Abstract
The assessment and control of microbial contamination in health care facilities is presently a mandatory and vital part of strategies to prevent and control hospital-acquired infections. This study aims to assess the bioburden with two passive sampling methods (30 ventilations grids swabs and 16 electrostatic dust collectors (EDCs)) at Clinical Pathology Services. The fungal burden was characterized through molecular tools, antifungal resistance, and the mycotoxins and cytotoxicity profile. Total bacteria presented the highest prevalence in both matrixes, whereas Gram-bacteria presented the lowest. Swabs presented a higher prevalence (27.6%) for fungal burden. Chrysonilia sitophila presented the highest prevalence in swabs, whereas for EDCs, C. sitophila and Mucor sp. were the most prevalent. Concerning Aspergillus genera on swabs, section Flavi was the one with the highest prevalence (58.02%), whereas, for EDCs, section Versicolores was the only section observed (100%). Aspergillus section Fumigati was detected in 10 swabs and 7 EDC samples and Aspergillus section Versicolores was detected in one EDC sample. Fungal growth on azole-supplemented media was observed in eight EDC samples. No mycotoxins were detected in any of the samples. A low cytotoxic effect was observed in two sites upon incubation of collected samples with A549 and SK cells and in two other sites upon incubation of collected samples with SK cells only. A medium cytotoxic effect was observed with one EDC sample upon incubation with A549 cells. This study reinforces the need of determination of the azole resistance profile for fungal species and allowed a preliminary risk characterization regarding the cytotoxicity. An intervention including the use of a ultraviolet with wavelength between 200 nm and 280 nm (UVC)—emitting device and an increased maintenance and cleaning of the central heating, ventilation, and air conditioning (HVAC) systems should be ensured to promote the reduction of microbial contamination.
1. Introduction
Hospital facilities are indoor environments with a high risk of cross infection between their patients and staff. Studies performed in European hospitals [1] reported that nosocomial infections add significantly to morbidity and mortality rates and that many of these infections are transmitted by airborne pathogens [2]. Several everyday activities from health care facilities can boost bio-aerosols dispersion [3,4,5,6], leading to infectious disease transmission. As such, the assessment and control of microbial contamination in health care facilities is currently a mandatory and vital part of the strategies to prevent and control hospital-acquired infections [7,8]. Aiming at addressing this challenging assessment, several studies have already reported the upmost importance of applying passive sampling methods [9,10,11,12,13]. These methods allow characterization of the contamination from a larger period of time (weeks to several months) [14].
The presence of distinct species of airborne microorganisms in clinical environments may concur for a wide range of negative health effects, both in patients, a potentially more vulnerable population, and in clinical staff, long-term exposed to resident bioburdens during work tasks. The toxicity and pathogenicity for humans of microorganisms varies greatly among genus and among species within the same genus. The emergence worldwide of drug-resistant human pathogenic microbial species, such as Methicillin-resistant Staphylococcus aureus (MRSA) and fungal species, such as Candida sp. and Aspergillus fumigatus, and the increasing reports of therapeutic failure against fungal infections [15,16,17] has revealed the need of surveillance of both bacterial and fungal resistance in health care organizations [18,19].
Although it is not currently well known to which degree the exposure to toxigenic airborne microbiota may contribute to actual adverse effects on human health, it is accepted, for instance, that fungi development in buildings is unhealthy and must be prevented [20]. Bacteria and fungi can promote allergic and infectious responses, and also inflammatory responses via inhalation of endotoxin or ß-glucans [21,22]. Additionally, it has been suggested by previous studies that modification of immune responses may be induced by long-term exposure to toxigenic fungi, leading to the development of neurological symptoms, fever, cough, and fatigue [23].
Regarding fungi, Aspergillus fumigatus is one of the most studied species in clinical environments due to their high prevalence as contaminants in clinical material, while being responsible for invasive fungal infections among immunocompromised patients [24]. Their great toxicity is due to the production of specific mycotoxins, such as aflatoxin B1, ochratoxin A, and gliotoxin. Indeed, in a review by Viegas et al. from 2018 [25], it was possible to conclude the relevance to monitor mycotoxins presence in clinical environments.
