Microbial Community Composition Analysis in Spring Aerosols at Urban and Remote Sites over the Tibetan Plateau
Abstract
1. Introduction
2. Materials and Methods
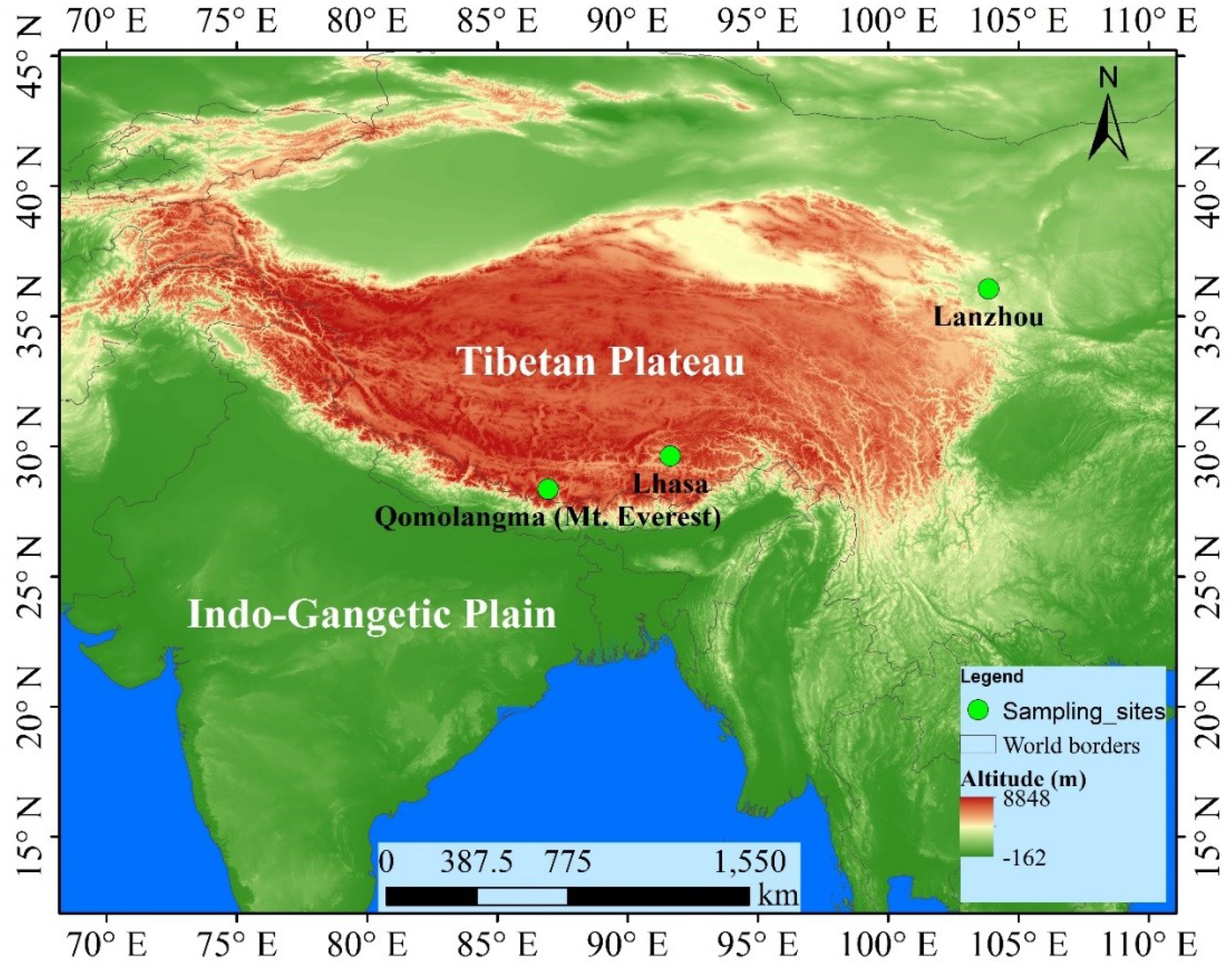
2.1. Sampling Site Description
2.2. Samples Collection
2.3. Microbiological Analysis
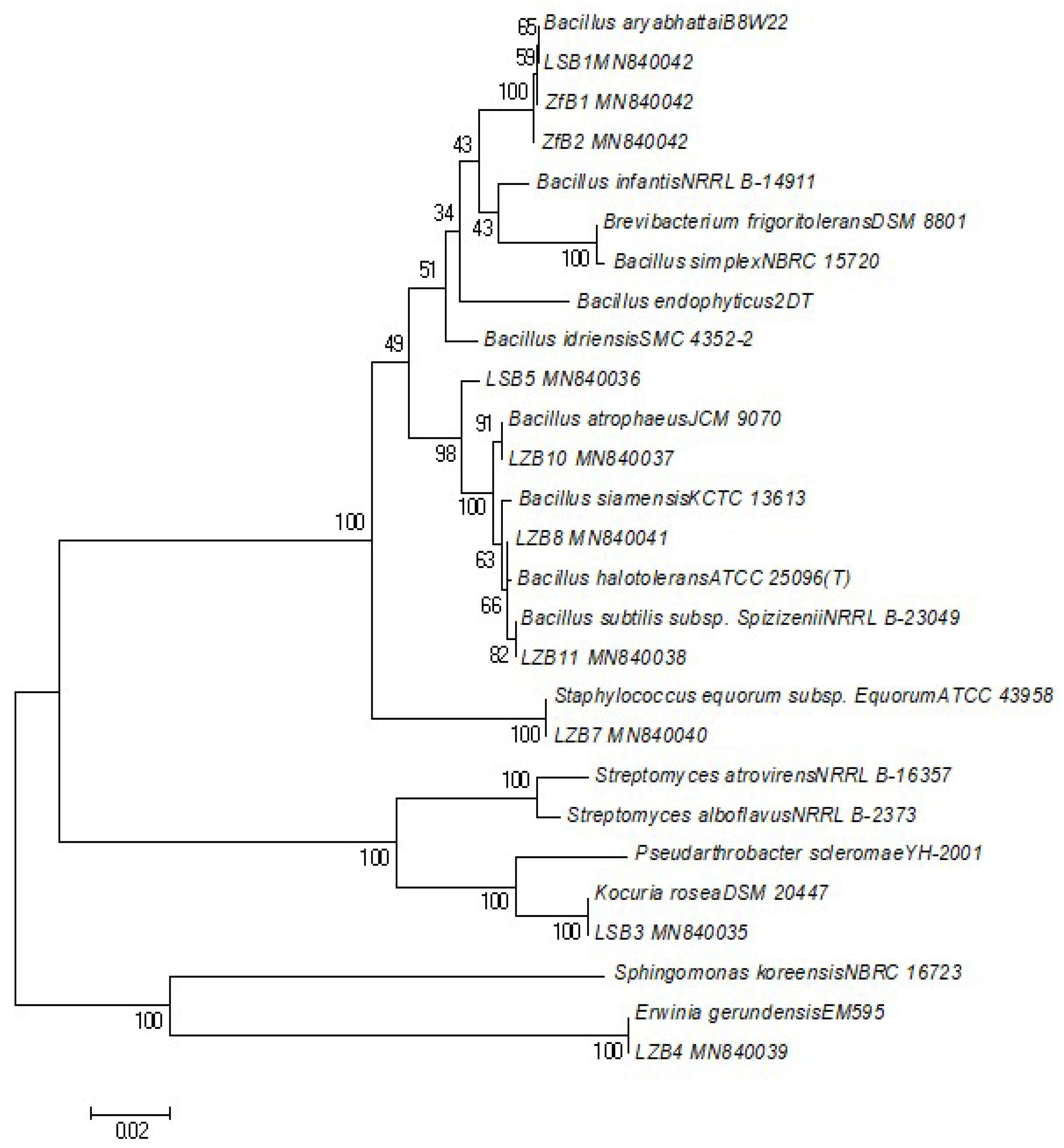
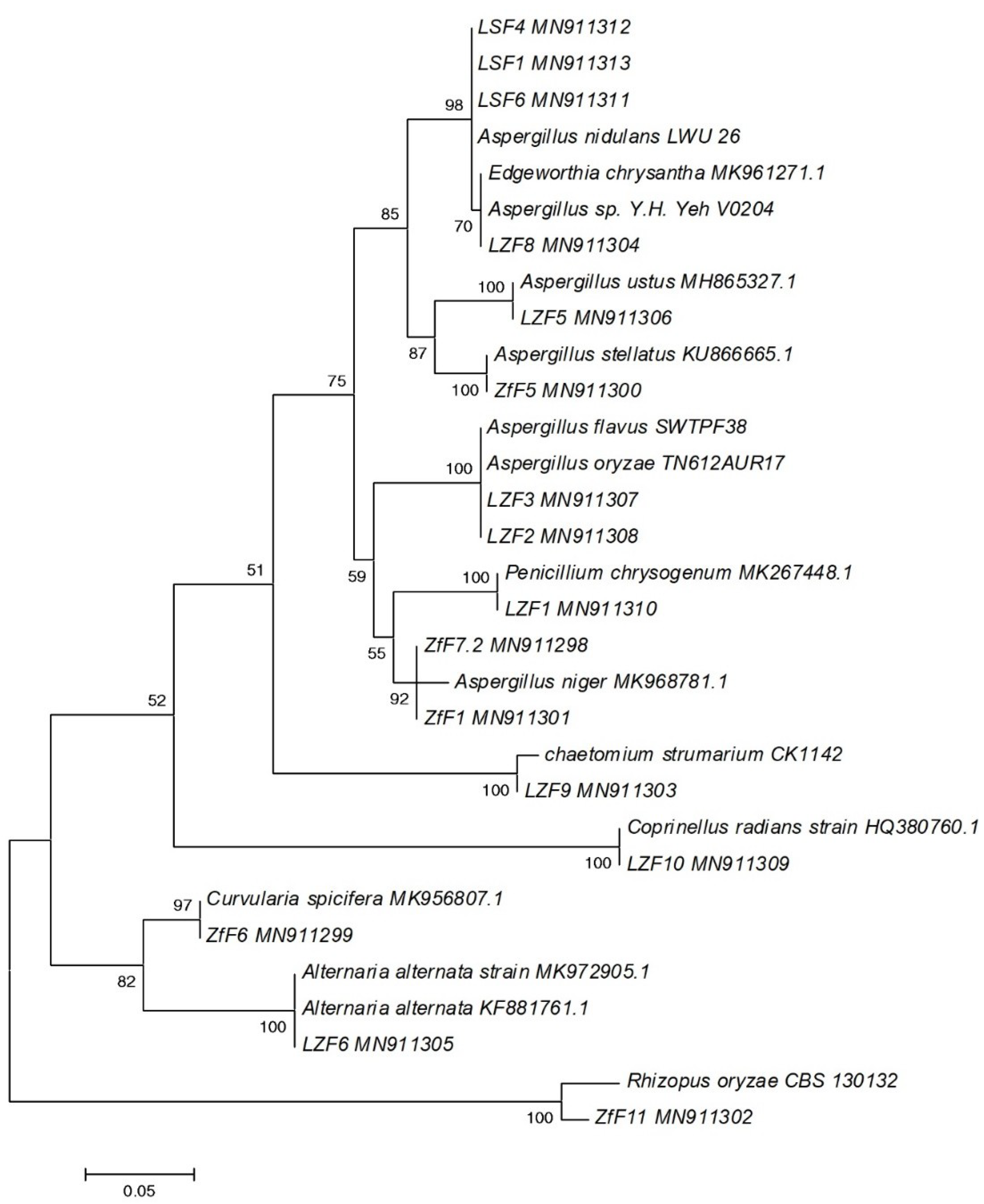
2.4. DNA Extraction and Phylogenetic Analysis
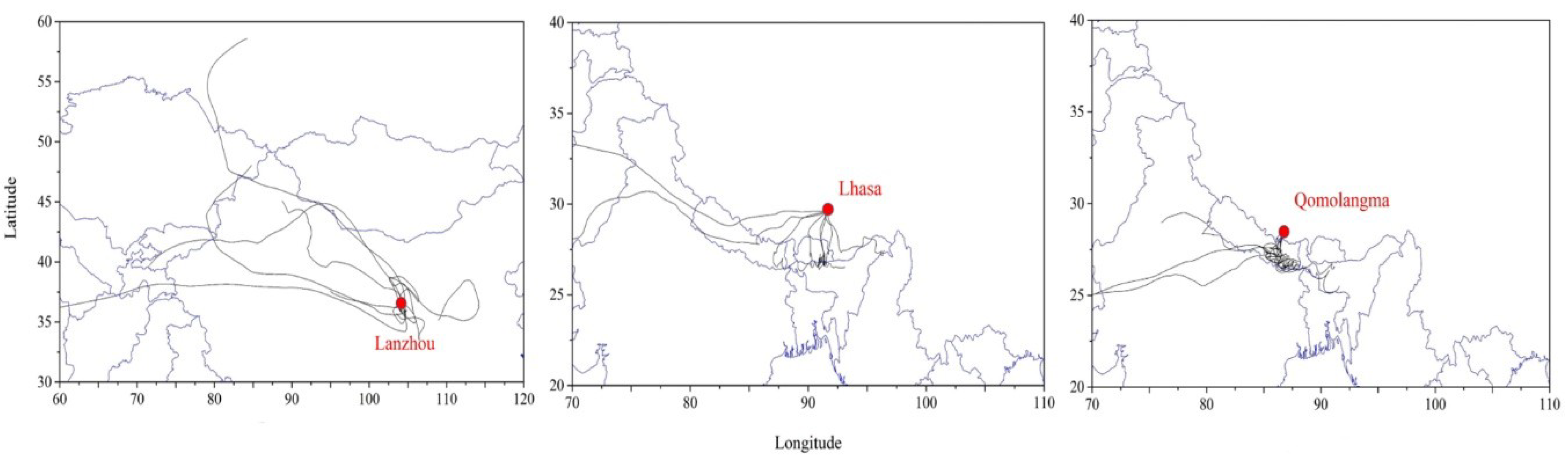
2.5. TSP Mass and Back Trajectory Analysis
3. Results
3.1. Meteorological Conditions during the Sampling Period
3.2. Airborne Microbial Community Abundance
3.3. Airborne Microbial Community Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maki, T.; Hara, K.; Iwata, A.; Lee, K.C.; Kawai, K.; Kai, K.; Iwasaka, Y. Variations in airborne bacterial communities at high altitudes over the Noto Peninsula (Japan) in response to Asian dust events. Atmos. Chem. Phys. 2017, 17, 11877–11897. [Google Scholar] [CrossRef]
- Amato, P.; Ménager, M.; Sancelme, M.; Laj, P.; Mailhot, G.; Delort, A.-M. Microbial population in cloud water at the Puy de Dôme: Implications for the chemistry of clouds. Atmos. Environ. 2005, 39, 4143–4153. [Google Scholar] [CrossRef]
- Bowers, R.M.; Lauber, C.L.; Wiedinmyer, C.; Hamady, M.; Hallar, A.G.; Fall, R.; Knight, R.; Fierer, N. Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl. Environ. Microbiol. 2009, 75, 5121–5130. [Google Scholar] [CrossRef] [PubMed]
- Sattler, B.; Puxbaum, H.; Psenner, R. Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 2001, 28, 239–242. [Google Scholar] [CrossRef]
- Hamady, M.; Walker, J.J.; Harris, J.K.; Gold, N.J.; Knight, R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 2008, 5, 235. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, M.; Zhen, Y.; Wu, L. Characterization of bioaerosol bacterial communities during hazy and foggy weather in Qingdao, China. J. Ocean Univ. China 2018, 17, 516–526. [Google Scholar] [CrossRef]
- Romano, S.; Di Salvo, M.; Rispoli, G.; Alifano, P.; Perrone, M.R.; Talà, A. Airborne bacteria in the Central Mediterranean: Structure and role of meteorology and air mass transport. Sci. Total Environ. 2019, 697, 134020. [Google Scholar] [CrossRef]
- Triadó-Margarit, X.; Caliz, J.; Reche, I.; Casamayor, E.O. High similarity in bacterial bioaerosol compositions between the free troposphere and atmospheric depositions collected at high-elevation mountains. Atmos. Environ. 2019, 203, 79–86. [Google Scholar] [CrossRef]
- Tang, K.; Huang, Z.; Huang, J.; Maki, T.; Zhang, S.; Shimizu, A.; Ma, X.; Shi, J.; Bi, J.; Wang, G.; et al. Characterization of atmospheric bioaerosols along the transport pathway of Asian dust during the Dust-Bioaerosol 2016 Campaign. Atmos. Chem. Phys. 2018, 18, 7131. [Google Scholar] [CrossRef]
- D’amato, G. Environmental urban factors (air pollution and allergens) and the rising trends in allergic respiratory diseases. Allergy 2002, 57, 30–33. [Google Scholar] [CrossRef]
- Menetrez, M.; Foarde, K.; Esch, R.; Dean, T.; Betancourt, D.; Moore, S.; Yeatts, K.; Svendsen, E.R. The measurement of ambient bioaerosol exposure. Aerosol Sci. Technol. 2007, 41, 884–893. [Google Scholar] [CrossRef]
- Kumar, A.; Attri, A.K. Characterization of fungal spores in ambient particulate matter: A study from the Himalayan region. Atmos. Environ. 2016, 142, 182–193. [Google Scholar] [CrossRef]
- Brown, J.K.; Hovmøller, M.S. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Elster, J.; Delmas, R.; Petit, J.-R.; Reháková, K. Composition of microbial communities in aerosol, snow and ice samples from remote glaciated areas (Antarctica, Alps, Andes). Biogeosci. Discuss. 2007, 4, 1779–1813. [Google Scholar] [CrossRef]
