A New Thioalkalivibrio sp. Strain Isolated from Petroleum-Contaminated Brackish Estuary Sediments: A New Candidate for Bio-Based Application for Sulfide Oxidation in Halo-Alkaline Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Total Petroleum Hydrocarbon Quantification in Sediments
2.2. Isolation of Thioalkalivibrio sp. 10fs10
2.3. Thioalkalivibrio sp. 10fs10 Genome Sequencing
2.4. Taxonomic Characterisation of the Thioalkalivibrio sp. 10fs10
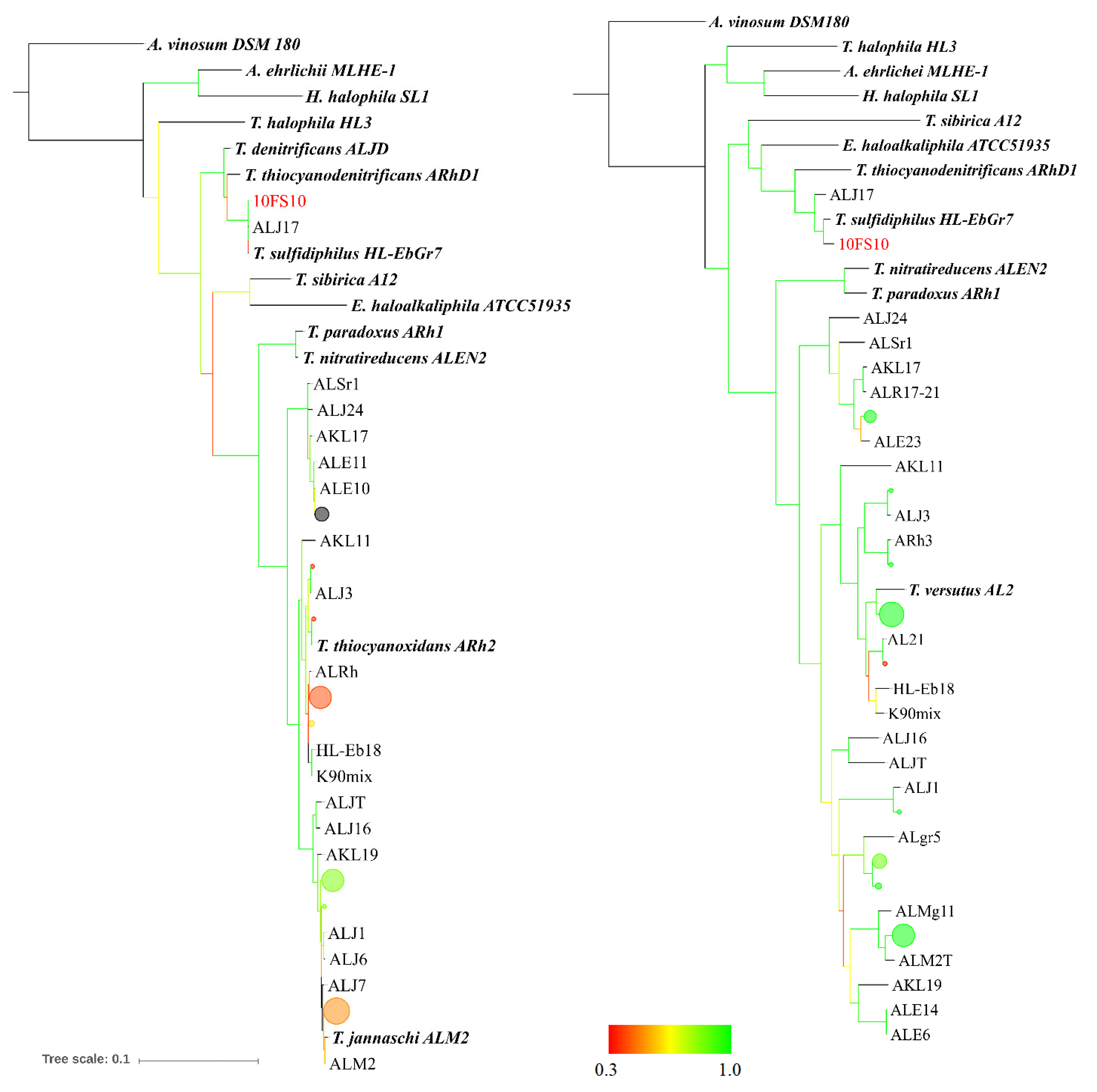
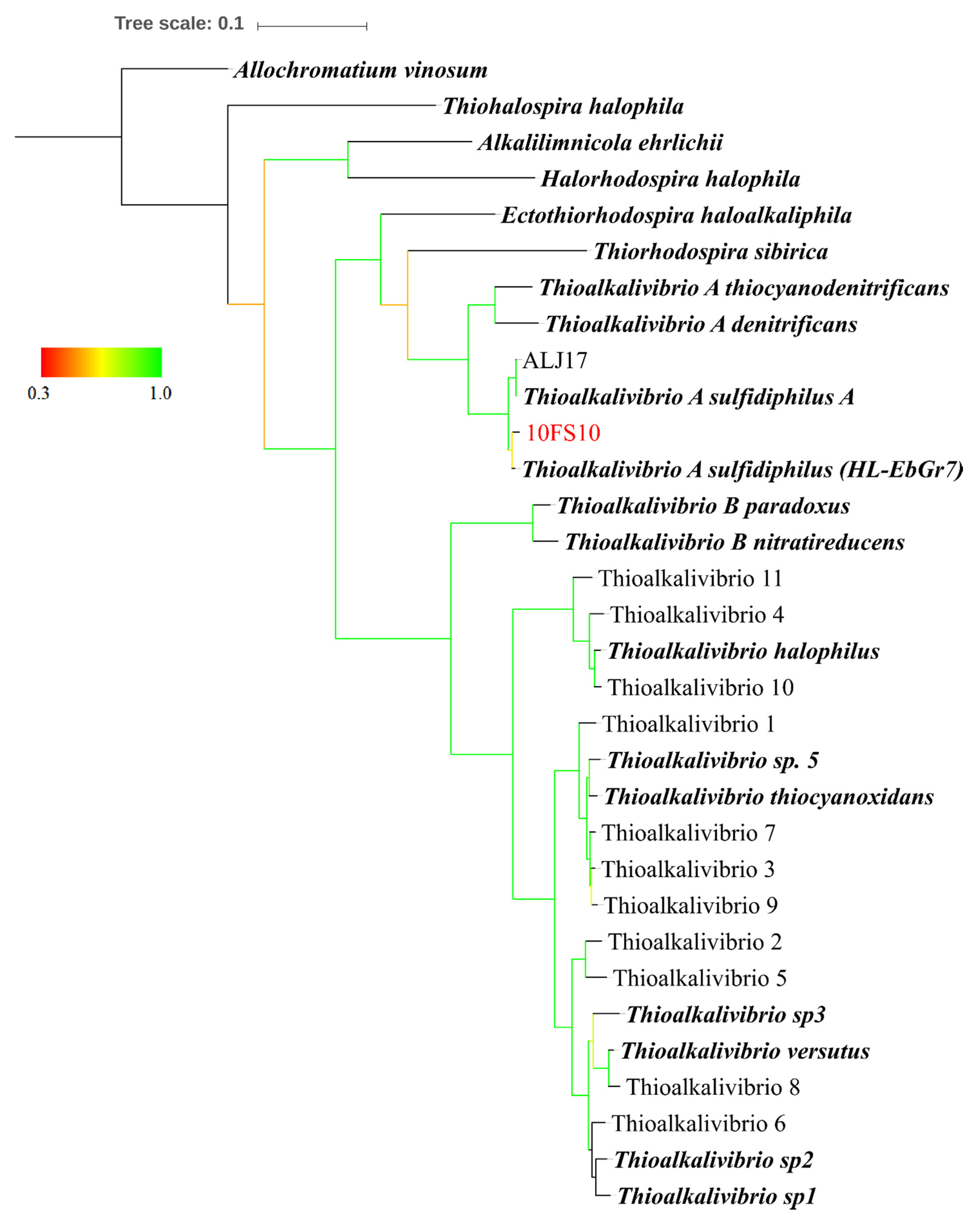
2.5. Maximum Likelihood Tree Based on Multi-Locus Sequence (MLS) Analysis
2.6. Classification of Thioalkalivibrio sp. 10fs10 by ANI-BBH, dDDH, and Genome Taxonomy Database
2.7. Thioalkalivibrio sp. 10fs10 Growth in Halo-Alkaline Conditions
2.8. Thioalkalivibrio sp. 10fs10 Hydrogen Sulfide Oxidation Kinetics
3. Results
3.1. Thioalkalivibrio sp. 10fs10 Strain Isolation and Phylogenetic Characterisation
3.2. Genome Properties
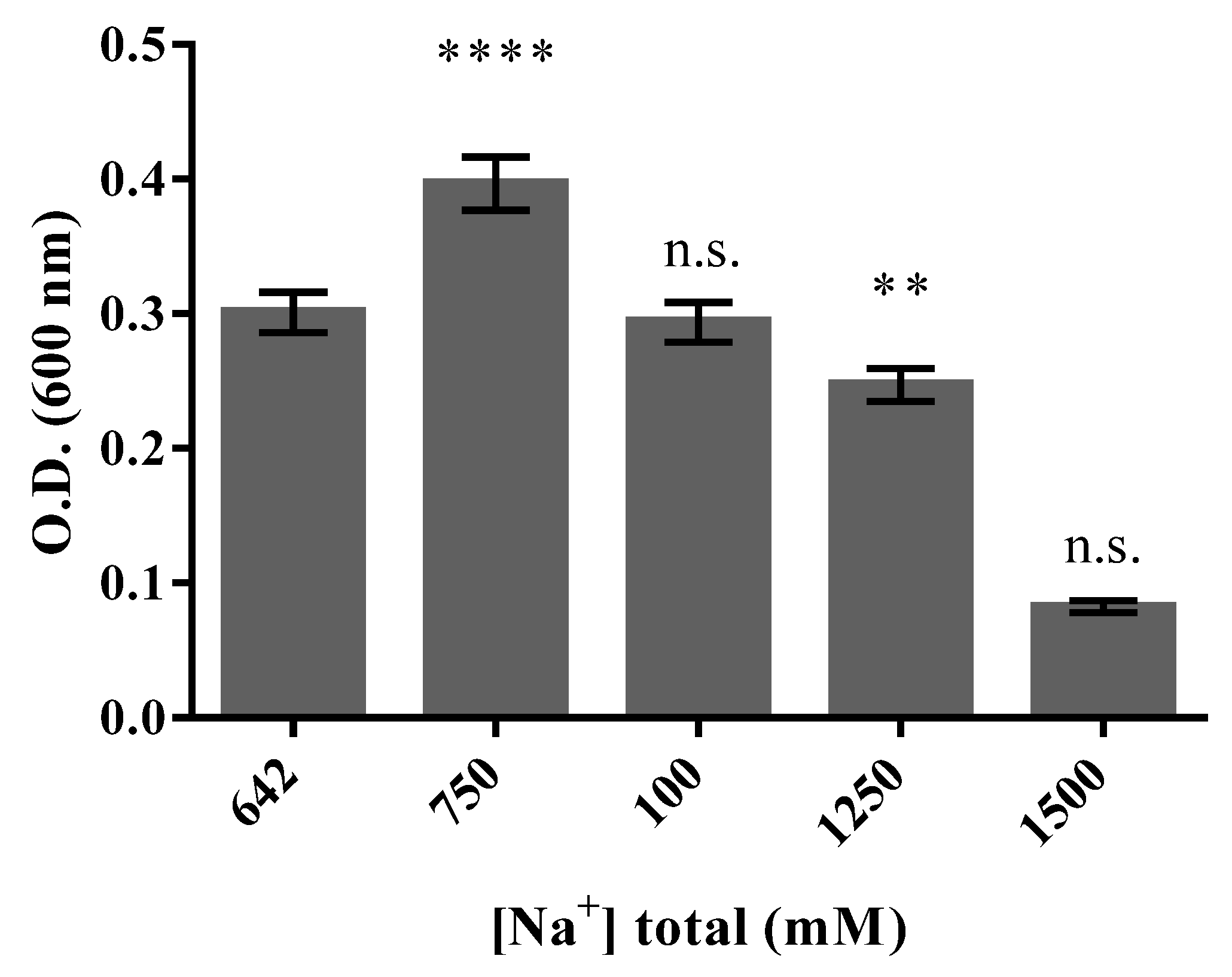
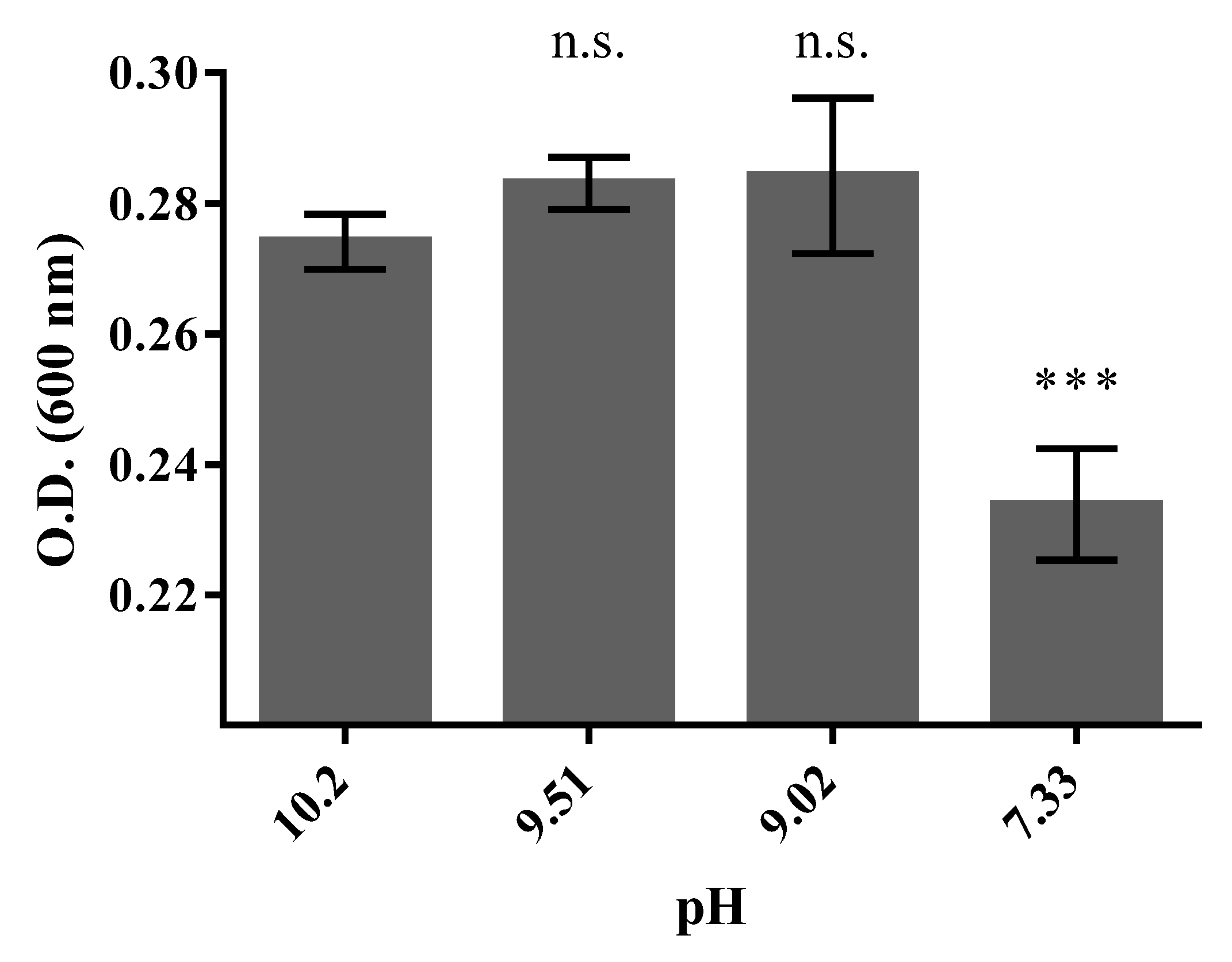
3.3. Halo-Alkaline Condition of Growth
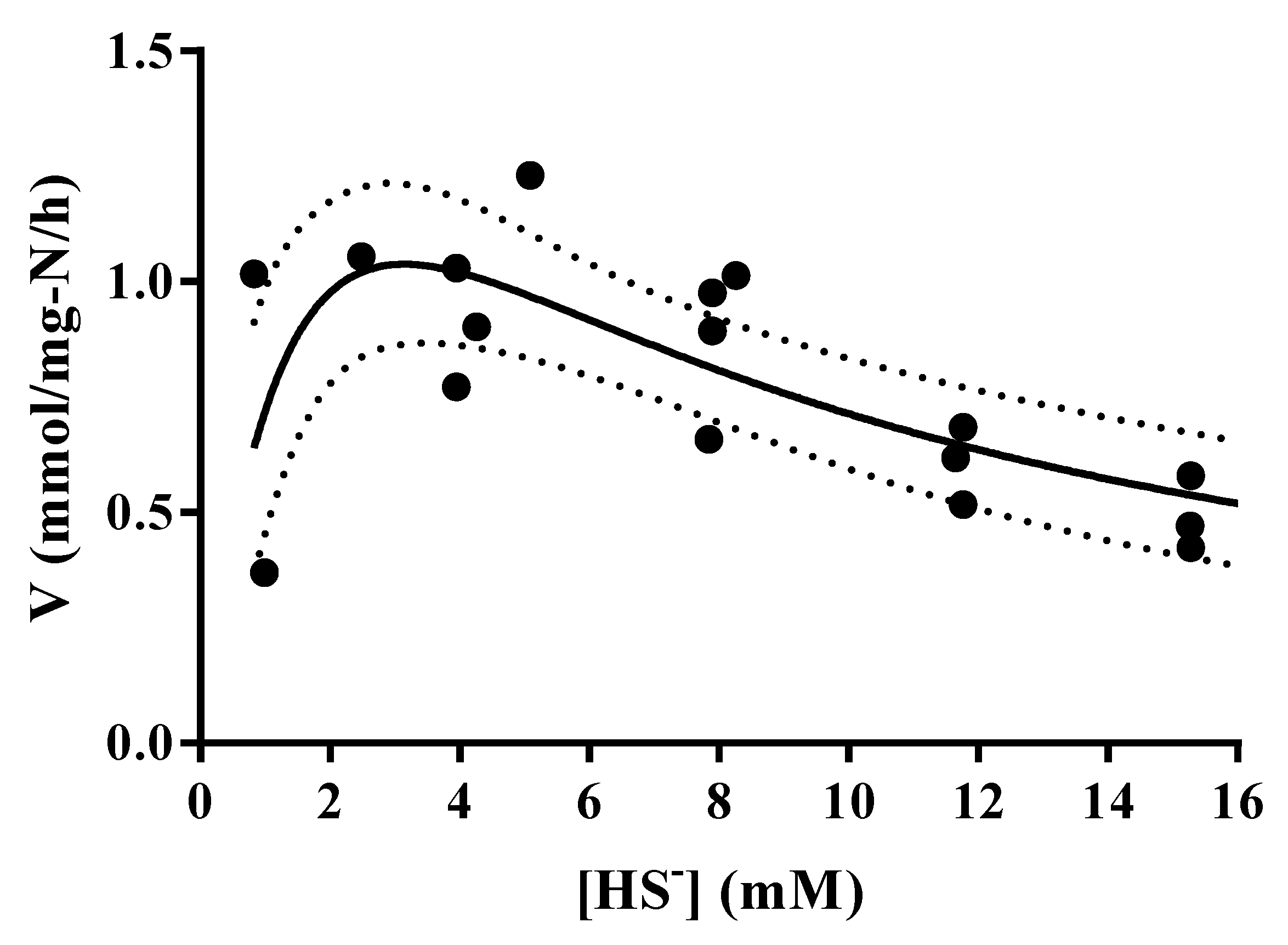
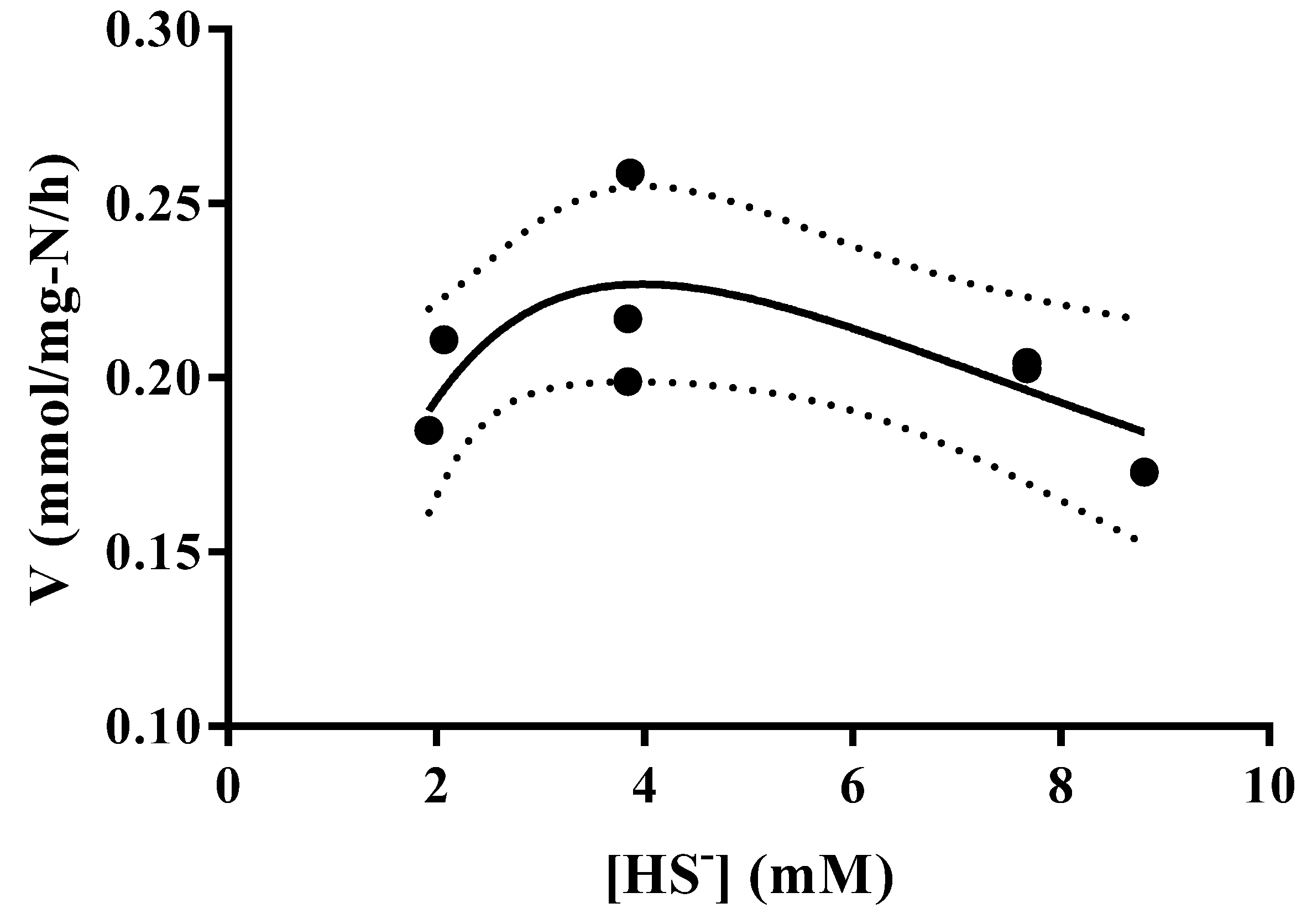
3.4. Thioalkalivibrio sp. 10fs10 Capacity of Hydrogen Sulfide Oxidation in Halo-Alkaline Growth Conditions
3.5. Thioalkalivibrio sp. 10fs10 Genome Sequencing and Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Becarelli, S.; Chicca, I.; Siracusa, G.; La China, S.; Gentini, A.; Lorenzi, R.; Munz, G.; Petroni, G.; Levin, D.B.; Di Gregorio, S. Hydrocarbonoclastic Ascomycetes to enhance co-composting of total petroleum hydrocarbon (TPH) contaminated dredged sediments and lignocellulosic matrices. N. Biotechnol. 2019, 50, 27–36. [Google Scholar] [CrossRef]
- Di Gregorio, S.; Gentini, A.; Siracusa, G.; Becarelli, S.; Azaizeh, H.; Lorenzi, R. Phytoremediation for improving the quality of effluents from a conventional tannery wastewater treatment plant. Int. J. Environ. Sci. Technol. 2015, 12, 1387–1400. [Google Scholar] [CrossRef] [Green Version]
