Prevalence of Arcobacter and Other Pathogenic Bacteria in River Water in Nepal
Abstract
1. Introduction
2. Materials and Methods
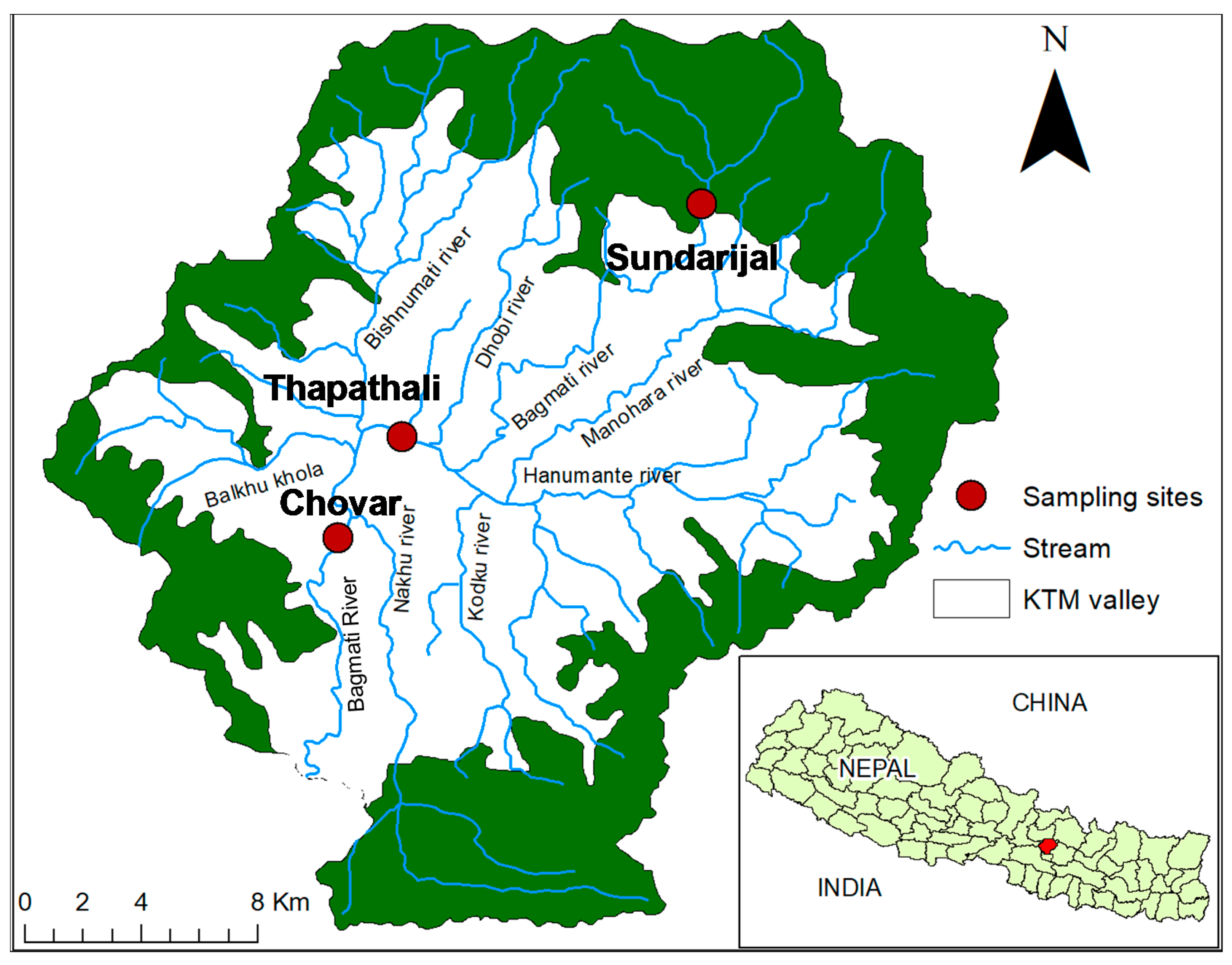
2.1. Collection of River Water Samples and Extraction of Bacterial DNA
2.2. NGS for the Characterization of Bacterial Communities
2.3. Quantification of Total Bacteria and Arcobacter
2.4. Statistical Analysis
3. Results
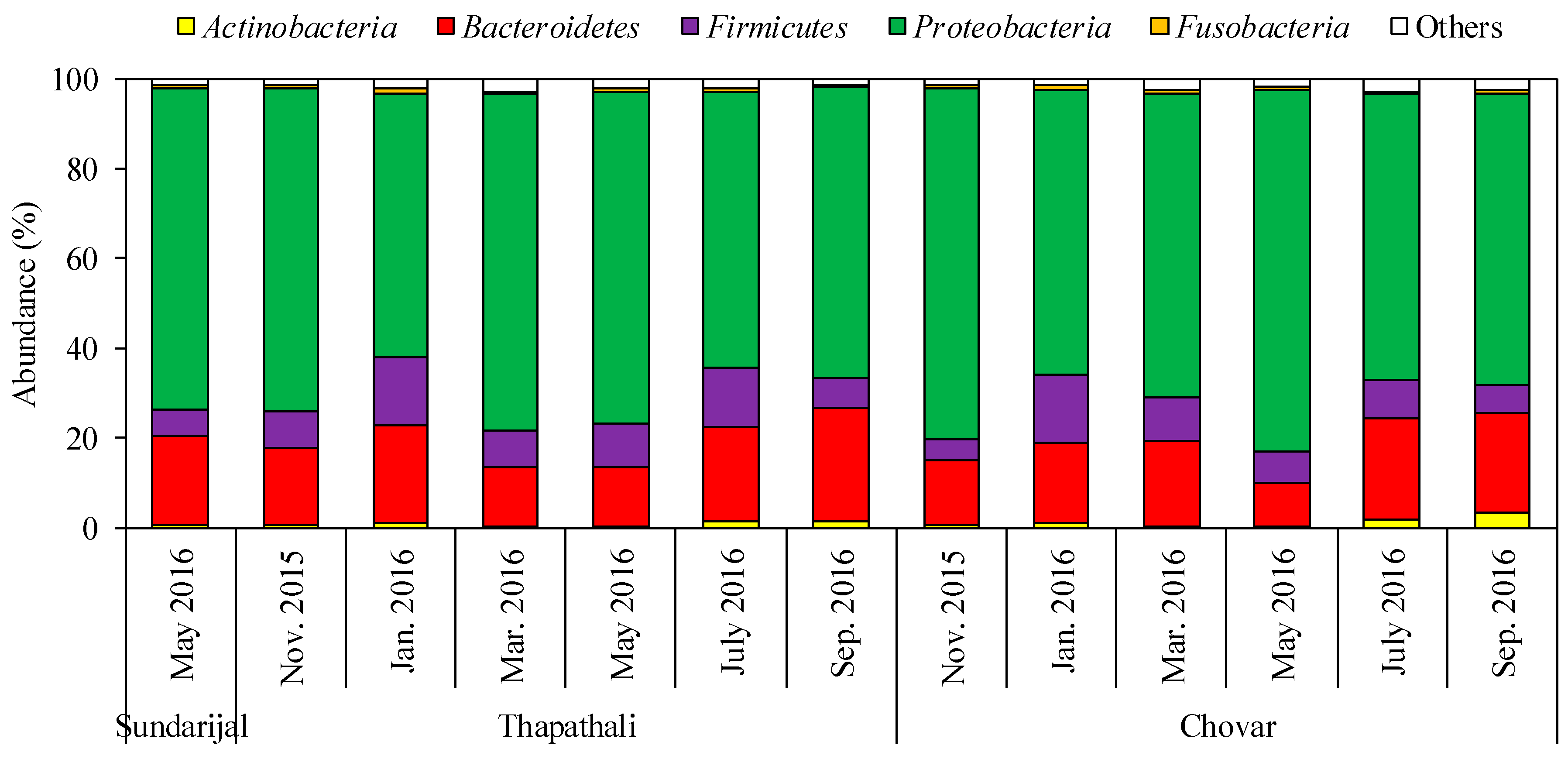
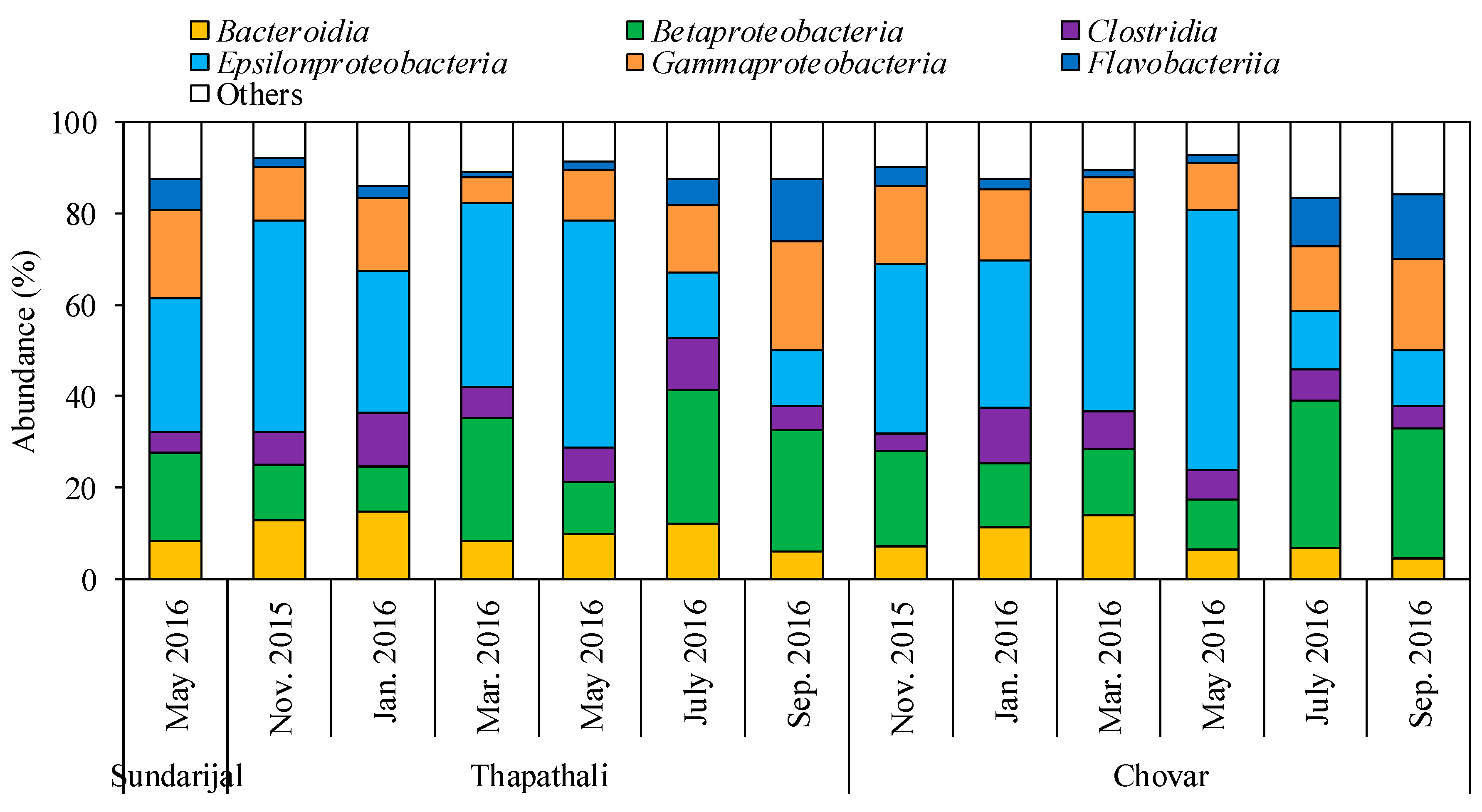
3.1. Characterization of Bacterial Community by NGS
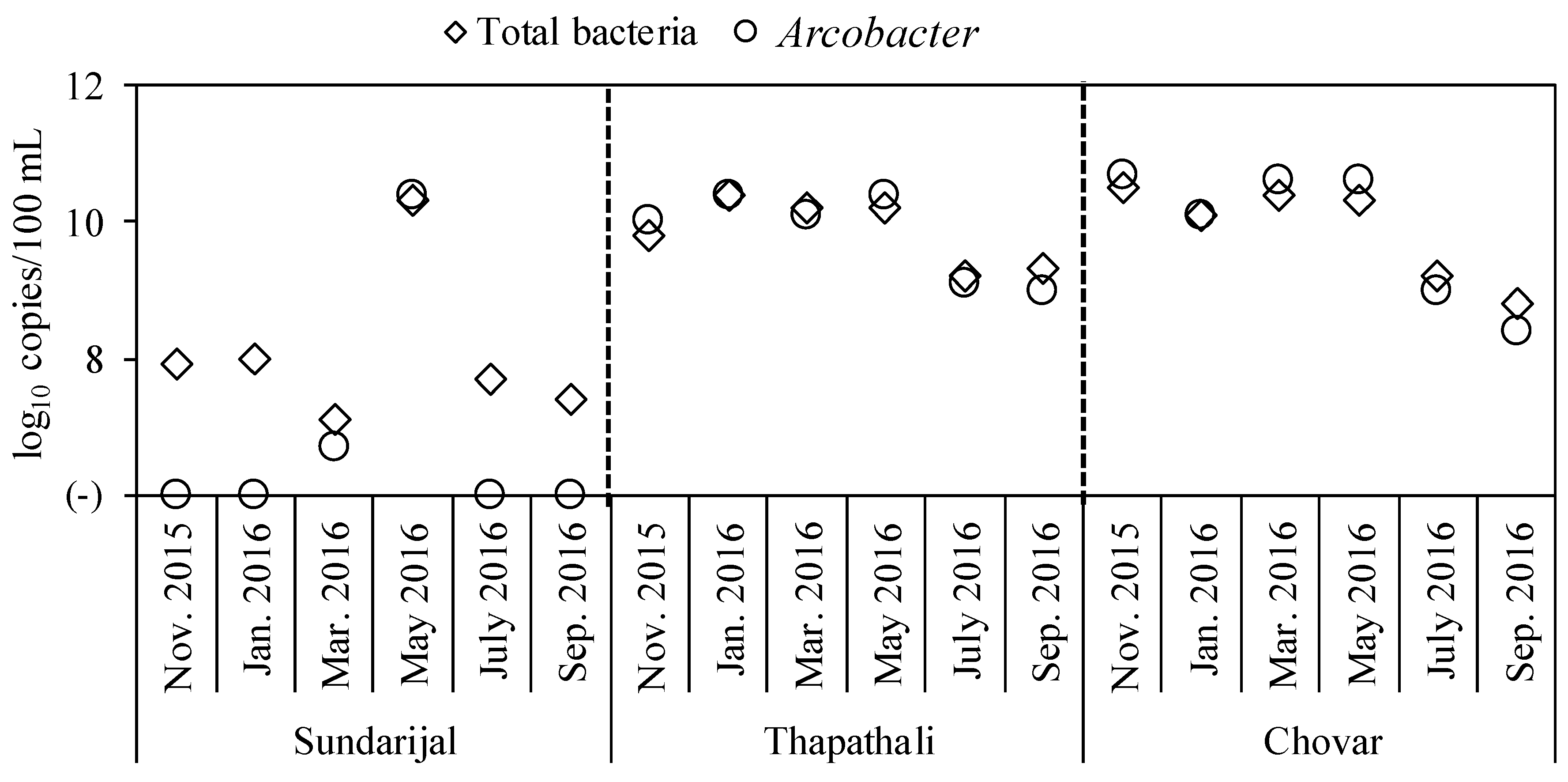
3.2. Occurrence of Total Bacteria and Arcobacter in River Water Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kannel, P.R.; Lee, S.; Lee, Y.-S.; Kanel, S.R.; Pelletier, G.J. Application of automated QUAL2Kw for water quality modeling and management in the Bagmati River, Nepal. Ecol. Model. 2007, 202, 503–517. [Google Scholar] [CrossRef]
- Karn, S.K.; Harada, H. Surface water pollution in three urban territories of Nepal, India, and Bangladesh. Environ. Manag. 2001, 28, 483–496. [Google Scholar]
- Bhatt, M.P.; McDowell, W.H.; Gardner, K.H.; Hartmann, J. Chemistry of the heavily urbanized Bagmati River system in Kathmandu Valley, Nepal: Export of organic matter, nutrients, major ions, silica, and metals. Environ. Earth Sci. 2014, 71, 911–922. [Google Scholar] [CrossRef]