- Fierer, N.; Liu, Z.; Rodríguez-Hernández, M.; Knight, R.; Henn, M.; Hernandez, M.T. Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol. 2008, 74, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Lighthart, B.; Shaffer, B.T. Viable bacterial aerosol particle size distributions in the midsummer atmosphere at an isolated location in the high desert chaparral. Aerobiologia 1995, 11, 19–25. [Google Scholar] [CrossRef]
- Agarwal, S.; Mandal, P.; Majumdar, D.; Aggarwal, S.G.; Srivastava, A. Characterization of bioaerosols and their relation with OC, EC and carbonyl VOCs at a busy roadside restaurants-cluster in New Delhi. Aerosol Air Qual. Res. 2016, 16, 3198–3211. [Google Scholar] [CrossRef]
- Du, P.; Du, R.; Ren, W.; Lu, Z.; Zhang, Y.; Fu, P. Variations of bacteria and fungi in PM2. 5 in Beijing, China. Atmos. Environ. 2018, 172, 55–64. [Google Scholar] [CrossRef]
- Gandolfi, I.; Bertolini, V.; Ambrosini, R.; Bestetti, G.; Franzetti, A. Unravelling the bacterial diversity in the atmosphere. Appl. Microbiol. Biotechnol. 2013, 97, 4727–4736. [Google Scholar] [CrossRef]
- Tanaka, D.; Sato, K.; Goto, M.; Fujiyoshi, S.; Maruyama, F.; Takato, S.; Shimada, T.; Satakotu, A.; Aoli, K.; Nakamura, S. Airborne microbial communities at high-altitude and suburban sites in Toyama, Japan suggest a new perspective for bioprospecting. Front. Bioeng. Biotechnol. 2019, 7, 12. [Google Scholar] [CrossRef]
- Jaenicke, R. Abundance of cellular material and proteins in the atmosphere. Science 2005, 308, 73. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; McCubbin, I.B.; Hallar, A.G.; Fierer, N. Seasonal variability in airborne bacterial communities at a high-elevation site. Atmos. Environ. 2012, 50, 41–49. [Google Scholar] [CrossRef]
- Awad, A.H.A. Vegetation: A source of air fungal bio-contaminant. Aerobiologia 2005, 21, 53–61. [Google Scholar] [CrossRef]
- Lin, W.-R.; Wang, P.-H.; Tien, C.-J.; Chen, W.-Y.; Yu, Y.-A.; Hsu, L.-Y. Changes in airborne fungal flora along an urban to rural gradient. J. Aerosol Sci. 2018, 116, 116–123. [Google Scholar] [CrossRef]
- Brągoszewska, E.; Pastuszka, J.S. Influence of meteorological factors on the level and characteristics of culturable bacteria in the air in Gliwice, Upper Silesia (Poland). Aerobiologia 2018, 34, 241–255. [Google Scholar] [CrossRef]
- Brągoszewska, E.; Mainka, A.; Pastuszka, J.S. Concentration and size distribution of culturable bacteria in ambient air during spring and winter in Gliwice: A typical urban area. Atmosphere 2017, 8, 239. [Google Scholar] [CrossRef]
- Genitsaris, S.; Stefanidou, N.; Katsiapi, M.; Kormas, K.A.; Sommer, U.; Moustaka-Gouni, M. Variability of airborne bacteria in an urban Mediterranean area (Thessaloniki, Greece). Atmos. Environ. 2017, 157, 101–110. [Google Scholar] [CrossRef]
- Hubad, B.; Lapanje, A. The efficient method for simultaneous monitoring of the culturable as well as nonculturable airborne microorganisms. PLoS ONE 2013, 8, e82186. [Google Scholar] [CrossRef]
- Yoo, K.; Lee, T.K.; Choi, E.J.; Yang, J.; Shukla, S.K.; Hwang, S.-I.; Park, J. Molecular approaches for the detection and monitoring of microbial communities in bioaerosols: A review. J. Environ. Sci. 2017, 51, 234–247. [Google Scholar] [CrossRef]
- Fang, Z.; Ouyang, Z.; Zheng, H.; Wang, X. Concentration and size distribution of culturable airborne microorganisms in outdoor environments in Beijing, China. Aerosol Sci. Technol. 2008, 42, 325–334. [Google Scholar] [CrossRef]
- Tsai, F.; Macher, J.; Hung, Y. Concentrations of airborne bacteria in 100 US office buildings. Proc. Indoor Air 2002, 15, 353–359. [Google Scholar]
