Enhanced Transcriptomic Resilience following Increased Alternative Splicing and Differential Isoform Production between Air Pollution Conurbations
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. PM Monitoring and Characterization
2.3. AP Transportation Time Assessment
2.4. Human Whole Blood Collection
2.5. Routine Blood Examination
2.6. Lung Function Test
2.7. Correlation of Haematological Indices and AQI, PM10, PM2.5
2.8. Isolation of Neutrophils and RNA Extraction
2.9. RNA Sequencing
2.10. Transcript Quantification
2.11. Correlation between Transcript Expression and PM2.5, PM10, AQI
2.12. ESE Identification
2.13. DET (Differentially Expressed Transcript) Identification
2.14. Variance Analysis of Physiological Phenotypes
2.15. Cytokine ELISA
2.16. Cytokine Array
3. Results
3.1. Selecting Beijing and Chengde as Investigation Sites
3.2. Physiological Responses upon PM Exposure and Data Generation
3.3. Alternative Splicing in BRs and CRs
3.4. AS Reshapes Glycolysis Landscape
3.5. AS Prolongs Neutrophil Lifespan and Enhances Migration in BRs
3.6. HIF-1 Mediated Impaired Glycolysis Mediates Neutrophil Dysfunction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Ambient Air Pollution: A Global Assessment of Exposure and Burden of Disease. 2016. Available online: https://apps.who.int/iris/handle/10665/250141-eng.pdf (accessed on 1 June 2020).
- Brook, R.D.; Rajagopalan, S.; Pope, C.A., 3rd; Brook, J.R.; Bhatnagar, A.; Diez-Roux, A.V.; Holguin, F.; Hong, Y.; Luepker, R.V.; Mittleman, M.A.; et al. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation 2010, 121, 2331–7238. [Google Scholar] [CrossRef]
- Zhao, J.; Li, M.; Wang, Z.; Chen, J.; Zhao, J.; Xu, Y.; Wei, X.; Wang, J.; Xie, J. Role of PM2.5 in the development and progression of COPD and its mechanisms. Respir. Res. 2019, 20, 120. [Google Scholar] [CrossRef]
- Feng, X.; Shao, L.; Xi, C.; Jones, T.; Zhang, D.; Bérubé, K. Particle-induced oxidative damage by indoor size-segregated particulate matter from coal-burning homes in the Xuanwei lung cancer epidemic area, Yunnan Province, China. Chemosphere 2020, 256, 127058. [Google Scholar] [CrossRef]
- Yan, Y.-H.; Chou, C.C.-K.; Wang, J.-S.; Tung, C.-L.; Li, Y.-R.; Lo, K.; Cheng, T.-J. Subchronic effects of inhaled ambient particulate matter on glucose homeostasis and target organ damage in a type 1 diabetic rat model. Toxicol. Appl. Pharmacol. 2014, 281, 211–220. [Google Scholar] [CrossRef]
- World Health Organization. Global Urban Ambient Air Pollution Database (Update 2016). Available online: http://www.who.int/phe/health_topics/outdoorair/databases/cities/en/ (accessed on 1 June 2019).
- Li, T.; Hu, R.; Chen, Z.; Li, Q.; Huang, S.; Zhu, Z.; Zhou, L.-F. Fine particulate matter (PM2.5): The culprit for chronic lung diseases in China. Chronic Dis. Transl. Med. 2018, 4, 176–186. [Google Scholar] [CrossRef]
- Price, H.D.; Jones, T.P.; Bérubé, K.A. Resolution of the mediators of in vitro oxidative reactivity in size-segregated fractions that may be masked in the urban PM10 cocktail. Sci. Total Environ. 2014, 485-486, 588–595. [Google Scholar] [CrossRef][Green Version]