There are several published guidelines regarding ventilation of health care facilities [26,27,28,29]. However, the majority focuses on specialist facilities, such as isolation rooms, operating theatres, and bronchoscopy wards, where the risk of infection is well characterized. In contrast, guidelines regarding the ventilation of general wards, patient rooms, intensive care wards, or even other specific areas are much scarcer and often dedicated to clinical cases and not to the indoor environment. The same gap was found in microbial contamination studies, as indicated in scientific databases [13,14]. Furthermore, almost all studies focused on patients’ health, disregarding clinical and laboratory health of staff [14]. However, the staff can transport and disseminate microbial contamination through all hospital facilities, thus affecting patients [13].
Several studies were performed aiming at testing and recommending new technologies to ensure air cleaning [30,31,32,33]. However, efforts should prioritize the exposure assessment to better characterize and control the risk. This will allow the selection of the best and more suitable technologies.
To the best of our knowledge, no former studies were performed in a Clinical Pathology Service of a central hospital, applying passive methods, such as a sampling approach. This study aims to assess the bioburden using two passive sampling methods (ventilations grids and electrostatic dust collectors). The fungal burden was also characterized through molecular tools for the toxigenic species, antifungal resistance, and the mycotoxins and cytotoxicity profile.
2. Materials and Methods
2.1. Indoor Environment Assessed and Sampling Locations
The present study was performed in June, 2019 in a Clinical Pathology Service from one central hospital with 802 total number of hospital beds, comprising all hospital services. The hospital was located in the Lisbon district. It was part of an enlarged exploratory study with financial support to establish protocols for assessing occupational exposure to bioburdens in clinical environments. The Clinical Pathology Service assessed included laboratory facilities, in which diagnosis of disease was performed based on body fluids (biological samples). In this area, only hospital staff were allowed in, with the exception of the collection rooms.
A walkthrough survey and checklist were used in order to prioritize the more critical workplaces/areas regarding bioburdens, as previously published [13]. The assessed areas were equipped with central heating, ventilation, and air conditioning (HVAC) systems, with air handing units and a ductwork system. Filtered outdoor air was supplied through diffusers, coupled with grids (ventilation grids) in the ceiling/above the door, and evacuated by corner outlets at the floor level. Regarding the cleaning procedures, floors and surfaces were cleaned daily after laboratory service and working hours and were performed by an external company.
The sampling procedures were focused in specific areas, namely, one collection room in the emergency area, the core laboratory, the microbiology laboratory, six collection rooms, the triage area, the mycobacteria lab, one office, the hematology lab, the immunology room, the worker’s bedroom, bathroom, and canteen, and the immunoserology laboratory. In each area. two different types of samples were collected: swab samples in the ventilation grid and electrostatic dust collector (EDC) samples (Table 1). All the samples were collected in the same period of time, during a normal working day.

Table 1.
Samples collected in each location of the Clinical Pathology Service.
2.2. Sampling and Characterization of the Viable Bioburden
Passive sampling methods were applied to determine the contamination levels of a greater period of time and also to avoid any kind of disruption of the clinical activities. Ventilation grids (with 3 dm2 of area) samples (a total of 30) were collected by swabbing the grids. Swabs were subject to microbial extraction following a specific protocol for extraction [13] and plated onto the selected media, namely malt extract agar (MEA) supplemented with chloramphenicol (0.05%) and dichloran glycerol agar (DG-18) were used for fungi; tryptic soy agar (TSA) supplemented with cycloheximide (0.1%) was used to assess the bacterial load, and violet red bile agar (VRBA) to assess Gram negative bacteria.
Each EDC (a total of 16) had a surface expose area of 0.00942 m2 and dust was allowed to settle down for 15 days before analysis. Settled dust collected by the EDC was weighted after sampling to allow mass determination of the collected dust. EDC samples were subject to extraction and bioburden, characterized by culture-based methods, as previously performed [14]. EDC extracts were seeded onto the same culture media applied for the swabs.