- Di Gregorio, S.; Siracusa, G.; Becarelli, S.; Mariotti, L.; Gentini, A.; Lorenzi, R. Isolation and characterization of a hydrocarbonoclastic bacterial enrichment from total petroleum hydrocarbon contaminated sediments: Potential candidates for bioaugmentation in bio-based processes. Environ. Sci. Pollut. Res. 2016, 23, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Lampis, S.; Ferrari, A.; Cunha-Queda, A.C.; Alvarenga, P.; Di Gregorio, S.; Vallini, G. Selenite resistant rhizobacteria stimulate SeO(3) (2-) phytoextraction by Brassica juncea in bioaugmented water-filtering artificial beds. Environ. Sci. Pollut. Res. Int. 2009, 16, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Banciu, H.L.; Muyzer, G. Functional microbiology of soda lakes. Curr. Opin. Microbiol. 2015, 25, 88–96. [Google Scholar] [CrossRef]
- Mu, T.; Zhou, J.; Yang, M.; Xing, J. Complete genome sequence of Thialkalivibrio versutus D301 isolated from Soda Lake in northern China, a typical strain with great ability to oxidize sulfide. J. Biotechnol. 2016, 227, 21–22. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.M.; Hahm, D.; Jung, Y.-J.; Park, S.H.; Chun, J.; Hong, S.G. Bacterial community of sediments from the Australian-Antarctic ridge. Polar Biol. 2014, 37, 587–593. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Lee, O.O.; Zeng, X.; Shao, Z.; Qian, P.Y. Microbial sulfur cycle in two hydrothermal chimneys on the Southwest Indian Ridge. MBio 2014, 5, e00980-13. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.B.; Chen, Y.Q.; Lan, C.Y.; Tam, N.F.Y.; Zan, Q.J.; Huang, L.N. Recovery of novel bacterial diversity from mangrove sediment. Mar. Biol. 2007, 150, 739–747. [Google Scholar] [CrossRef]