- Adams, R.C. Training Response Inhibition to Reduce Food Consumption. Ph.D. Thesis, Cardiff University, Cardiff, UK, February 2014. [Google Scholar]
- Chuang, H.-C.; Jones, T.; Chen, T.-T.; Bérubé, K. Cytotoxic effects of incense particles in relation to oxidative stress, the cell cycle and F-actin assembly. Toxicol. Lett. 2013, 220, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.-C.; Be, K.B.; Lung, S.-C.C.; Bai, K.-J.; Jones, T. Investigation into the oxidative potential generated by the formation of particulate matter from incense combustion. J. Hazard. Mater. 2013, 244–245, 142–150. [Google Scholar] [CrossRef]
- Huttunen, K.; Wlodarczyk, A.J.; Tirkkonen, J.; Mikkonen, S.; Täubel, M.; Krop, E.; Jacobs, J.; Pekkanen, J.; Heederik, D.; Zock, J.P.; et al. Oxidative capacity and haemolytic activity of settled dust from moisture-damaged schools. Indoor Air 2019, 29, 299–307. [Google Scholar] [CrossRef]
- Shao, L.; Hou, C.; Geng, C.; Liu, J.; Hu, Y.; Wang, J.; Jones, T.; Zhao, C.; Bérubé, K. The oxidative potential of PM 10 from coal, briquettes and wood charcoal burnt in an experimental domestic stove. Atmos. Environ. 2016, 127, 372–381. [Google Scholar] [CrossRef]
- Shao, L.; Hu, Y.; Shen, R.; Schäfer, K.G.; Wang, J.; Wang, J.; Schnelle-Kreis, J.; Zimmermann, R.; Bérubé, K.; Suppan, P. Seasonal variation of particle-induced oxidative potential of airborne particulate matter in Beijing. Sci. Total Environ. 2017, 579, 1152–1160. [Google Scholar] [CrossRef]
- Jones, T.; Be, K.B. The bioreactivity of the sub-10μm component of volcanic ash: Soufrière Hills volcano, Montserrat. J. Hazard. Mater. 2011, 194, 128–134. [Google Scholar] [CrossRef]
- Chen, T.-T.; Chuang, K.-J.; Chiang, L.-L.; Chen, C.-C.; Yeh, C.-T.; Wang, L.-S.; Gregory, C.; Jones, T.; Bérubé, K.; Lee, C.-N.; et al. Characterization of the interactions between protein and carbon black. J. Hazard. Mater. 2014, 264, 127–135. [Google Scholar] [CrossRef]
- Moreno, T.; Martins, V.; Querol, X.; Jones, T.; Bérubé, K.; Minguillón, M.C.; Amato, F.; Capdevila, M.; de Miguel, E.; Centelles, S.; et al. A new look at inhalable metalliferous airborne particles on rail subway platforms. Sci. Total Environ. 2015, 505, 367–375. [Google Scholar] [CrossRef]
- Pope, C.A., 3rd; Burnett, R.T.; Thurston, G.D.; Thun, M.J.; Calle, E.E.; Krewski, D.; Godleski, J.J. Cardiovascular mortality and long-term exposure to particulate air pollution-epidemiological evidence of general pathophysiological pathways of disease. Circulation 2004, 109, 71–77. [Google Scholar] [CrossRef]
- Injarabian, L.; Devin, A.; Ransac, S.; Marteyn, B.S. Neutrophil metabolic shift during their lifecycle: Impact on their survival and activation. Int. J. Mol. Sci. 2020, 21, 287. [Google Scholar] [CrossRef]
- Wright, H.; Moots, R.J.; Bucknall, R.C.; Edwards, S.W. Neutrophil function in inflammation and inflammatory diseases. Rheumatology 2010, 49, 1618–1631. [Google Scholar] [CrossRef]
- Weiss, D.J.; Klausner, J.S. Neutrophil-induced Erythrocyte Injury: A potential cause of erythrocyte destruction in the anemia associated with inflammatory disease. Vet. Pathol. 1988, 25, 450–455. [Google Scholar] [CrossRef]