- Kang, S.; Zhang, Q.; Qian, Y.; Ji, Z.; Li, C.; Cong, Z.; You, Q.; Panday, A.K.; Rupakheti, M.; Chen, D.; et al. Linking atmospheric pollution to cryospheric change in the Third Pole region: Current progress and future prospects. Natl. Sci. Rev. 2019, 6, 796–809. [Google Scholar] [CrossRef]
- Cong, Z.; Kang, S.; Kawamura, K.; Liu, B.; Wan, X.; Wang, Z.; Gao, S.; Fu, P. Carbonaceous aerosols on the south edge of the Tibetan Plateau: Concentrations, seasonality and sources. Atmos. Chem. Phys. 2015, 15, 1573–1584. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; Wu, R.; Hu, Z.; Yang, K.; Li, M.; Ma, W.; Zhong, L.; Sun, F.; Chen, X.; et al. Recent advances on the study of atmosphere-land interaction observations on the Tibetan Plateau. Hydrol. Earth Syst. Sci. 2009, 13, 1103–1111. [Google Scholar] [CrossRef]
- Wan, X.; Kang, S.; Xin, J.; Liu, B.; Wen, T.; Wang, P.; Wang, Y.; Cong, Z. Chemical composition of size-segregated aerosols in Lhasa city, Tibetan Plateau. Atmos. Res. 2016, 174, 142–150. [Google Scholar] [CrossRef]
- Cong, Z.; Kang, S.; Luo, C.; Li, Q.; Huang, J.; Gao, S.; Li, X. Trace elements and lead isotopic composition of PM10 in Lhasa, Tibet. Atmos. Environ. 2011, 45, 6210–6215. [Google Scholar] [CrossRef]
- Huang, J.; Kang, S.; Wang, S.; Wang, L.; Zhang, Q.; Guo, J.; Wang, K.; Zhang, G.; Tripathee, L. Wet deposition of mercury at Lhasa, the capital city of Tibet. Sci. Total Environ. 2013, 447, 123–132. [Google Scholar] [CrossRef]
- Guo, J.; Kang, S.; Huang, J.; Zhang, Q.; Tripathee, L.; Sillanpää, M. Seasonal variations of trace elements in precipitation at the largest city in Tibet, Lhasa. Atmos. Res. 2015, 153, 87–97. [Google Scholar] [CrossRef]
- Zhang, Y.; Kang, S. Characteristics of carbonaceous aerosols analyzed using a multiwavelength thermal/optical carbon analyzer: A case study in Lanzhou City. Sci. China Earth Sci. 2019, 62, 389–402. [Google Scholar] [CrossRef]
- Yu, X.; Zhu, B.; Fan, S.; Yin, Y.; Bu, X. Ground-based observation of aerosol optical properties in Lanzhou, China. J. Environ. Sci. 2009, 21, 1519–1524. [Google Scholar] [CrossRef]
- Qiang, Z. The influence of terrain and inversion layer on pollutant transfer over Lanzhou City. China Environ. Sci. 2001, 21, 230–234. [Google Scholar]
- Bielawska-Drózd, A.; Cieślik, P.; Bohacz, J.; Korniłłowicz-Kowalska, T.; Żakowska, D.; Bartoszcze, M.; Wlizło-Skowronek, B.; Winnicka, I.; Brytan, M.; Kubiak, L. Microbiological analysis of bioaerosols collected from Hospital Emergency Departments and ambulances. Ann. Agric. Environ. Med. 2018, 25, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Dacarro, C.; Picco, A.; Grisoli, P.; Rodolfi, M. Determination of aerial microbiological contamination in scholastic sports environments. J. Appl. Microbiol. 2003, 95, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Srivastava, R. Identification of indoor airborne microorganisms in residential rural houses of Uttarakhand, India. Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 146–152. [Google Scholar]
- Srivastava, A.; Singh, M.; Jain, V. Identification and characterization of size-segregated bioaerosols at Jawaharlal Nehru University, New Delhi. Nat. Hazards 2012, 60, 485–499. [Google Scholar] [CrossRef]
- Madsen, A.M.; Zervas, A.; Tendal, K.; Nielsen, J.L. Microbial diversity in bioaerosol samples causing ODTS compared to reference bioaerosol samples as measured using Illumina sequencing and MALDI-TOF. Environ. Res. 2015, 140, 255–267. [Google Scholar] [CrossRef]