- Sundarakrishnan, B.; Pushpanathan, M.; Jayashree, S.; Rajendhran, J.; Sakthivel, N.; Jayachandran, S.; Gunasekaran, P. Assessment of microbial richness in pelagic sediment of Andaman Sea by bacterial tag encoded FLX titanium amplicon pyrosequencing (bTEFAP). Indian J. Microbiol. 2012, 52, 544–550. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Lo, I.M.C.; Yan, D.Y.S. An integrated bioremediation process for petroleum hydrocarbons removal and odor mitigation from contaminated marine sediment. Water Res. 2015, 83, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; van den Bosch, P.L.F.; Abbas, B.; Janssen, A.J.H.; Muyzer, G. Microbiological analysis of the population of extremely haloalkaliphilic sulfur-oxidizing bacteria dominating in lab-scale sulfide-removing bioreactors. Appl. Microbiol. Biotechnol. 2008, 80, 965–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Bosch, P.L. Biological Sulfide Oxidation by Natron-Alkaliphilic Bacteria: Application in Gas Desulfurization. Doctoral Dissertation, Wageningen Universiteit, Wageningen, The Netherlands, 2008. [Google Scholar]
- de Graaff, M.; Klok, J.B.M.; Bijmans, M.F.M.; Muyzer, G.; Janssen, A.J.H. Application of a 2-step process for the biological treatment of sulfidic spent caustics. Water Res. 2012, 46, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Smet, E.; Lens, P.; Van Langenhove, H. Treatment of waste gases contaminated with odorous sulfur compounds. Crit. Rev. Environ. Sci. Technol. 1998, 28, 89–117. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dombek, P.E.; Johnson, L.K.; Zimmerley, S.T.; Sadowsky, M.J. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 2000, 66, 2572–2577. [Google Scholar] [CrossRef] [Green Version]
- Versalovic, J.; Koeuth, T.; Lupski, J.R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Varghese, N.J.; Mukherjee, S.; Ivanova, N.; Konstantinidis, K.T.; Mavrommatis, K.; Kyrpides, N.C.; Pati, A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015, 43, 6761–6771. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appia-Ayme, C.; Little, P.J.; Matsumoto, Y.; Leech, A.P.; Berks, B.C. Cytochrome Complex Essential for Photosynthetic Oxidation of both Thiosulfate and Sulfide in Rhodovulum sulfidophilum. J. Bacteriol. 