- Meinderts, S.M.; Oldenborg, P.A.; Beuger, B.M.; Klei, T.; Johansson, J.; Kuijpers, T.W.; Matozaki, T.; Huisman, E.J.; de Haas, M.; van den Berg, T.K.; et al. Human and murine splenic neutrophils are potent phagocytes of IgG-opsonized red blood cells. Blood Adv. 2017, 1, 875–886. [Google Scholar] [CrossRef]
- Peretz, A.; Peck, E.C.; Bammler, T.K.; Beyer, R.P.; Sullivan, J.H.; Trenga, C.A.; Srinouanprachnah, S.; Farin, F.M.; Kaufman, J.D. Diesel exhaust inhalation and assessment of peripheral blood mononuclear cell gene transcription effects: An exploratory study of healthy human volunteers. Inhal. Toxicol. 2007, 19, 1107–1119. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Duan, F.; Qin, M.; Wu, F.; Sheng, W.; Yang, L.; Liu, J.; He, K. Transcriptomic analyses of the biological effects of airborne PM2.5 exposure on human bronchial epithelial cells. PLoS ONE 2015, 10, e0138267. [Google Scholar] [CrossRef]
- Líbalová, H.; Uhlířová, K.; Kléma, J.; Machala, M.; Šrám, R.J.; Ciganek, M.; Topinka, J. Global gene expression changes in human embryonic lung fibroblasts induced by organic extracts from respirable air particles. Part. Fibre Toxicol. 2012, 9, 1. [Google Scholar] [CrossRef]
- Watterson, T.L.; Sorensen, J.; Martin, R.; Coulombe, R.A. Effects of PM2.5 collected from cache valley utah on genes associated with the inflammatory response in human lung cells. J. Toxicol. Environ. Health Part A 2007, 70, 1731–1744. [Google Scholar] [CrossRef]
- Zhang, H.; Yuan, H.; Liu, X.; Yu, J.; Jiao, Y. Impact of synoptic weather patterns on 24 h-average PM2.5 concentrations in the North China Plain during 2013–2017. Sci. Total Environ. 2018, 627, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Shi, Z.; Jones, T.; Li, J.; Whittaker, A.; Bérubé, K. Bioreactivity of particulate matter in Beijing air: Results from plasmid DNA assay. Sci. Total Environ. 2006, 367, 261–272. [Google Scholar] [CrossRef]
- Shi, Z.; Shao, L.; Jones, T.; Whittaker, A.; Lu, S.; BeruBe, K.; He, T.; Richards, R. Characterization of airborne individual particles collected in an urban area, a satellite city and a clean air area in Beijing, 2001. Atmos. Environ. 2003, 37, 4097–4108. [Google Scholar] [CrossRef]
- Lu, S.; Shao, L.; Wu, M.; Jones, T.P.; Merolla, L.; Richard, R.J. Correlation between plasmid DNA damage induced by PM10 and trace metals in inhalable particulate matters in Beijing air. Sci. China Ser. D Earth Sci. 2006, 49, 1323–1331. [Google Scholar] [CrossRef]
- Shao, L.; Li, J.; Zhao, H.; Yang, S.; Li, H.; Li, W.; Jones, T.; Sexton, K.; Be, K.B. Associations between particle physicochemical characteristics and oxidative capacity: An indoor PM10 study in Beijing, China. Atmos. Environ. 2007, 41, 5316–5326. [Google Scholar] [CrossRef]
- Jones, T.; Wlodarczyk, A.; Koshy, L.; Brown, P.; Longyi, S.; Be, K.B. The geochemistry and bioreactivity of fly-ash from coal-burning power stations. Biomarkers 2009, 14, 45–48. [Google Scholar] [CrossRef]
- Shi, Z.; Shao, L.; Li, H.; Whittaker, A.G.; Jones, T.P.; BeruBe, K.; Richards, R.J. Physicochemical characterization of the PM10 in ambient air of northwestern Beijing urban area during heating-period. Huan Jing Ke Xue 2002, 23, 30–34. [Google Scholar]