- Piterina, A.V.; Bartlett, J.; Pembroke, J.T. Molecular analysis of bacterial community DNA in sludge undergoing autothermal thermophilic aerobic digestion (ATAD): Pitfalls and improved methodology to enhance diversity recovery. Diversity 2010, 2, 505–526. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Lan, P.T.N.; Hayashi, H.; Sakamoto, M.; Benno, Y. Phylogenetic analysis of cecal microbiota in chicken by the use of 16S rDNA clone libraries. Microbiol. Immunol. 2002, 46, 371–382. [Google Scholar] [CrossRef]
- Tripathee, L.; Kang, S.; Rupakheti, D.; Cong, Z.; Zhang, Q.; Huang, J. Chemical characteristics of soluble aerosols over the central Himalayas: Insights into spatiotemporal variations and sources. Environ. Sci. Pollut. Res. Int. 2017, 24, 24454–24472. [Google Scholar] [CrossRef]
- Draxler, R.; Stunder, B.; Rolph, G.; Stein, A.; Taylor, A. HYSPLIT4 User’s Guide Version 4-Last Revision: February 2016; HYSPLIT Air Resources Laboratory: College Park, MD, USA, 2016. [Google Scholar]
- Gao, M.; Yan, X.; Qiu, T.; Han, M.; Wang, X. Variation of correlations between factors and culturable airborne bacteria and fungi. Atmos. Environ. 2016, 128, 10–19. [Google Scholar] [CrossRef]
- González-Rocha, G.; Muñoz-Cartes, G.; Canales-Aguirre, C.B.; Lima, C.A.; Domínguez-Yévenes, M.; Bello-Toledo, H.; Hernandez, C.E. Diversity structure of culturable bacteria isolated from the Fildes Peninsula (King George Island, Antarctica): A phylogenetic analysis perspective. PLoS ONE 2017, 12, e0179390. [Google Scholar] [CrossRef]
- Li, Y.; Lu, R.; Li, W.; Xie, Z.; Song, Y. Concentrations and size distributions of viable bioaerosols under various weather conditions in a typical semi-arid city of Northwest China. J. Aerosol Sci. 2017, 106, 83–92. [Google Scholar] [CrossRef]
- Liu, H.-M.; Lin, Y.-H.; Tsai, M.-Y.; Lin, W.-H. Occurrence and characterization of culturable bacteria and fungi in metalworking environments. Aerobiologia 2010, 26, 339–350. [Google Scholar] [CrossRef]
- Dannemiller, K.C.; Weschler, C.J.; Peccia, J. Fungal and bacterial growth in floor dust at elevated relative humidity levels. Indoor Air 2017, 27, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Qiu, T.; Jia, R.; Han, M.; Song, Y.; Wang, X. Concentration and size distribution of viable bioaerosols during non-haze and haze days in Beijing. Environ. Sci. Pollut. Res. 2015, 22, 4359–4368. [Google Scholar] [CrossRef]
- Heo, K.J.; Kim, H.B.; Lee, B.U. Concentration of environmental fungal and bacterial bioaerosols during the monsoon season. J. Aerosol Sci. 2014, 77, 31–37. [Google Scholar] [CrossRef]
- Hurtado, L.; Rodríguez, G.; López, J.; Castillo, J.; Molina, L.; Zavala, M.; Quintana, P.J. Characterization of atmospheric bioaerosols at 9 sites in Tijuana, Mexico. Atmos. Environ. 2014, 96, 430–436. [Google Scholar] [CrossRef]
- Lee, T.; Grinshpun, S.A.; Martuzevicius, D.; Adhikari, A.; Crawford, C.M.; Reponen, T. Culturability and concentration of indoor and outdoor airborne fungi in six single-family homes. Atmos. Environ. 2006, 40, 2902–2910. [Google Scholar] [CrossRef]
- Wang, W.; Ma, Y.; Ma, X.; Wu, F.; Ma, X.; An, L.; Feng, H. Seasonal variations of airborne bacteria in the Mogao Grottoes, Dunhuang, China. Int. Biodeterior. Biodegr. 2010, 64, 309–315. [Google Scholar] [CrossRef]
- Ghimire, P.S.; Tripathee, L.; Chen, P.; Kang, S. Linking the conventional and emerging detection techniques for ambient bioaerosols: A review. Rev. Environ. Sci. Biotechnol. 2019, 18, 495–523. [Google Scholar] [CrossRef]
- Yan, D.; Zhang, T.; Su, J.; Zhao, L.L.; Wang, H.; Fang, X.M.; Zhang, Y.-Q.; Liu, H.-Y.; Yu, L.-Y. Structural variation in the bacterial community associated with airborne particulate matter in Beijing, China, during hazy and nonhazy days. Appl. Environ. Microbiol. 2018, 84, e00004–e00018. [Google Scholar] [CrossRef]