2001, 183, 6107–6118. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, C.G.; Rother, D.; Bardischewsky, F.; Quentmeier, A.; Fischer, J. Oxidation of Reduced Inorganic Sulfur Compounds by Bacteria: Emergence of a Common Mechanism? Appl. Environ. Microbiol. 2001, 67, 2873–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosshard, H.R.; Davidson, M.W.; Knaff, D.B.; Millett, F. Complex formation and electron transfer between mitochondrial cytochrome c and flavocytochrome c552 from Chromatium vinosum. J. Biol. Chem. 1986, 261, 190–193. [Google Scholar] [PubMed]
- Beller, H.R.; Chain, P.S.G.; Letain, T.E.; Chakicherla, A.; Larimer, F.W.; Richardson, P.M.; Coleman, M.A.; Wood, A.P.; Kelly, D.P. The Genome Sequence of the Obligately Chemolithoautotrophic, Facultatively Anaerobic Bacterium Thiobacillus denitrificans. J. Bacteriol. 2006, 188, 1473–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockdreher, Y.; Venceslau, S.S.; Josten, M.; Sahl, H.G.; Pereira, I.A.C.; Dahl, C. Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: Evidence for sulfur transfer from DsrEFH to DsrC. PLoS ONE 2012, 7, e40785. [Google Scholar] [CrossRef]
- Cort, J.R.; Selan, U.; Schulte, A.; Grimm, F.; Kennedy, M.A.; Dahl, C. Allochromatium vinosum DsrC: Solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. J. Mol. Biol. 2008, 382, 692–707. [Google Scholar] [CrossRef] [Green Version]
- Dahl, C. Sulfur metabolism in phototrophic bacteria. In Mod. Top. Phototrophic Prokaryotes; Springer International Publishing: Cham, Switzerland, 2017; pp. 27–66. [Google Scholar] [CrossRef]
- Grein, F.; Pereira, I.A.C.; Dahl, C. Biochemical characterization of individual components of the Allochromatium vinosum DsrMKJOP transmembrane complex aids understanding of complex function in vivo. J. Bacteriol. 2010, 192, 6369–6377. [Google Scholar] [CrossRef] [Green Version]