EDC samples were screened for antifungal resistance on Sabouraud dextrose agar (SDA) media, supplemented with 4 mg/L itraconazole (ITRA), 1 mg/L voriconazole (VORI), or 0.5 mg/L posaconazole (POSA), with non-supplemented SDA as the control as performed previously [13,14].
2.3. Molecular Detection of Aspergillus Sections
Molecular identification of the different fungal species/strains was obtained by real-time PCR (qPCR), using the automation system (CFX)-Connect PCR System (Bio-Rad). Reactions were performed, using positive controls of DNA amplification, as indicated previously [13]. Reactions included 1× iQ Supermix (Bio-Rad), 0.5 μM of each primer (Table 2), and 0.375 μM of TaqMan probe in a total volume of 20 μL. For each gene amplified, a non-template control and a positive control, consisting of DNA obtained from a reference, belonged to the culture collection of the Reference Unit for Parasitic and Fungal Infections, Department of Infectious Diseases of National Institute of Health Dr. Ricardo Jorge. These strains have been sequenced for Internal transcribed spacer (ITS), B-tubulin, and Calmodulin.

Table 2.
Sequence of primers and TaqMan probes used for real-time PCR.
2.4. Mycotoxins Analysis
EDC samples (0.05 g) were extracted by shaking 2.5 mL of mixture of acetonitrile (ACN) with: water (H2O): acetic acid (AcOH) (79:20:1) for 90 min. After centrifugation (5 min, 5000 rpm), each sample (2 mL) was evaporated under nitrogen and reconstituted in a mobile phase (0.2 mL; A:B 7:3). Mycotoxins detection was developed through the use of a high-performance liquid chromatograph (HPLC) Nexera (Shimadzu, Tokyo, Japan), with a mass detector 5500 QTrap (Sciex, Foster City, CA, USA) (Table 3). Mycotoxins were separated on a chromatographic column Gemini C18 (150 × 4.6 mm, 5 μm) (Phenomenex, Torrance, CA, USA). The flow rate was 1 mL/min and the injection volume was 5 μL. Mobile phases were: a methanol/water/acetic acid 10/89/1 (v/v/v) and B methanol/water/acetic acid 97/2/1 (v/v/v) (both phases contained 5 mmol/L ammonium acetate) with the following gradient: 0% B up to 2 min, 50% B from 2 to 5 min, 100% B from 5 to 14 min, 100% B up to 18 min, then 0% B to 22.5 min. The Tandem Mass Spectrometry MS/MS was performed in scheduled multiple reaction monitoring (sMRM) mode in both in negative and positive polarities in one chromatographic run per sample. The Characterization of Electrospray Ionization (ESI)-source parameters were as follows: curtain gas 30 psi, collision gas medium, ionspray voltage 4500 V (negative polarity) and 5500 V (positive polarity), temperature 550 °C, ion source gas1 80 psi, ion source gas2 80 psi.

Table 3.
Detection and quantification values for each mycotoxin (ng/g).
2.5. Cell Culture and MTT Assay
Cell viability of was determined after incubation of a swine kidney monolayer (SK) and adenocarcinomic human alveolar basal epithelial (A549) cell lines with EDC samples. Cells were maintained in tissue culture flasks (TPP) medium (minimum essential medium (MEM) with Earle’s salts, Sigma-Aldrich, St. Louis, MO, USA), supplemented with antibiotics (Penicillin, Streptomycin (Sigma-Aldrich, St. Louis, MO, USA)) and fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C in a humidified atmosphere with 5% CO2. For the assay, cells were harvested with trypsin/ Ethylenediaminetetraacetic acid solution (EDTA) (1:10, v:v) in phosphate buffered saline (PBS) and plated in flat bottom 96-microtiter plates (TPP) in 100 μL culture medium at densities of about 2.5 × 105 cells/mL.