- Yan, D.; Zhang, T.; Su, J.; Zhao, L.L.; Wang, H.; Fang, X.M.; Zhang, Y.-Q.; Liu, H.-Y.; Yu, L.-Y. Diversity and composition of airborne fungal community associated with particulate matters in Beijing during haze and non-haze days. Front. Microbiol. 2016, 7, 487. [Google Scholar] [CrossRef] [PubMed]
- Mouli, P.; Mohan, S.; Reddy, S. Assessment of microbial(bacteria) Concentrations of ambient air at semi-arid urban region: Influence of meteorological factors. Appl. Ecol. Environ. Res. 2005, 3, 139–149. [Google Scholar] [CrossRef]
- Frankel, M.; Bekö, G.; Timm, M.; Gustavsen, S.; Hansen, E.W.; Madsen, A.M. Seasonal variations of indoor microbial exposures and their relation to temperature, relative humidity, and air exchange rate. Appl. Environ. Microbiol. 2012, 78, 8289–8297. [Google Scholar] [CrossRef] [PubMed]
- Garrett, M.H.; Hooper, B.M.; Cole, F.M.; Hooper, M.A. Airborne fungal spores in 80 homes in the Latrobe Valley, Australia: Levels, seasonality and indoor-outdoor relationship. Aerobiologia 1997, 13, 121–126. [Google Scholar] [CrossRef]
- Haas, D.; Habib, J.; Galler, H.; Buzina, W.; Schlacher, R.; Marth, E.; Reinthaler, F. Assessment of indoor air in Austrian apartments with and without visible mold growth. Atmos. Environ. 2007, 41, 5192–5201. [Google Scholar] [CrossRef]
- Wei, K.; Zheng, Y.; Li, J.; Shen, F.; Zou, Z.; Fan, H.; Li, X.; Wu, C.Y.; Yao, M. Microbial aerosol characteristics in highly polluted and near-pristine environments featuring different climatic conditions. Sci. Bull. 2015, 60, 1439–1447. [Google Scholar] [CrossRef]
- Vaïtilingom, M.; Deguillaume, L.; Vinatier, V.; Sancelme, M.; Amato, P.; Chaumerliac, N.; Delort, A.M. Potential impact of microbial activity on the oxidant capacity and organic carbon budget in clouds. Proc. Natl. Acad. Sci. USA 2013, 110, 559–564. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, Q.; Wang, Z.; Fu, P.; Li, J.; Yang, T.; Yin, Y. Investigation of the sources and evolution processes of severe haze pollution in Beijing in January 2013. J. Geophys. Res. Atmos. 2014, 119, 4380–4398. [Google Scholar] [CrossRef]
- Tong, Y.; Lighthart, B. The annual bacterial particle concentration and size distribution in the ambient atmosphere in a rural area of the Willamette Valley, Oregon. Aerosol Sci. Technol. 2000, 32, 393–403. [Google Scholar] [CrossRef]
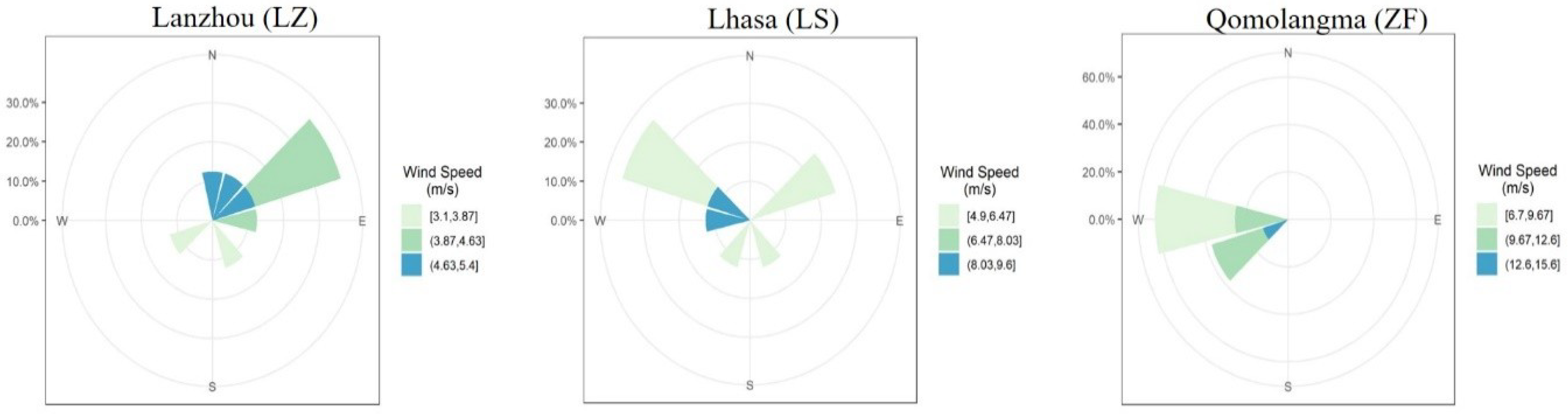

| Temperature (°C) | Pressure (hPa) | RHU (%) | Wind Speed (m/s) | Wind Direction | TSP (µg m−3) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | LZ | LS | ZF | LZ | LS | ZF | LZ | LS | ZF | LZ | LS | ZF | LZ | LS | ZF | LZ | LS | ZF |
| 5/1/2019 | 21.1 | 22.7 | 15.8 | 831.1 | 661.9 | 605.5 | 32.9 | 89.9 | 49.9 | 3.5 | 5.6 | 10.9 | SW | WNW | WSW | 58.01 | 129.26 | 90.35 |
| 5/4/2019 | 25.1 | 19.4 | 18.6 | 832.3 | 660.9 | 603.8 | 22.9 | 32.9 | 18.9 | 4.4 | 4.9 | 6.7 | E | ENE | W | 72.05 | 96.49 | 40.39 |
| 5/7/2019 | 21 | 25.5 | 15.6 | 834.8 | 658 | 603.5 | 31.9 | 99.9 | 99.9 | 4.7 | 5.9 | 10.3 | NE | SSW | W | 1530.54 | 170.77 | 61.49 |
| 5/10/2019 | 18.1 | 24.2 | 18.4 | 833.4 | 658.2 | 605 | 36.9 | 59.9 | 69.9 | 4.5 | 9.2 | 11.8 | NE | W | W | 1521.63 | 153.86 | 72.15 |
| 5/13/2019 | 23.3 | 19 | 12.7 | 834.5 | 658.9 | 604 | 19.9 | 18.9 | 15.9 | 4.5 | 9.6 | 8.9 | ENE | WNW | W | 673.59 | 199.31 | 30.36 |
| 5/16/2019 | 25.7 | 20 | 16.7 | 830.7 | 660.9 | 604.9 | 23.9 | 21.9 | 12.9 | 5.4 | 5.1 | 10.3 | NNE | SE | WSW | 60.99 | 68.72 | 32.43 |
| 5/19/2019 | 18.7 | 22.2 | 16.2 | 839.7 | 659.3 | 604.8 | 21.9 | 16.9 | 99.9 | 5.4 | 5.3 | 8.5 | N | WNW | W | 225.84 | 104.36 | 33.93 |
| 5/22/2019 | 28.1 | 18.6 | 16.8 | 833.8 | 663.1 | 605.7 | 69.9 | 25.9 | 89.9 | 3.1 | 5.1 | 15.6 | SE | NE | WSW | 40.84 | 116.68 | 130.05 |
