, Davidovitch N, Ronen Z, Moran-Gilad J. Quantification and Characterization of Antimicrobial Resistance in Greywater Discharged to the Environment. Water. 2020; 12(5):1460. https://doi.org/10.3390/w12051460
Chicago/Turabian StylePorob, Seema, Hillary A. Craddock, Yair Motro, Orly Sagi, Michael Gdalevich, Zubaida Ezery, Nadav Davidovitch, Zeev Ronen, and Jacob Moran-Gilad. 2020. "Quantification and Characterization of Antimicrobial Resistance in Greywater Discharged to the Environment" Water 12, no. 5: 1460. https://doi.org/10.3390/w12051460
APA StylePorob, S., Craddock, H. A., Motro, Y., Sagi, O., Gdalevich, M., Ezery, Z., Davidovitch, N., Ronen, Z., & Moran-Gilad, J. (2020). Quantification and Characterization of Antimicrobial Resistance in Greywater Discharged to the Environment. Water, 12(5), 1460. https://doi.org/10.3390/w12051460