Colorimetric assay for assessing cell metabolic activity (MTT) assay was used to assess cellular viability after incubation with EDC samples through cleavage of MTT tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide). The formation of formazan crystals was measured by spectrophotometrical absorbance of the samples using the ELISA microplate reader (ELISA LEDETECT 96, biomed Dr. Wieser GmbH) at a wavelength of 510 nm.Data analysis was performed with MikroWin 2013SC software (Labsis, Neunkirchen-Seelscheid, Germany). The threshold toxicity level was defined as the lowest concentration of the extract that causes a drop in sample absorption to values <50% of cell division activity, as evaluated by the dilution method.
2.6. Statistical Analysis
The data were analyzed in statistical software, SPSS 22.0 for Windows. The results were considered significant at the 5% significance level. The Shapiro–Wilk test was used to test the normality of the data. Frequency analysis (n; %), and the calculation of the minimum and maximum medians were performed for the qualitative and quantitative data, respectively. The Spearman correlation coefficient was used to study the relationship between fungal and bacterial contamination and cytotoxicity.
3. Results
3.1. Bioburden Characterization
Total bacteria (TSA) presented the highest prevalence in both matrixes (99.99% Swabs; 100% EDC), whereas Gram-Bacteria (VRBA) presented the lowest (0.01% Swabs; 0% EDC) (Table 4).

Table 4.
Bioburden distribution on swabs and EDC.
Concerning the fungal burden, the highest prevalence for swabs was observed on MEA (27.6%), whereas EDC presented the highest prevalence on DG18 (91.9%) (Table 4).
Concerning the number of different fungal species in each matrix, swabs presented 16 different species in both media, whereas EDC presented nine different species in MEA media and seven in DG18 media.
Chrysonilia sitophila presented the highest prevalence in swabs in both media (52.50% MEA; 60.93% DG18), followed by Penicillium sp. in MEA (18.43%) and Cladosporium sp. in DG18 (25.02%) (Figure 1). In EDC, Cladosporium sp. presented the highest prevalence in DG18 media (45.98%), whereas, on MEA, the highest prevalence belonged to C. sitophila and Mucor sp. (44.52%) (Figure 1).
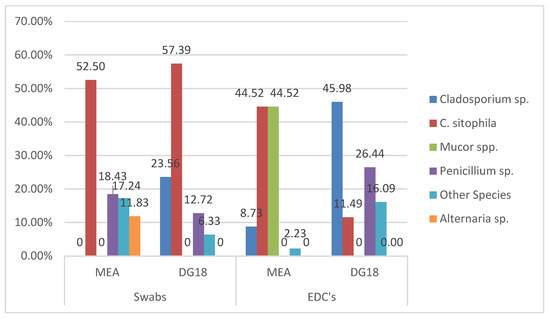
Figure 1.
Fungal distribution on both environmental matrices.
Among Aspergillus genera, section Flavi was the one with the highest prevalence on MEA (58.02%) and Nigri section on DG18 (55.41%). Referring to the EDC samples, section Versicolores was the section that presented the highest prevalence in both media (100% MEA; 75% DG18). However, the Aspergillus section Circumdati (25%) was also found in DG18 (Table 5).

Table 5.
Aspergillus section distribution on swabs and EDC.
Fungal growth on azole-supplemented media was observed in eight EDC samples on ITRA, 10 samples on VORI, and three samples on POSA. The most frequent species were: C. sitophila on SAB (97.51%), Cladosporium sp. on ITRA (27.27%) and VORI (68.00%), and Penicillium sp. on POSA (66.67%) (Table 6). Cladosporium sp. was able to grow in more than one azole in two EDC samples, namely from “Core lab” (ITRA+VORI) and from “Microbiology” (ITRA+VORI+POSA).

Table 6.
Fungal azole resistance distribution.