- Ghaju Shrestha, R.; Tanaka, Y.; Malla, B.; Bhandari, D.; Tandukar, S.; Inoue, D.; Sei, K.; Sherchand, J.B.; Haramoto, E. Next-generation sequencing identification of pathogenic bacterial genes and their relationship with fecal indicator bacteria in different water sources in the Kathmandu Valley, Nepal. Sci. Total Environ. 2017, 601–602, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Yamada, K.; Nishida, K. Prevalence of protozoa, viruses, coliphages and indicator bacteria in groundwater and river water in the Kathmandu Valley, Nepal. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Kannel, P.R.; Lee, S.; Kanel, S.R.; Khan, S.P.; Lee, Y.-S. Spatial–temporal variation and comparative assessment of water qualities of urban river system: A case study of the river Bagmati (Nepal). Environ. Monit. Assess. 2007, 129, 433–459. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Shrestha, S.; Shindo, J.; Sherchand, J.B.; Haramoto, E. Virological quality of irrigation water sources and pepper mild mottle virus and tobacco mosaic virus as index of pathogenic virus contamination level. Food Environ. Virol. 2018, 10, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Haramoto, E.; Shindo, J. Assessing the infection risk of enteropathogens from consumption of raw vegetables washed with contaminated water in Kathmandu Valley, Nepal. J. Appl. Microbiol. 2017, 123, 1321–1334. [Google Scholar] [CrossRef]