- Shi, Z.; Shao, L.; Jones, T.P.; Lu, S. Microscopy and mineralogy of airborne particles collected during severe dust storm episodes in Beijing, China. J. Geophys. Res. Atmos. 2005, 110, 1–10. [Google Scholar] [CrossRef]
- Whittaker, A.G.; Jones, T.P.; Shao, L.; Shi, Z.; BérubéK, A.; Richards, R.J. Mineral dust in urban air: Beijing, China. Miner. Mag. 2003, 67, 173–182. [Google Scholar] [CrossRef]
- Yuan, G.; Yang, W. Evaluating China’s air pollution control policy with extended AQI indicator system: Example of the Beijing-Tianjin-Hebei region. Sustainability 2019, 11, 939. [Google Scholar] [CrossRef]
- Miller, M.R.; Crapo, R.; Hankinson, J.; Brusasco, V.; Burgos, F.; Casaburi, R.; Coates, A.; Enright, P.; Van Der Grinten, C.P.M.; Gustafsson, P.; et al. General considerations for lung function testing. Eur. Respir. J. 2005, 26, 153–161. [Google Scholar] [CrossRef]
- Piorunek, T.; Kostrzewska, M.; Cofta, S.; Batura-Gabryel, H.; Andrzejczak, P.; Bogdanski, P.; Wysocka, E. Impulse oscillometry in the diagnosis of airway resistance in Chronic Obstructive Pulmonary Disease. Adv. Exp. Med. Biol. 2015, 838, 47–52. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Cartegni, L.; Wang, J.; Zhu, Z.; Zhang, M.Q.; Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003, 31, 3568–3571. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Zhang, T.; Rong, Z.; Hu, L.; Gu, Z.; Wu, Q.; Dong, S.; Liu, Q.; Lin, Z.; Deutschova, L.; et al. Population transcriptomes reveal synergistic responses of DNA polymorphism and RNA expression to extreme environments on the Qinghai-Tibetan Plateau in a predatory bird. Mol. Ecol. 2017, 26, 2993–3010. [Google Scholar] [CrossRef]
- Zhou, X.; Lindsay, H.; Robinson, M.D. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. 2014, 42, e91. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Li, J.; Tibshirani, R. Finding consistent patterns: A nonparametric approach for identifying differential expression in RNA-seq data. Stat. Methods Med. Res. 2013, 22, 519–536. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Mor, I.; Cheung, E.C.; Vousden, K.H. Control of glycolysis through regulation of PFK1: Old friends and recent additions. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 211–216. [Google Scholar] [CrossRef]
- Martinez-Contreras, R.; Cloutier, P.; Shkreta, L.; Fisette, J.-F.; Revil, T.; Chabot, B. hnRNP Proteins and splicing control. Adv. Exp. Med. Biol. 2007, 623, 123–147. [Google Scholar] [CrossRef]
- Zhou, Z.; Fu, X.-D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef]
- Jian, X.; Boerwinkle, E.; Liu, X. In silico tools for splicing defect prediction: A survey from the viewpoint of end users. Genet. Med. 2014, 16, 497–503. [Google Scholar] [CrossRef]
- Chen, C.D.; Kobayashi, R.; Helfman, D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta -tropomyosin gene. Genes Dev. 1999, 13, 593–606. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef]