Among the four Aspergillus sections investigated by qPCR in swab samples from ventilation grids and in the EDC, only two were detected. Aspergillus section Fumigati was detected in 10 swab samples (10 out of 15; 66.67%) and 7 EDC samples (7 out of 12; 58.33%) (Table 7). Interestingly, Fumigati was not detected by culture-based methods in any of the samples (EDC and ventilation grid swabs).

Table 7.
Molecular detection results from the EDC and ventilation grids swab samples.
The Aspergillus section Versicolores was detected in one EDC sample by qPCR (1 out of 12; 8.33%) (Table 7); although it was detected in more samples by culture-based methods (3 out of 12 EDC samples; 25%, and 4 out of 15 swab samples; 26.67%), none of which corresponded to the sample detected at the molecular level (the workers’ bedroom EDC sample).
3.2. Cytotoxicity Analysis
EDC samples were submitted to a cytotoxicity evaluation in two distinct cell lines A549 and SK cells via application of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay. The MTT assay assesses cell viability by means of reducing the yellow tetrazolium salt to insoluble formazan. A semi-quantitative scale for cytotoxicity grading was adopted: low cytotoxic effect (+) for percentage of extinction values ranging from 80% to 90%, medium cytotoxic effect (++) for values ranging from 60% to 79%, and a high effect (+++) for values below 60%. The absence of cytotoxicity was determined when the extinction values were at ≥90. A low cytotoxic effect was observed in two locals with A549 and SK cells (“Collection room-emergency area” and “Core lab”) and in other two locals with SK cells only (“Immunoserology lab-PCR room” and “Worker’s bathroom”). The medium cytotoxic effect was observed with one EDC sample from “Canteen” on A549 cells. Although isolated analysis of two EDC samples from “Core lab” also revealed a medium cytotoxic effect on SK or A549 cells, in average, and “Core lab” presented a low cytotoxicity effect in vitro (Figure 2).
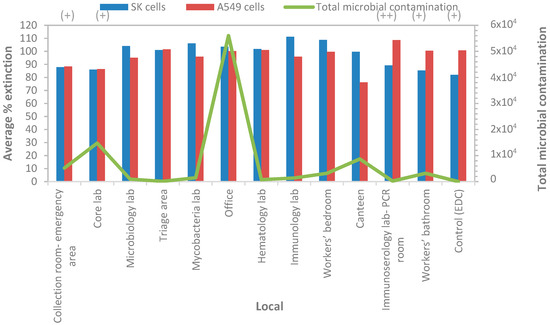
Figure 2.
Cumulative microbial contamination (TSA, VRBA, MEA, and DG18) and cell viability (average percentage of extinction, MTT assay) in EDC samples. (+), low cytotoxic effect; (++), medium cytotoxic effect.
Regarding mycotoxins analysis, in all the samples analyzed, there were no mycotoxins detected.
3.3. Comparison and Correlation Analysis
Comparing the various collection sites from the swabs, it was found that the room-emergency area and office sites were the ones with the highest fungi counts on MEA (CFU) and on DG18 (CFU), whereas triage areas, offices, hematology labs, immunology rooms, and immunoserology lab-PCR rooms were the ones with the highest total bacteria counts (TSA (CFU)). Concerning Gram-bacteria (VRBA (CFU)), none of the sites revealed their presence (Figure 3). In EDC, all sites had identical counts of both fungi and bacteria, except for the office, which had higher counts of fungi on MEA (Figure 4).
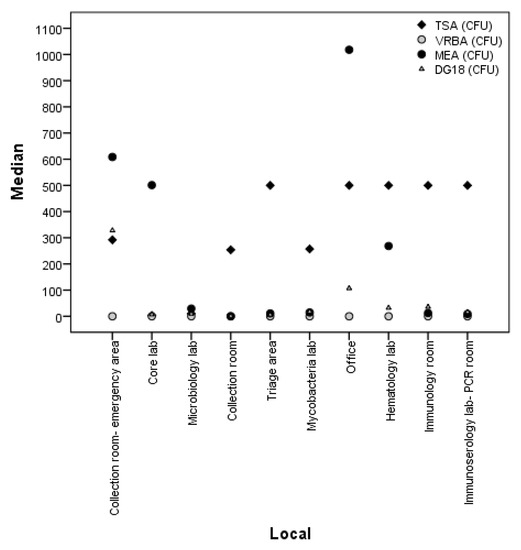
Figure 3.