- Tandukar, S.; Sherchand, J.B.; Bhandari, D.; Sherchan, S.P.; Malla, B.; Ghaju Shrestha, R.; Haramoto, E. Presence of human enteric viruses, protozoa, and indicators of pathogens in the Bagmati River, Nepal. Pathogens 2018, 7, 38. [Google Scholar] [CrossRef]
- Banting, G.; Figueras Salvat, M.J. Arcobacter. In Global Water Pathogens Project; Rose, J.B., Jiménez-Cisneros, B., Eds.; Michigan State University: E. Lansing, MI, USA; UNESCO: Paris, France, 2017; pp. 3–25. Available online: http://www.waterpathogens.org/book/arcobacter (accessed on 9 July 2019).
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.-T.D.; Lee, J. Global distribution and prevalence of Arcobacter in food and water. Zoonoses Public Health 2015, 62, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Collado, L.; Inza, I.; Guarro, J.; Figueras, M.J. Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 2008, 10, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Ghaju Shrestha, R.; Tanaka, Y.; Malla, B.; Tandukar, S.; Bhandari, D.; Inoue, D.; Sei, K.; Sherchand, J.B.; Haramoto, E. Development of a quantitative PCR assay for Arcobacter spp. and its application to environmental water samples. Microbes Environ. 2018, 33, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Moreno, Y.; Botella, S.; Alonso, J.L.; Ferrús, M.A.; Hernández, M.; Hernández, J. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and Fluorescent In Situ Hybridization. Appl. Environ. Microbiol. 