- Dai, J.; Sun, C.; Yao, Z.; Chen, W.; Yu, L.; Long, M. Exposure to concentrated ambient fine particulate matter disrupts vascular endothelial cell barrier function via the IL-6/HIF-1α signaling pathway. FEBS Open Biol. 2016, 6, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Cramer, T.; Yamanishi, Y.; Clausen, B.E.; Förster, I.; Pawlinski, R.; Mackman, N.; Haase, V.H.; Jaenisch, R.; Corr, M.; Nizet, V.; et al. HIF-1α Is Essential for Myeloid Cell-Mediated Inflammation. Cell 2003, 112, 645–657. [Google Scholar] [CrossRef]
- Egners, A.; Erdem, M.; Cramer, T. The response of macrophages and neutrophils to hypoxia in the context of cancer and other inflammatory diseases. Mediat. Inflamm. 2016, 2016, 2053646. [Google Scholar] [CrossRef]
- Sun, L.; Fu, J.; Lin, S.H.; Sun, J.L.; Xia, L.; Lin, C.H.; Liu, L.; Zhang, C.; Yang, L.; Xue, P.; et al. Particulate matter of 2.5 μm or less in diameter disturbs the balance of TH17/regulatory T cells by targeting glutamate oxaloacetate transaminase 1 and hypoxia-inducible factor 1α in an asthma model. J. Allergy Clin. Immunol. 2020, 145, 402–414. [Google Scholar] [CrossRef]
- Li, H.; Cai, J.; Chen, R.; Zhao, Z.; Ying, Z.; Wang, L.; Chen, J.; Hao, K.; Kinney, P.L.; Chen, H.; et al. Particulate matter exposure and stress hormone levels: A randomized, double-blind, crossover trial of air purification. Circulation 2017, 136, 618–627. [Google Scholar] [CrossRef]
- Yi, M.; Ban, Y.; Tan, Y.; Xiong, W.; Li, G.; Xiang, B. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer. Mol. Metab. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Kim, S.-G.; Manes, N.; El-Maghrabi, M.R.; Lee, Y.-H. Crystal structure of the hypoxia-inducible form of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): A possible new target for cancer therapy. J. Biol. Chem. 2006, 281, 2939–2944. [Google Scholar] [CrossRef] [PubMed]
- Minchenko, O.H.; Ogura, T.; Opentanova, I.L.; Minchenko, D.O.; Esumi, H. Splice isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-4: Expression and hypoxic regulation. Mol. Cell. Biochem. 2005, 280, 227–234. [Google Scholar] [CrossRef]
- Semenza, G.L.; Jiang, B.-H.; Leung, S.W.; Passantino, R.; Concordet, J.-P.; Maire, P.; Giallongo, A. Hypoxia response elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef]
- Tanguay, R.L.; Gallie, D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996, 16, 146–156. [Google Scholar] [CrossRef]
- Zhao, R.Z.; Jiang, S.; Zhang, L.; Yu, Z.B. Mitochondrial electron transport chain, ROS generation and uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef]
- Sanders, L.H.; Greenamyre, T.J. Oxidative damage to macromolecules in human Parkinson disease and the rotenone model. Free Radic. Biol. Med. 2013, 62, 111–120. [Google Scholar] [CrossRef]
- Minchenko, O.; Leshchinsky, I.; Opentanova, I.; Sang, N.; Srinivas, V.; Armstead, V.; Caro, J. Hypoxia-inducible factor-1-mediated expression of the 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 2002, 277, 6183–6187. [Google Scholar] [CrossRef]
- Cui, X.G.; Han, Z.T.; He, S.H.; Wu, X.D.; Chen, T.R.; Shao, C.H.; Chen, D.L.; Su, N.; Chen, Y.M.; Wang, T.; et al. HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget 2017, 8, 24840–24852. [Google Scholar] [CrossRef]