- Sander, J.; Engels-Schwarzlose, S.; Dahl, C. Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 2006, 186, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Lübbe, Y.J.; Youn, H.-S.; Timkovich, R.; Dahl, C. Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c-diamide synthase, for sulfur oxidation. FEMS Microbiol. Lett. 2006, 261, 194–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, T.; Dahl, C. A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 2018, 12, 2479–2491. [Google Scholar] [CrossRef] [Green Version]
- Kappler, U. Bacterial sulfite-oxidizing enzymes. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, B.; Kuever, J. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5′-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 2007, 153, 3478–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahl, C. Insertional gene inactivation in a phototrophic sulfur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology 1996, 142, 3363–3372. [Google Scholar] [CrossRef] [Green Version]
- Parey, K.; Demmer, U.; Warkentin, E.; Wynen, A.; Ermler, U.; Dahl, C. Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum. PLoS ONE 2013, 8, e74707. [Google Scholar] [CrossRef]
- Brändén, M.; Sanden, T.; Brzezinski, P.; Widengren, J. Localized proton microcircuits at the biological membrane-water interface. Proc. Natl. Acad. Sci. USA 2006, 103, 19766–19770. [Google Scholar] [CrossRef] [Green Version]
- Haines, T.H.; Dencher, N.A. Cardiolipin: A proton trap for oxidative phosphorylation. FEBS Lett. 2002, 528, 35–39. [Google Scholar] [CrossRef] [Green Version]
- Ascenzi, P.; Coletta, M.; Wilson, M.T.; Fiorucci, L.; Marino, M.; Polticelli, F.; Sinibaldi, F.; Santucci, R. Cardiolipin-cytochrome c complex: Switching cytochrome c from an electron-transfer shuttle to a myoglobin- and a peroxidase-like heme-protein. IUBMB Life 2015, 67, 98–109. [Google Scholar] [CrossRef]