2003, 69, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Maruyama, S.; Kabeya, H.; Boonmar, S.; Nimsuphan, B.; Nagai, A.; Kozawa, K.; Nakajima, T.; Mikami, T.; Kimura, H. Isolation and phylogenetic analysis of Arcobacter spp. in ground chicken meat and environmental water in Japan and Thailand. Microbiol. Immunol. 2004, 48, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Schmitz, B.W.; Caton, K.; Zhang, B.; Zabaleta, J.; Garai, J.; Taylor, C.M.; Romanchishina, T.; Gerba, C.P.; Pepper, I.L.; et al. Assessing the spatial and temporal variability of bacterial communities in two Bardenpho wastewater treatment systems via Illumina MiSeq sequencing. Sci. Total Environ. 2019, 657, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Takai, K.; Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 2000, 66, 5066–5072. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Aihara, Y.; Bhattarai, A.P.; Bista, N.; Rajbhandari, S.; Kondo, N.; Kazama, F.; Nishida, K.; Shindo, J. Dynamics of domestic water consumption in the urban area of the Kathmandu Valley: Situation analysis pre and post 2015 Gorkha earthquake. Water 2017, 9, 222. [Google Scholar] [CrossRef]
- Rutkowski, T.; Raschid-Sally, L.; Buechler, S. Wastewater irrigation in the developing world—Two case studies from the Kathmandu Valley in Nepal. Agric. Water Manag. 2007, 88, 83–91. [Google Scholar] [CrossRef]
- Bajracharya, R.; Tamrakar, N.K. Environmental status of Manahara River, Kathamandu, Nepal. Bull. Dep. Geol. 2007, 10, 21–32. [Google Scholar] [CrossRef]