- Greijer, A.E.; van der Wall, E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 2004, 57, 1009–1014. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, Y.; Zhang, X.; Chen, L. Part mutual information for quantifying direct associations in networks. Proc. Natl. Acad. Sci. USA 2016, 113, 5130–5135. [Google Scholar] [CrossRef]
- Anderson, J.M.; Rodriguez, A.; Chang, D.T. Foreign body reaction to biomaterials. Semin. Immunol. 2008, 20, 86–100. [Google Scholar] [CrossRef]
- McCracken, J.M.; Allen, L.-A.H. Regulation of human neutrophil apoptosis and lifespan in health and disease. J. Cell Death 2014, 7, 15–23. [Google Scholar] [CrossRef]
- Volpe, E.; Sambucci, M.; Battistini, L.; Borsellino, G. Fas–Fas ligand: Checkpoint of T cell functions in multiple sclerosis. Front. Immunol. 2016, 7, 382. [Google Scholar] [CrossRef] [PubMed]
- Selders, G.S.; Fetz, A.E.; Radic, M.; Bowlin, G.L. An overview of the role of neutrophils in innate immunity, inflammation and host-biomaterial integration. Regen. Biomater. 2017, 4, 55–68. [Google Scholar] [CrossRef]
- Hou, G.; Valujskikh, A.; Bayer, J.; Stavitsky, A.B.; Metz, C.; Heeger, P.S. In vivo blockade of macrophage migration inhibitory factor prevents skin graft destruction after indirect allorecognition1. Transplantation 2001, 72, 1890–1897. [Google Scholar] [CrossRef]
- Demir, Y.; Chen, Y.; Metz, C.; Renz, H.; Heeger, P.S. Cardiac allograft rejection in the absence of macrophage migration inhibitory factor. Transplantation 2003, 76, 244–247. [Google Scholar] [CrossRef]
- Huang, X.R.; Hui, C.W.C.; Chen, Y.-X.; Chun, B.; Wong, B.; Fung, P.C.; Metz, C.; Cho, C.H.; Hui, W.M.; Bucala, R.; et al. Macrophage migration inhibitory factor is an important mediator in the pathogenesis of gastric inflammation in rats. Gastroenterology 2001, 121, 619–630. [Google Scholar] [CrossRef]
- Ferro, E.A.V.; Mineo, J.R.; Ietta, F.; Bechi, N.; Romagnoli, R.; Silva, D.A.O.; Sorda, G.; Bevilacqua, E.; Paulesu, L.R. Macrophage migration inhibitory factor is up-regulated in human first-trimester placenta stimulated by soluble antigen of Toxoplasma gondii, resulting in increased monocyte adhesion on villous explants. Am. J. Pathol. 2008, 172, 50–58. [Google Scholar] [CrossRef]
- Jovanović, M.; Vićovac, L. Interleukin-6 stimulates cell migration, invasion and integrin expression in HTR-8/SVneo cell line. Placenta 2009, 30, 320–328. [Google Scholar] [CrossRef]
- Lei, C.-T.; Su, H.; Ye, C.; Tang, H.; Gao, P.; Wan, C.; He, F.-F.; Wang, Y.-M.; Zhang, C. The classic signalling and trans-signalling of interleukin-6 are both injurious in podocyte under high glucose exposure. J. Cell. Mol. Med. 2018, 22, 251–260. [Google Scholar] [CrossRef]
- Maston, L.D.; Jones, D.T.; Giermakowska, W.; Resta, T.C.; Ramiro-Diaz, J.M.; Howard, T.A.; Jernigan, N.L.; Herbert, L.; Maurice, A.A.; Bosc, L.V.G. Interleukin-6 trans-signaling contributes to chronic hypoxia-induced pulmonary hypertension. Pulm. Circ. 2018, 8, 2045894018780734. [Google Scholar] [CrossRef]
- Stringer, K.A.; Tobias, M.; O’Neill, H.C.; Franklin, C.C. Cigarette smoke extract-induced suppression of caspase-3-like activity impairs human neutrophil phagocytosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L1572–L1579. [Google Scholar] [CrossRef] [PubMed]