- Liu, J.; Xue, Y.; Wang, Q.; Wei, Y.; Swartz, T.H.; Hicks, D.B.; Ito, M.; Ma, Y.; Krulwich, T.A. The activity profile of the NhaD-type Na+(Li+)/H+ antiporter from the soda lake haloalkaliphile Alkalimonas amylolytica is adaptive for the extreme environment. J. Bacteriol. 2005, 187, 7589–7595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, A.C.; Meier-Kolthoff, J.P.; Overmars, L.; Richter, M.; Woyke, T.; Sorokin, D.Y.; Muyzer, G. Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS ONE 2017, 12, e0173517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Berben, T.; Overmars, L.; Sorokin, D.Y.; Muyzer, G. Diversity and distribution of sulfur oxidation-related genes in Thioalkalivibrio, a genus of chemolithoautotrophic and haloalkaliphilic sulfur-oxidizing bacteria. Front. Microbiol. 2019, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Sorokin, D.Y.; Mavromatis, K.; Lapidus, A.; Foster, B.; Sun, H.; Ivanova, N.; Pati, A.; D’haeseleer, P.; Woyke, T.; et al. Complete genome sequence of Thioalkalivibrio sp. K90mix. Stand Genom. Sci. 2011, 5, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biegel, E.; Schmidt, S.; González, J.M.; Müller, V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol. Life Sci. 2011, 68, 613–634. [Google Scholar] [CrossRef]
- Strittmatter, A.W.; Liesegang, H.; Rabus, R.; Decker, I.; Amann, J.; Andres, S.; Henne, A.; Fricke, W.F.; Martinez-Arias, R.; Bartels, D.; et al. Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 2009, 11, 1038–1055. [Google Scholar] [CrossRef] [Green Version]
- Curatti, L.; Brown, C.S.; Ludden, P.W.; Rubio, L.M. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii. PNAS 2005, 102, 6291–6296. [Google Scholar] [CrossRef] [Green Version]
- Müller, V.; Aufurth, S.; Rahlfs, S. The Na+ cycle in Acetobacterium woodii: Identification and characterization of a Na+ translocating F1F0-ATPase with a mixed oligomer of 8 and 16 kDa proteolipids. Biochim. Biophys. Acta Bioenerg. 2001, 1505, 108–120. [Google Scholar] [CrossRef] [Green Version]