- Bajracharya, R.; Nakamura, T.; Shakya, B.M.; Kei, N.; Shrestha, S.D.; Tamrakar, N.K. Identification of river water and groundwater interaction at central part of the Kathmandu valley, Nepal using stable isotope tracers. Int. J. Adv. Sci. Tech. Res. 2018, 8, 29–41. [Google Scholar] [CrossRef]
- Devkota, D.C.; Watanabe, K. Impact of solid waste on water quality of Bishnumati River and surrounding areas in Kathmandu, Nepal. J. Nepal Geol. Soc. 2005, 31, 19–24. [Google Scholar] [CrossRef][Green Version]
| Genus | Abundance Ratio (%) | ||
|---|---|---|---|
| Sundarijal (n = 1) | Thapathali (n = 6) | Chovar (n = 6) | |
| Acidovorax | 2.2 | 1.0 ± 0.8 | 1.1 ± 0.6 |
| Acinetobacter | 10.3 | 5.3 ± 5.3 | 5.0 ± 3.9 |
| Arcobacter | 28.6 | 31.3 ± 15.8 | 31.8 ± 17.2 |
| Bacteroides | 1.7 | 2.6 ± 0.9 | 2.3 ± 1.1 |
| Blautia | 0.4 | 0.8 ± 0.4 | 0.7 ± 0.6 |
| Burkholderia | 0.3 | 0.5 ± 0.6 | 0.5 ± 0.2 |
| Chromobacterium | 0.4 | 0.4 ± 0.3 | 0.6 ± 0.5 |
| Chryseobacterium | 1.4 | 0.9 ± 0.6 | 1.5 ± 1.0 |
| Clostridium | 0.3 | 0.7 ± 0.2 | 0.7 ± 0.3 |
| Comamonas | 2.1 | 1.1 ± 1.4 | 1.0 ± 0.7 |
| Escherichia | 1.1 | 0.4 ± 0.2 | 0.3 ± 0.1 |
| Flavobacterium | 4.7 | 3.0 ± 4.3 | 3.4 ± 3.9 |
| Mycobacterium | 0.1 | 0.2 ± 0.1 | 0.4 ± 0.8 |
| Parabacteroides | 0.4 | 0.8 ± 0.7 | 1.1 ± 0.9 |
| Plesiomonas | 2.2 | 1.2 ± 0.6 | 1.5 ± 0.7 |
| Prevotella | 5.9 | 6.8 ± 2.4 | 4.6 ± 1.5 |
| Pseudomonas | 1.6 | 1.6 ± 0.9 | 1.8 ± 1.0 |
| Sphingobacterium | 1.9 | 0.1 ± 0.0 | 0.2 ± 0.1 |