- McGovern, N.N.; Cowburn, A.S.; Porter, L.; Walmsley, S.R.; Summers, C.; Thompson, A.; Anwar, S.; Willcocks, L.C.; Whyte, M.; Condliffe, A.M.; et al. Hypoxia selectively inhibits respiratory burst activity and killing of Staphylococcus aureus in human neutrophils. J. Immunol. 2011, 186, 453–463. [Google Scholar] [CrossRef]
- Drannik, A.G.; Pouladi, M.; Robbins, C.; Goncharova, S.I.; Kianpour, S.; Stampfli, M. Impact of cigarette smoke on clearance and inflammation after Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 2004, 170, 1164–1171. [Google Scholar] [CrossRef]
- Sapey, E.; Stockley, J.A.; Greenwood, H.; Ahmad, A.; Bayley, D.; Lord, J.; Insall, R.; Stockley, R.A. Behavioral and structural differences in migrating peripheral neutrophils from patients with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2011, 183, 1176–1186. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; A Park, S.; Park, I.; Kim, P.; Cho, N.H.; Hyun, J.W.; Hyun, Y.-M. PM2.5 Exposure in the respiratory system induces distinct inflammatory signaling in the lung and the liver of mice. J. Immunol. Res. 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Sun, Q.; Jiang, S.; Xu, X. Particulate air pollution on systemic inflammatory response: Roles of monocytes and neutrophils. Eur. Respir. J. 2012, 40, 4783. [Google Scholar] [CrossRef]
- Hoenderdos, K.; Condliffe, A. The neutrophil in Chronic Obstructive Pulmonary Disease. Too little, too late or too much, too soon? Am. J. Respir. Cell Mol. Biol. 2013, 48, 531–539. [Google Scholar] [CrossRef]
- Alberts, N.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Wang, A.; Huen, S.C.; Luan, H.H.; Yu, S.; Zhang, C.; Gallezot, J.-D.; Booth, C.J.; Medzhitov, R. Opposing effects of fasting Metabolism on tissue tolerance in bacterial and viral inflammation. Cell 2016, 166, 1512–1525.e12. [Google Scholar] [CrossRef]
- Bowden, S.D.; Rowley, G.; Hinton, J.C.D.; Thompson, A. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar typhimurium. Infect. Immun. 2009, 77, 3117–3126. [Google Scholar] [CrossRef] [PubMed]
- Xavier, M.N.; Winter, M.G.; Spees, A.M.; den Hartigh, A.B.; Nguyen, K.; Roux, C.M.; Silva, T.M.; Atluri, V.L.; Kerrinnes, T.; Keestra, A.M.; et al. PPARγ-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 2013, 14, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Caballero, H.; Rao, X.; Sun, Q.; Warmoes, M.O.; Lin, P.; Sussan, T.E.; Park, B.; Fan, T.W.-M.; Maiseyeu, A.; Rajagopalan, S.; et al. Air pollution-derived particulate matter dysregulates hepatic Krebs cycle, glucose and lipid metabolism in mice. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
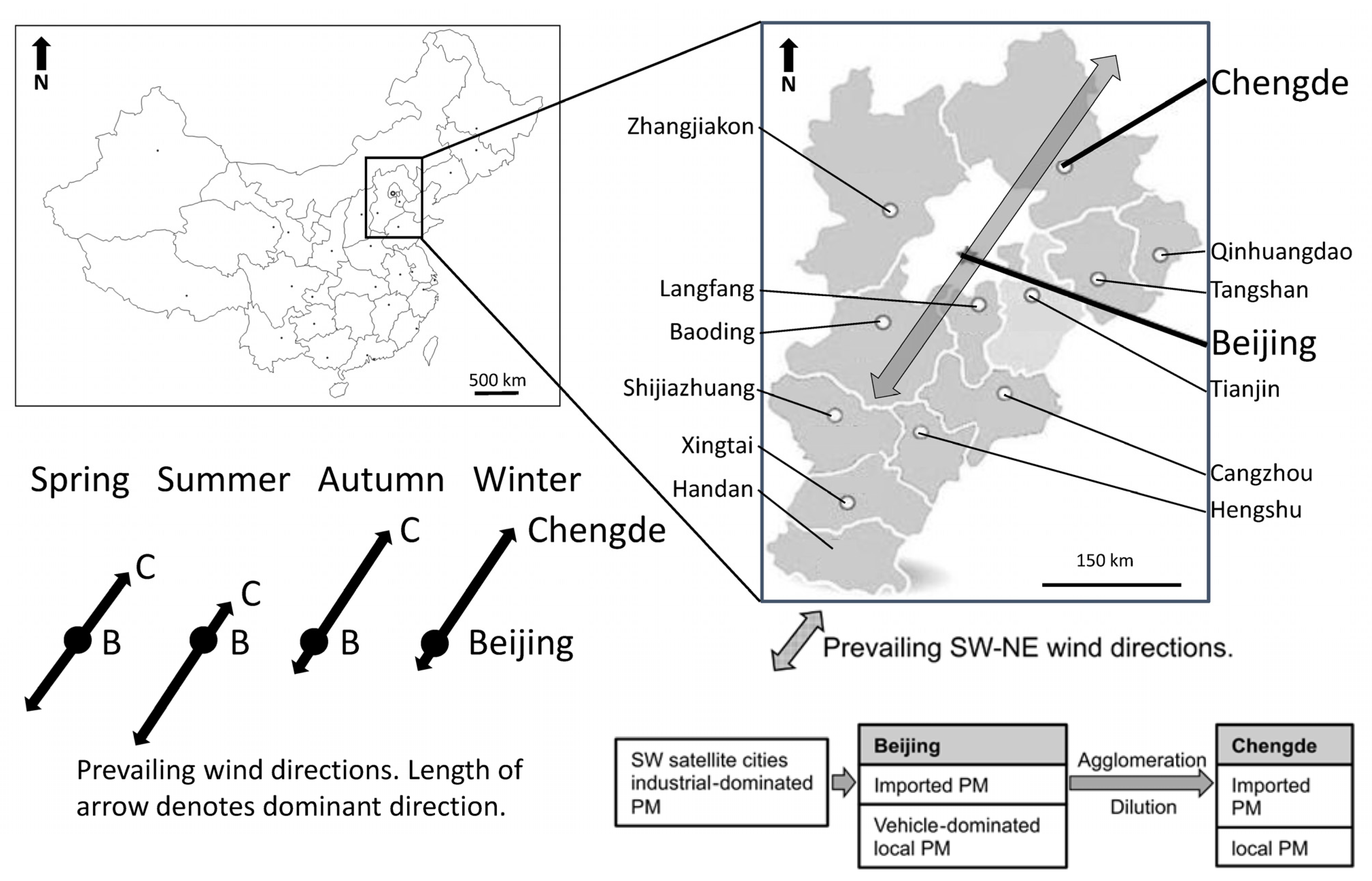
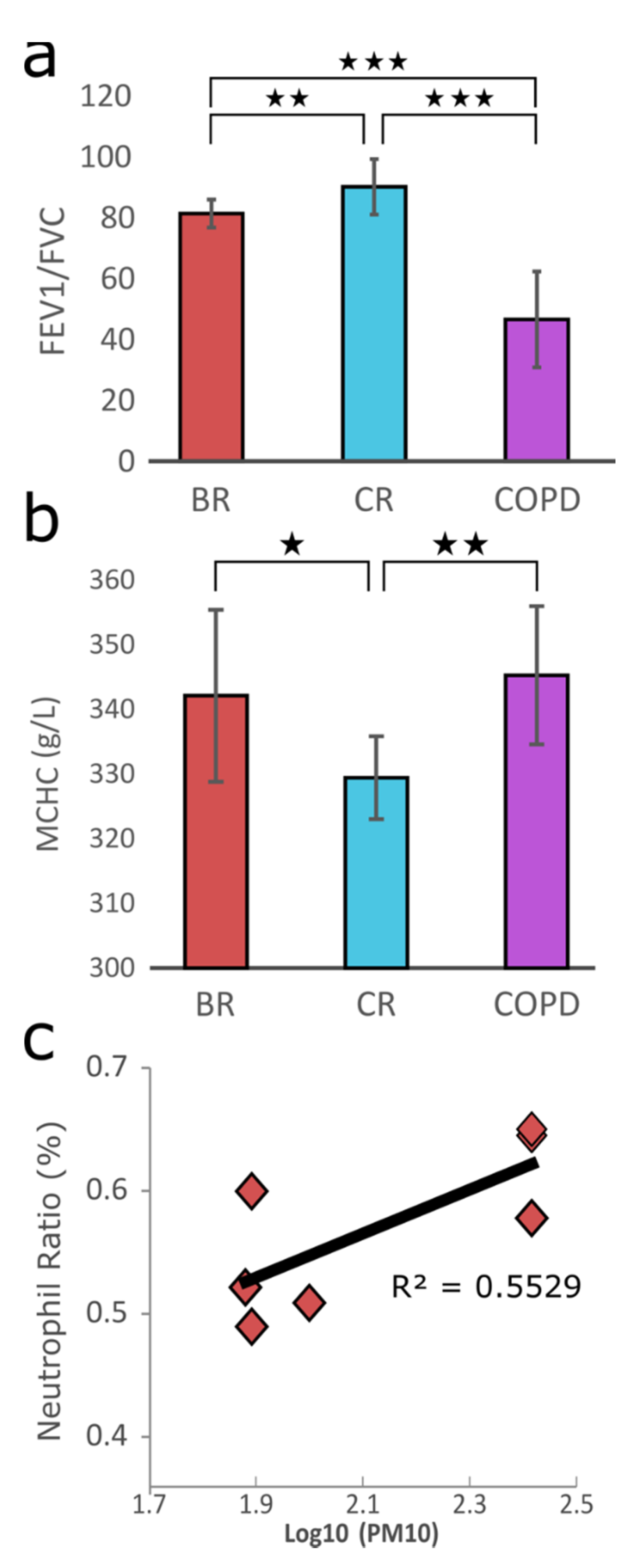
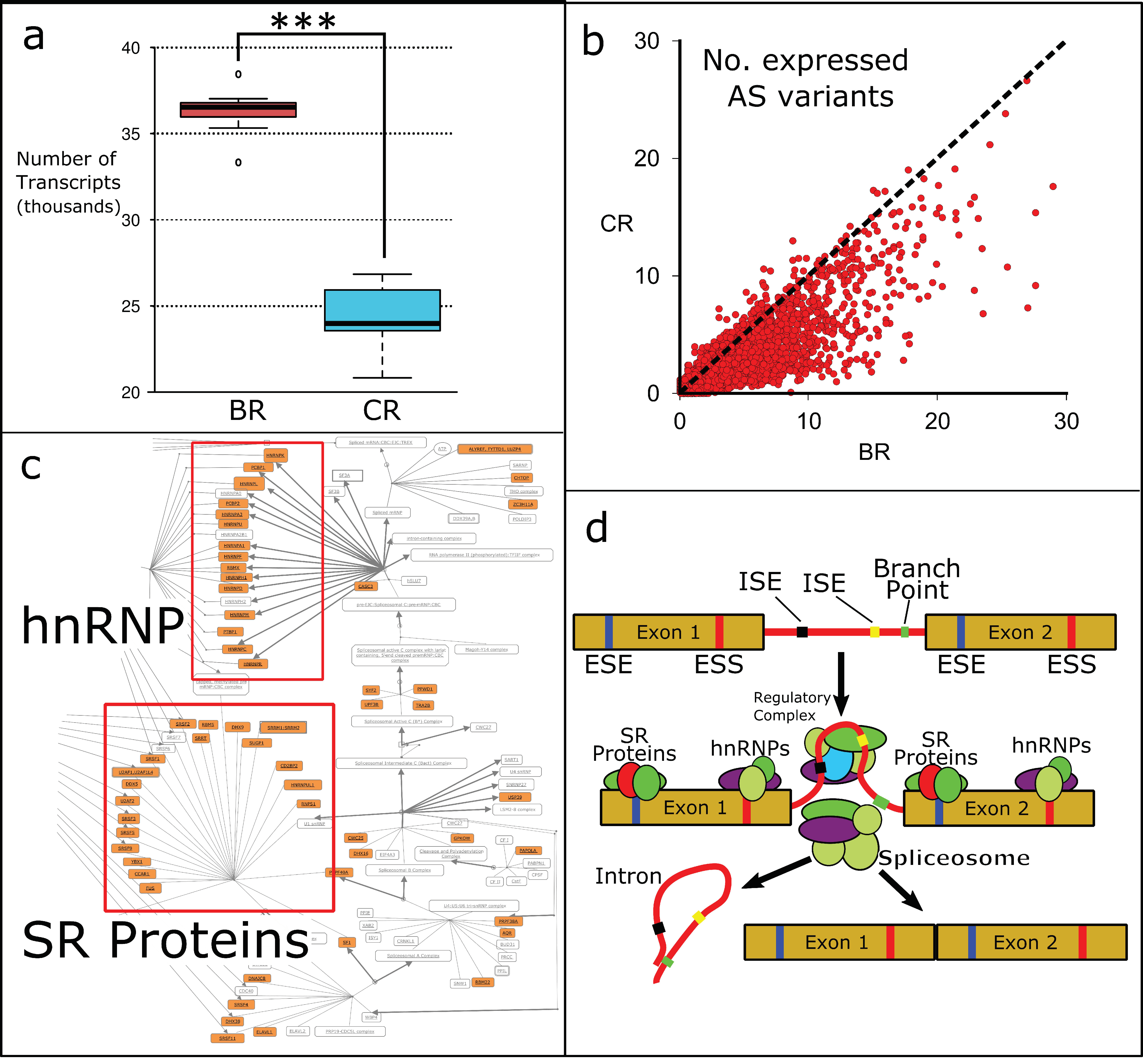
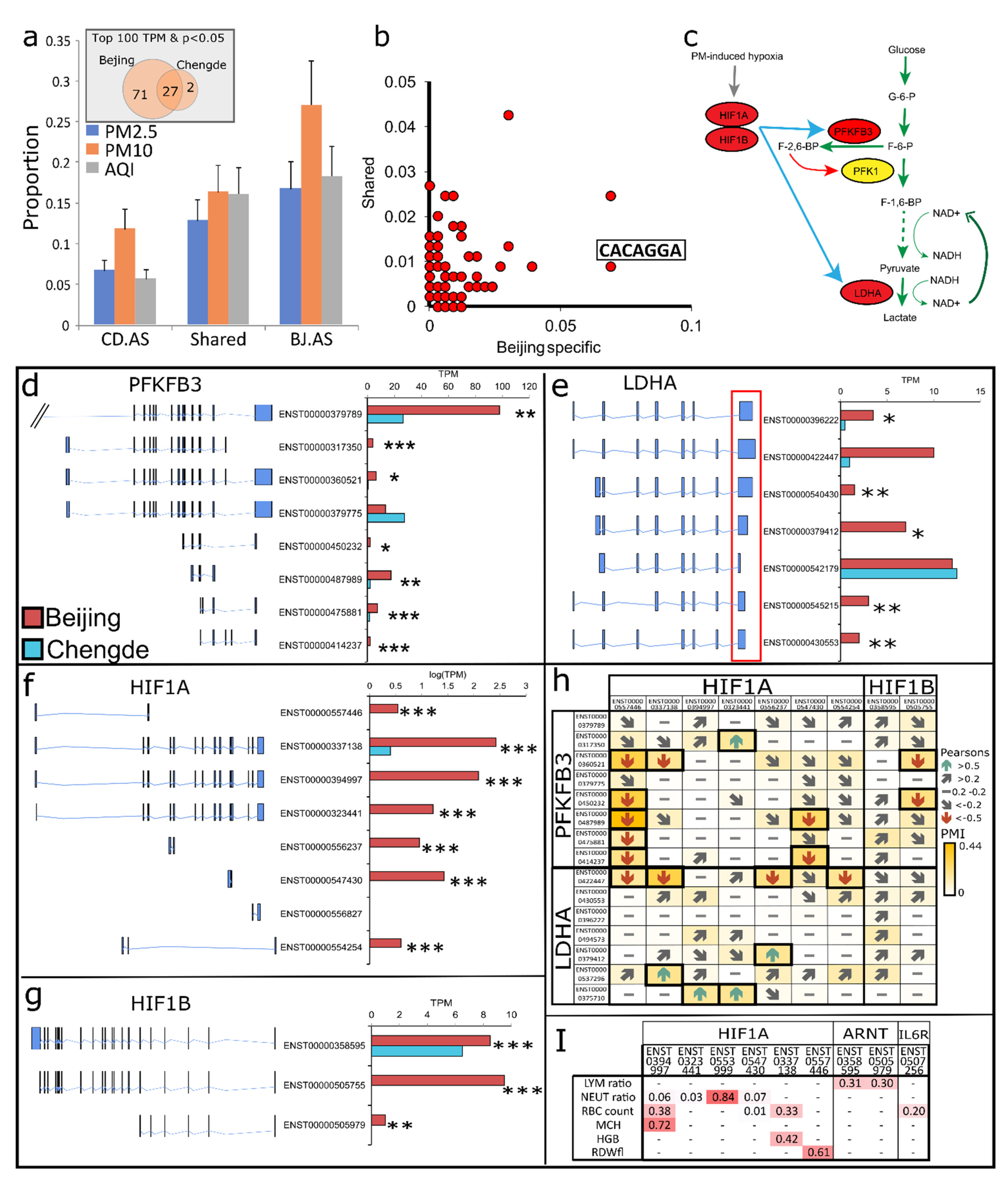
| City | Sample ID | Sex | Sample Type | Age (yr) | Height (cm) | Weight (kg) |
|---|---|---|---|---|---|---|
| Chengde | 201705CD0001 | Female | RNA + plasma | 23 | 163 | 52 |
| 201705CD0002 | Female | RNA + plasma | 29 | 169 | 52 | |
| 201705CD0003 | Female | RNA + plasma | 26 | 168 | 48 | |