Comparison of fungal and bacterial load between collection sites in swabs.
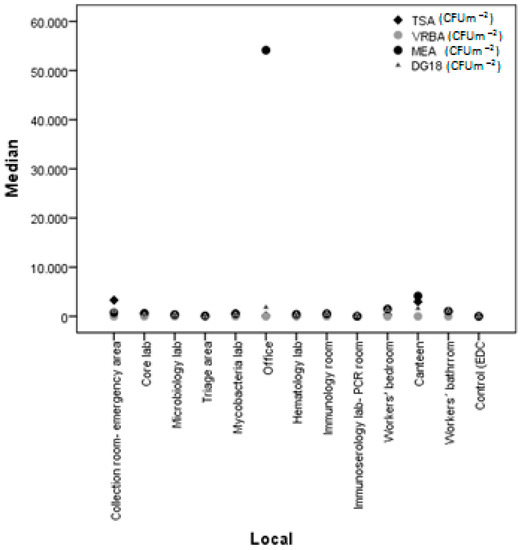
Figure 4.
Comparison of fungal and bacterial load between collection sites in EDC.
The following correlations were detected on swabs: (i) total bacteria (TSA) counts on swabs (CFU) and TSA counts on EDC (CFU m−2) (rs = −0.719, p = 0.006), which meant that higher TSA counts (CFU) on swabs were related to lower TSA counts (CFU m−2) on EDC; (ii) TSA counts on swabs (CFU) and A549 cells (rs = 0.610, p = 0.027), revealing that higher TSA counts (CFU) on swabs were related to higher A549 cell viability, thus, lower cytotoxicity; (iii) Fungi counts on MEA (CFU) on swabs and fungi counts on DG18 (CFU) on swabs (rs = 0.628, p < 0.0001), revealing that higher MEA counts on swabs (CFU) were related to higher DG18 counts (CFU) on swabs; (iv) MEA counts (CFU) on swabs and MEA counts (CFU m−2) on EDC (rs = 0.742, p = 0.004) and with DG18 counts (CFU m−2) on EDC (rs = 0.645, p = 0.017), indicating that higher MEA counts (CFU) on swabs were related with higher MEA counts (CFU m−2) on EDC and higher DG18 counts (CFU m−2) on EDCs (Table 8).

Table 8.
Study of the relationship between microbial contamination and cell viability (MTT assay) (Spearman correlation coefficient results).
The following correlations were detected on EDC: (i) TSA counts on EDC (CFU m−2), MEA counts (CFU m−2), and DG18 counts (CFU m−2), both in EDC (rs = 0.509, p = 0.037 and rs = 0.490, p = 0.046, respectively), which means that higher TSA counts (CFU m−2) on EDC were related to higher MEA counts (CFU m−2) and higher DG18 counts (CFU m−2) on EDC; (ii) TSA counts (CFU m−2) on EDC and A549 cells (rs = −0.729, p = 0.001), revealing that higher TSA counts (CFU m−2) on EDC were related to lower A549 cell viability, thus, higher cytotoxicity; (iii) MEA counts (CFU m−2) and DG18 counts (CFU m−2) on EDC (rs = 0.861, p < 0.0001), indicating that higher MEA counts (CFU m−2) on EDC were related to higher DG18 counts (CFU m−2) on EDC (Table 8).