- Westphal, L.; Wiechmann, A.; Baker, J.; Minton, N.P.; Müller, V. The Rnf complex is an energy-coupled transhydrogenase essential to reversibly link cellular NADH and ferredoxin pools in the acetogen Acetobacterium woodii. J. Bacteriol. 2018, 200, e00357-18. [Google Scholar] [CrossRef] [Green Version]
- Banciu, H.; Sorokin, D.Y.; Kleerebezem, R.; Muyzer, G.; Galinski, E.A.; Kuenen, J.G. Growth kinetics of haloalkaliphilic, sulfur-oxidizing bacterium Thioalkalivibrio versutus strain ALJ 15 in continuous culture. Extremophiles 2004, 8, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Sorokin, D.Y.; Mavromatis, K.; Lapidus, A.; Clum, A.; Ivanova, N.; Pati, A.; d’Haeseleer, P.; Woyke, T.; Kyrpides, N.C. Complete genome sequence of Thioalkalivibrio sulfidophilus HL-EbGr7. Stand Genom. Sci. 2011, 4, 23–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Genomes | ANI-BBH | dDDH | |||||
|---|---|---|---|---|---|---|---|
| Reference genome | ANI 1->2 | ANI 2->1 | AF 1->2 | AF 2->1 | Estimated DDH value (%) | PdDDH>70% | PdDDH>79% |
| Tv. sulfidiphilus HL-EbGR7 | 95.52 | 95.52 | 78.39 | 80.31 | 56.8–62.4 | 51.07% | 12.52% |
| Tv. sulfidiphilus ALJ17 | 94.33 | 94.33 | 82.68 | 73.67 | 49.1–54.4 | 24.30% | 5.08% |
| Genome Properties | Value |
|---|---|
| Total scaffolds | 17 |
| Length | 3.42 Mbp |
| G + C content | 64.95% |
| Total DNA coding region | 3243 |
| Protein coding region | 3175 |
| rRNA genes | 3 (5S rRNA, 16S rRNA, 23s rRNA) |
| tRNA | 52 |
| tmRNA | 1 |
| Protein coding region with Pfam | 2966 |
| Protein coding region with COG | 2241 |
| Protein coding region with KEGG | 1656 |
| Transmembrane protein coding gene | 781 |
| CRISPR repeats | 2 |
| Air Flow | 0.25 NL/min | 0.09 NL/min |
|---|---|---|
| Vmax (mmol/mg-N/h) | 2.648 | 0.745 |
| KM (mM) | 2.438 | 4.547 |
| Ki (mM) | 4.053 | 3.491 |
| Parameters | Low Air Flow (0.018 NL/min) | High Air Flow (0.25 NL/min) |
|---|---|---|
| (HS-) initial (mM) | 5.21 | 6 |
| Vox spec (mmol/(mg-N·h)) | 0.057 | 0.318 |
| Biomass (mg-N/L) | 7.31 | 6.51 |
| ΔpH | Negligible | −0.1 |
| Elemental sulfur | Yes | no |