| Location | Latitude (°N) and Longitude (°E) | Elevation (AMSL) | Site | Method Used for Study | Microbial Community | References |
|---|---|---|---|---|---|---|
| Toyama Prefecture, Japan | 36°41′54′′ N, 137°11′13′′ E | 23 m | Suburban | Polycarbonate filters, Illumina sequencing | Alpha-Beta-Gammaproteobacteria, Acidimicrobia, Planctomycetia, Bacillus, Solibacteres, Flavobacteria | [20] |
| Huairou, Beijing, China | 40°24′29″ N, 116°40′ 28″ E | 40–60 m | Peri-urban | Quartz filters, Illumina sequencing | Streotophyta, Bacillus, Clostridium, Kocuria, Staphylococcus, MethylobacteriumSarcinomyces, Trichothecium, Acromonium, Chaetomium, Aspergillus, Penicillium | [18] |
| New Delhi city, India | 28°12’ N–28°53’ N, 77°50’ E–77°23’ E | 218 m | Urban | Quartz filters, automated DNA sequencing | Bacillus, AcenitobactorAspergillus, Cladosporium, Alternaria, Fusarium, Penicillium, Trichoderma, Mucor | [17] |
| Jawali, India | 31°2’ N–32°5’ N 75°0’ E–77°45’ E | 600 m | Rural | Quartz filters, Light microscopy | Basidiospora, Ascospora, Fusarium, Ganoderma, Alternaria, Curvularia | [12] |
| Erenhot | 43.668, 111.953 | 957 m | Urban | Polycarbonate filters, Illumina sequencing | Chloroacidobacter, Saprospirae, Actinobacteria, Alphaproteobacteria, BacilliAgaricomycetes, Dothideomycetes, Sordariomycetes, Eurotiomycetes | [9] |
| Qingdao, China | 36°16′ N, 120° 50′ E | 1133 m | Urban | Six stage cascade impactor, DGGE band sequencing | Alphaproteobacteria, Betaproteobacteria, Bacillus | [6] |
| Lanzhou | 36°3′1″ N, 103°51′33″ E | 1520 m | Urban | Quartz filters, Illumina sequencing | Proteobacteria, BacillusEurotiomycetes, Malvales, Dothideomycete, Sordariales, Agaricales | This study |
| Salento’s peninsula, Italy | 40.3° N; 18.1° E | 1895 m | Suburban | PTFE filters, Illumina sequencing | Proteobacteria, Cyanobacteria, Actinobacteria, Pseudomonas, Enterobacter, Vibrio, Streptomyces | [7] |
| Mt. Jodo, Japan | 36°34′00′′ N, 137°36′21′′ E | 2839 m | High altitude, pristine | Polycarbonate filters, Illumina sequencing | Alpha/Beta, Gammaproteobacteria, Acidimicrobia, Planctomycetia, Bacillus, Solibacteres, Flavobacteria | [20] |
| Sierra Nevada, Spain | 37°03’ N, 3°23’ W | 2896 m | High altitude, Rocky and meadows | Passive automatic sampler | Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria | [8] |
| Noto Peninsula, Japan | Uchinada (36°67 N, 136°64 E) to Hakui (36°92 N, 136°76 E) | 500–3000 m | Coastal | Polycarbonate filters, Illumina sequencing | Cyanobacteria, Actinobacteria, Bacillus, Alpha, Beta, Gammaproteobacteria | [1] |
| Mt. Werner, Colorado, USA | 40.45° N, 106.73° W | 3200 m | High altitude, pristine | Cellulose nitrate filters, Sanger sequencing | Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Bacillus, Eurotiomycetes, Sordariomycetes, Dothideomycete | [3] |
| Lhasa | 29°38′ N, 91°38′ E | 3640 m | High altitude, Peri-urban | Quartz filters, Illumina sequencing | Bacillus, Kocuria, Zygomycetes, Eurotiomycetes, Malvales | This study |
| Qomolangma | 28.36°N, 86.95°E | 4276 m | High altitude, Pristine | Quartz filters, Illumina sequencing | Bacillus, Eurotiomycetes, Malvales, Dothideomycete, Agaricales, Zygomycetes | This study |
| Sample Name | Strain | Matched Accession Number | Similarity% | Length (bp) | Pathogenesis | Gram Stain | Colony Characteristics | GC Content% |
|---|---|---|---|---|---|---|---|---|
| LZB4 | Erwinia gerundensis | KJ004603.1 | 99.39 | 1331 | Plant pathogen | Gram-negative, rod shaped | Yellowish, circular | 56.12 |
| LZB7 | Staphylococcus equorum | MN229550.1 | 100 | 1393 | Produce cheese and meat order, May inhibit Listeria’s growth | Gram-positive cocci | Opaque, white entire margin | 50.54 |