| Bacteria and Viruses Tested # | Indicators | r Value with Arcobacter |
|---|---|---|
| Fecal indicator bacteria | E. coli | 0.86 * |
| Human-fecal markers | BK polyomaviruses | 0.72 * |
| JC polyomaviruses | 0.54 | |
| Human Bacteroidales | 0.93 * | |
| Index viruses | Tobacco mosaic virus | 0.36 |
| Pepper mild mottle virus | 0.60 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrestha, R.G.; Tandukar, S.; Bhandari, D.; Sherchan, S.P.; Tanaka, Y.; Sherchand, J.B.; Haramoto, E. Prevalence of Arcobacter and Other Pathogenic Bacteria in River Water in Nepal. Water 2019, 11, 1416. https://doi.org/10.3390/w11071416
Shrestha RG, Tandukar S, Bhandari D, Sherchan SP, Tanaka Y, Sherchand JB, Haramoto E. Prevalence of Arcobacter and Other Pathogenic Bacteria in River Water in Nepal. Water. 2019; 11(7):1416. https://doi.org/10.3390/w11071416
Chicago/Turabian StyleShrestha, Rajani Ghaju, Sarmila Tandukar, Dinesh Bhandari, Samendra P. Sherchan, Yasuhiro Tanaka, Jeevan B. Sherchand, and Eiji Haramoto. 2019. "Prevalence of Arcobacter and Other Pathogenic Bacteria in River Water in Nepal" Water 11, no. 7: 1416. https://doi.org/10.3390/w11071416
APA StyleShrestha, R. G., Tandukar, S., Bhandari, D., Sherchan, S. P., Tanaka, Y., Sherchand, J. B., & Haramoto, E. (2019). Prevalence of Arcobacter and Other Pathogenic Bacteria in River Water in Nepal. Water, 11(7), 1416. https://doi.org/10.3390/w11071416