| Gene | 10fs10 | HL-EbGr7 | ALJ17 |
|---|---|---|---|
| soxA | 3 | 4 | 4 |
| soxX | 3 | 4 | 4 |
| soxY | 2 | 1 | not found |
| soxZ | 2 | 1 | 1 |
| soxB | 1 | 1 | 1 |
| soxC | 0 | 0 | 0 |
| soxD | 0 | 0 | 0 |
| fccA | 1 | 3 | 2 |
| fccB | 1 | 3 | 2 |
| dsrA | 1 | 1 | 1 |
| dsrB | 1 | 1 | 1 |
| dsrC | 1 | 1 | 1 |
| aprA | 1 | 1 | 1 |
| aprB | 1 | 1 | 1 |
| sat | 2 | 1 | 1 |
| hdrA | 1 | 1 | 1 |
| hdrB | 2 | 1 | 1 |
| hdrC | 2 | 1 | 1 |
| sorA | 1 | 1 | 1 |
| sor B | 1 | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becarelli, S.; La China, S.; Lapidus, A.; Prijibelski, A.; Polev, D.; Petroni, G.; Di Gregorio, S. A New Thioalkalivibrio sp. Strain Isolated from Petroleum-Contaminated Brackish Estuary Sediments: A New Candidate for Bio-Based Application for Sulfide Oxidation in Halo-Alkaline Conditions. Water 2020, 12, 1385. https://doi.org/10.3390/w12051385
Becarelli S, La China S, Lapidus A, Prijibelski A, Polev D, Petroni G, Di Gregorio S. A New Thioalkalivibrio sp. Strain Isolated from Petroleum-Contaminated Brackish Estuary Sediments: A New Candidate for Bio-Based Application for Sulfide Oxidation in Halo-Alkaline Conditions. Water. 2020; 12(5):1385. https://doi.org/10.3390/w12051385
Chicago/Turabian StyleBecarelli, Simone, Salvatore La China, Alla Lapidus, Andrey Prijibelski, Dmitrii Polev, Giulio Petroni, and Simona Di Gregorio. 2020. "A New Thioalkalivibrio sp. Strain Isolated from Petroleum-Contaminated Brackish Estuary Sediments: A New Candidate for Bio-Based Application for Sulfide Oxidation in Halo-Alkaline Conditions" Water 12, no. 5: 1385. https://doi.org/10.3390/w12051385
APA StyleBecarelli, S., La China, S., Lapidus, A., Prijibelski, A., Polev, D., Petroni, G., & Di Gregorio, S. (2020). A New Thioalkalivibrio sp. Strain Isolated from Petroleum-Contaminated Brackish Estuary Sediments: A New Candidate for Bio-Based Application for Sulfide Oxidation in Halo-Alkaline Conditions. Water, 12(5), 1385. https://doi.org/10.3390/w12051385