| LZB8 | Bacillus halotolerans | MK517597.1 | 99.93 | 1390 | Unknown | Gram-positive bacteria, rod shaped | Opaque, smooth, creamy colored | 54.82 |
| LZB10 | Bacillus atrophaeus | NR_024689.1 | 99.86 | 1410 | Unknown | Gram-positive bacteria, rod shaped | Opaque, smooth, creamy colored | 55.04 |
| LZB11 | Bacillus subtilis | NR_116187.1 | 99.86 | 1408 | Unknown | Gram-positive bacteria, rod shaped | Dull surface, thick/opaque, creamy colored, wrinkled (sometimes) | 54.9 |
| LSB1 | Bacillus aryabhattai | NR_115953.1 | 99.79 | 1415 | Unknown | Gram-positive bacteria, rod shaped | Opaque, smooth, creamy colored | 53.57 |
| LSB3 | Kocuria rosea | NR_044871.1 | 99.85 | 1370 | Infections in immunocompromised patients | Gram-positive cocci | Pinkish, smooth, shiny, circular | 57.23 |
| LSB5 | Bacillus altitudinis | NR_042337.1 | 100 | 1382 | Plant soft-rot causing pathogen | Gram-positive bacteria, rod shaped | White, regular margin | 54.99 |
| ZFB1 | Bacillus aryabhattai | MK860027.1 | 100 | 1398 | Unknown | Gram-positive bacteria, rod shaped | Opaque, smooth, creamy colored | 53.58 |
| ZFB2 | Bacillus aryabhattai | NR_115953.1 | 99.79 | 1414 | Unknown | Gram-positive bacteria, rod shaped | Opaque, smooth, creamy colored | 53.54 |
| Sample Name | Strain | Matched Accession Number | Similarity% | Length (bp) | Pathogenesis | Colony Characteristics | GC Content (%) |
|---|---|---|---|---|---|---|---|
| LZF1 | Penicillium chrysogenum | MK267448.1 | 100 | 563 | Rare human pathogen, human allergen, source of antibiotics | Blue to blue–green conidia and the mold exudes a yellow pigment | 57.02 |
| LZF2 | Aspergillus flavus | MG575511.1 | 100 | 564 | Plant pathogen, opportunistic human and animal pathogen, causing aspergillosis in immunocompromised individuals | Powdery masses of yellow-green spores on the upper surface and reddish-gold on the lower surface | 58.33 |
| LZF3 | Aspergillus flavus | MG991646.1 | 100 | 570 | Plant pathogen, opportunistic human and animal pathogen, causing aspergillosis in immunocompromised individuals | Powdery masses of yellow-green spores on the upper surface and reddish-gold on the lower surface | 58.07 |
| LZF4 | Edgeworthia chrysantha | MK961271.1 | 100 | 543 | Not known. Possess anti-inflammatory and analgesic activity | Possesses pain brush type flowering | 58.93 |
| LZF5 | Aspergillus ustus | MH865327.1 | 100 | 551 | Human pathogen causing onychomycosis and otitis media, rarely found to cause endocarditis, pneumonia, disseminated disease, opportunistic pathogen in immunocompromised | Dull brown with a purplish to gray brown or dark brown colonies | 58.08 |
| LZF6 | Alternaria alternata | MH865327.1 | 100 | 549 | Opportunistic pathogen causing leaf spots, rots and blights on many plant parts | Black to olivaceous-black or greyish and are suede-like to floccose | 46.27 |
| LZF8 | Aspergillus sp. | MH141246.1 | 99.82 | 550 | Most commonly human, animal and plant pathogen, cause disease on many grain crops and some variants synthesize mycotoxins and aflatoxins | Powdery masses of yellow-green spores on the upper surface and yellowish on the lower surface | 59.09 |
| LZF9 | Chaetomium sp. | KJ935022.1 | 99.82 | 544 | Human allergens and opportunistic agents of ungual mycosis and neurological infections. Source of cellulose degrading enzymes | Cottony and white in color initially. Mature colonies become gray to olive in color | 57.17 |
| LZF10 | Coprinellus radians | HQ380760.1 | 100 | 664 | Unknown | Scattered yellowish-orange mat | 49.4 |
| LSF2 | Rhizopus oryzae | MH865594.1 | 100 | 582 | Opportunistic pathogen of humans causing mucormycosis. It is also used economically in the production of the enzymes, glucoamylase and lipase | Colonies are white initially, becoming brownish with age | 40.55 |