| 201705CD0004 | Female | RNA + plasma | 25 | 160 | 50 | |
| 201705CD0005 | Female | RNA + plasma | 25 | 159 | 55 | |
| 201705CD0006 | Male | RNA + plasma | 26 | 170 | 53 | |
| 201705CD0007 | Male | RNA + plasma | 28 | 175 | 90 | |
| 201705CD0008 | Male | RNA + plasma | 26 | 178 | 100 | |
| 201705CD0009 | Male | RNA + plasma | 31 | 173 | 81 | |
| 201705CD0010 | Male | RNA + plasma | 30 | 183 | 80 | |
| Beijing | 201705IOZ0001 | Female | RNA + plasma | 26 | 173 | 65 |
| 201705IOZ0002 | Female | RNA + plasma | 24 | 167 | 60 | |
| 201603IOZ0014 | Male | RNA + plasma | 22 | 178 | 82 | |
| 201603IOZ0011 | Male | RNA + plasma | 29 | 167 | 63 | |
| 201603IOZ0002 | Female | RNA + plasma | 25 | 160 | 50 | |
| 201603IOZ0009 | Male | RNA + plasma | 33 | 160 | 58 | |
| 201603IOZ0012 | Male | RNA + plasma | 27 | 176 | 80 | |
| 201603IOZ0006 | Male | RNA + plasma | 38 | 169 | 67 | |
| 201703IOZ0013 | Male | RNA + plasma | 27 | 180 | 65 | |
| 201711IOZ0012 | Male | RNA + plasma | 39 | 178 | 78 |
| City | Sample ID | Sex | Sample Type | Age (yr) | Height (cm) | Weight (kg) |
|---|---|---|---|---|---|---|
| Beijing | 201612cy0001 | Male | RNA + plasma | 58 | 168 | 78 |
| 201612cy0002 | Male | RNA + plasma | 58 | 165 | 46 | |
| 201612cy0003 | Male | RNA + plasma | 70 | 160 | 57 | |
| 201612cy0004 | Male | RNA + plasma | 60 | 165 | 61 | |
| 201703cy0006 | Male | RNA + plasma | 56 | 160 | 58 | |
| 201703cy0007 | Male | RNA + plasma | 56 | 161 | 61 | |
| 201703cy0008 | Male | RNA + plasma | 82 | 163 | 61 | |
| 201703cy0009 | Male | RNA + plasma | 64 | 157 | 50 | |
| 201711cy0010 | Male | RNA + plasma | 67 | 172 | 88 | |
| 201711cy0011 | Male | RNA + plasma | 77 | 165 | 48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, S.; Feng, X.; Pass, D.; Adams, R.A.; Wang, Y.; Dong, X.; Lin, Z.; Jiang, C.; Jones, T.P.; BéruBé, K.A.; et al. Enhanced Transcriptomic Resilience following Increased Alternative Splicing and Differential Isoform Production between Air Pollution Conurbations. Atmosphere 2021, 12, 959. https://doi.org/10.3390/atmos12080959
Pan S, Feng X, Pass D, Adams RA, Wang Y, Dong X, Lin Z, Jiang C, Jones TP, BéruBé KA, et al. Enhanced Transcriptomic Resilience following Increased Alternative Splicing and Differential Isoform Production between Air Pollution Conurbations. Atmosphere. 2021; 12(8):959. https://doi.org/10.3390/atmos12080959
Chicago/Turabian StylePan, Shengkai, Xiaokai Feng, Daniel Pass, Rachel A. Adams, Yusong Wang, Xuemin Dong, Zhenzhen Lin, Chunguo Jiang, Tim P. Jones, Kelly A. BéruBé, and et al. 2021. "Enhanced Transcriptomic Resilience following Increased Alternative Splicing and Differential Isoform Production between Air Pollution Conurbations" Atmosphere 12, no. 8: 959. https://doi.org/10.3390/atmos12080959
APA StylePan, S., Feng, X., Pass, D., Adams, R. A., Wang, Y., Dong, X., Lin, Z., Jiang, C., Jones, T. P., BéruBé, K. A., & Zhan, X. (2021). Enhanced Transcriptomic Resilience following Increased Alternative Splicing and Differential Isoform Production between Air Pollution Conurbations. Atmosphere, 12(8), 959. https://doi.org/10.3390/atmos12080959