| LSF3 | Rhizopus oryzae | MH865576.1 | 100 | 602 | Opportunistic pathogen of humans causing mucormycosis. It is also used economically in the production of the enzymes, glucoamylase and lipase | Colonies are white initially, becoming brownish with age | 40.86 |
| LSF6 | Emericella dentata | MH032749.1 | 100 | 527 | Unknown | Colonies are white and fluffy initially | 59.58 |
| LSF1 | Emericella rugulosa | EU289912.1 | 99.82 | 541 | Unknown | White to blackish sparse colony | 59.7 |
| LSF4 | Edgeworthia chrysantha | MK806488.1 | 100 | 539 | Not known. Possess anti-inflammatory and analgesic activity | Possesses pain brush type flowering | 59.55 |
| LSF8 | Aspergillus niger | MK256745.1 | 100 | 573 | Black mold of onions and ornamental plants, peanuts and grapes. Serious lung disease, aspergillosis in human. Produce important enzymes. | Granular to cottony, velvety or powdery; usually white at first and black at age. | 58.46 |
| ZFF2 | Aspergillus niger | MK258199.1 | 100 | 577 | Black mold of onions and ornamental plants, peanuts and grapes. Serious lung disease, aspergillosis in human. Produce important enzymes | Granular to cottony, velvety or powdery; usually white at first and black at age. | 58.06 |
| ZFF6 | Curvularia spicifera | MK956807.1 | 100 | 543 | Facultative pathogen or beneficial partner of many plant species | White to pinkish gray wooly colonies | 46.96 |
| ZFF11 | Rhizopus oryzae | MK742815.1 | 100 | 501 | Opportunistic pathogen of humans causing mucormycosis. It is also used economically in the production of the enzymes, glucoamylase and lipase | Colonies are white initially, becoming brownish with age | 38.92 |
| ZFF7.2 | Aspergillus tubingensis | MF186869.1 | 100 | 578 | Involved in food spoilage of fruits and wheat and industrial fermentation and a rare human pathogen. | Granular to cottony, velvety or powdery white-black colonies | 57.96 |
| ZFF1 | Aspergillus niger | MK256745.1 | 100 | 572 | Black mold of onions and ornamental plants, peanuts and grapes. Serious lung disease, aspergillosis in humans. Produce important enzymes | Granular to cottony, velvety or powdery; usually white at first and black at age. | 58.57 |
| ZFF4 | Edgeworthia chrysantha | MK961271.1 | 100 | 540 | Not known. Possess anti-inflammatory and analgesic activity | Possesses pain brush type flowering | 59.44 |
| ZFF5 | Aspergillus stellatus | KU866665.1 | 99.82 | 541 | Unknown | Colonies are initially while and later smooth orange to reddish brown | 58.04 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma Ghimire, P.; Kang, S.; Sajjad, W.; Ali, B.; Tripathee, L.; Chen, P. Microbial Community Composition Analysis in Spring Aerosols at Urban and Remote Sites over the Tibetan Plateau. Atmosphere 2020, 11, 527. https://doi.org/10.3390/atmos11050527
Sharma Ghimire P, Kang S, Sajjad W, Ali B, Tripathee L, Chen P. Microbial Community Composition Analysis in Spring Aerosols at Urban and Remote Sites over the Tibetan Plateau. Atmosphere. 2020; 11(5):527. https://doi.org/10.3390/atmos11050527
Chicago/Turabian StyleSharma Ghimire, Prakriti, Shichang Kang, Wasim Sajjad, Barkat Ali, Lekhendra Tripathee, and Pengfei Chen. 2020. "Microbial Community Composition Analysis in Spring Aerosols at Urban and Remote Sites over the Tibetan Plateau" Atmosphere 11, no. 5: 527. https://doi.org/10.3390/atmos11050527
APA StyleSharma Ghimire, P., Kang, S., Sajjad, W., Ali, B., Tripathee, L., & Chen, P. (2020). Microbial Community Composition Analysis in Spring Aerosols at Urban and Remote Sites over the Tibetan Plateau. Atmosphere, 11(5), 527. https://doi.org/10.3390/atmos11050527