4. Discussion
The sampling approach applied on this study allows us to recognize the advantages of using more than one sampling method [13]. Indeed, different counts and biodiversity (swabs: 16 different species in both media, EDC: 9 different species in MEA, and 7 in DG18) were observed in each environmental sample, allowing us to obtain a more accurate bioburden characterization. Additionally, using different agar media allowed bacteriota discrimination between total bacteria and Gram-bacteria. Regarding fungi, this approach has allowed the detection of a wider diversity concerning the Aspergillus genera, as well as the identification of different resistant profiles within the identified fungi. As such, the combination of these factors has promoted differentiation of the fungal species detected. Thus, it will be possible to introduce targeted cleaning procedures in specific surfaces, such as walls and floors, consistent with the fact that hospital surfaces might be a source for dissemination of microorganisms transported to the hands of patients or staff [37]. Indeed, hospital environments could be a source of outbreaks of resistant organisms [38], and experts suggest indirect transmission through the environment, besides direct person-to-person transmission [39].
The efficacy of biocides in eliminating microbial contaminants within healthcare facilities has been studied in the last years [40,41,42,43,44]. The widespread and increasing use of products containing low concentrations of biocidal agents or presenting low activity raises the concern of microorganisms’ selection through development of acquired resistance and selection of less susceptible microorganisms following exposure to biocidals. Although this phenomenon is mainly described for bacteria, the same principle applies to fungal development of resistance to antifungal agents used in the environment and cross-resistance to antifungal drugs. The rational of using biocidals in healthcare facilities should, therefore, be supported by the evidence on their added value and targeted to specific applications, for which the risks have been established. For that reason, the characterization of a microbial resistance profile in the healthcare facilities’ environment is of upmost importance.
Therefore, more than one sampling approach and more than one culture media should be the trend to be applied by exposure assessors (industrial hygienists or Indoor Air Quality assessors) when analyzing/sampling health care facilities [13].
Molecular tools were very efficient in detecting Aspergillus section Fumigati, whereas, for section Versicolores, the culture-based methods were more valuable. These differences highlight the importance of using culture-based methods for assessing the viable component of bioburden [13], combined with real-time PCR assay for targeting the indicators of harmful fungal contamination [45]. Indeed, culture-based methods can reveal less abundant fungi in a specific occupational environment when compared with more refined molecular tools (e.g., high-throughput sequencing) that provide a more exhaustive diversity profile [46]. However, although in less counts, the presence of some Aspergillus sections (opportunistic pathogens) may represent a health risk for patients and workers who are exposed on a daily basis [46]. Of note is the increased detection of Aspergillus section Fumigati, responsible by 80% of the cases of invasive aspergillosis, the most common invasive fungal infection [47].
Concerning the fact that mycotoxins null results, this can be due to several aspects, including the fact that the fungi are not stimulated to produce mycotoxins due to environmental reasons. However, since toxigenic species were found in the samples collected, this situation can change if some environmental variable changes. Therefore, the fact that no mycotoxins were measured does not allow discarding the need of future campaigns that will also monitor mycotoxins. Indeed, previous reports have already detected mycotoxins in health care centers, particularly in air and HVAC filter samples [13], justifying this need.
No Aspergillus sections were observed in azole screening media. Nevertheless, the Circumdati section was observed in Sabouraud. The burden of Aspergillus species in azole supplemented media might be underestimated due to presence of fast-growing species, such as C. sitophila on ITRA and VORI, and requires further investigation. In fact, molecular tools revealed the presence of other Aspergillus sections (Fumigati and Versicolores) in EDC not detected by culture. The fact that two samples (from Core lab and Microbiology) showed the same fungal species (Cladosporium and Penicillium) in two different azole media might suggest the development of multi-resistance to azoles, although this requires confirmation by minimum inhibitory concentration (MIC) determination by a reference method, such as The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Previous studies from our group performed in health care centers also revealed (in settled dust samples) the co-occurrence in the same sample of Penicillium growth in ITRA and VORI [48]. Another study on indoor air quality also describes fungal growth in the three azoles for Penicillium and Cladosporium [13] and raises the question of the need of determination of the azole resistance profile for fungal species that might represent some health risk.
EDC sampling allowed a preliminary risk characterization regarding the cytotoxicity in human alveolar basal epithelial cells as a model for in vitro inhalation toxicology [49,50] and in swine kidney monolayer cells as a model for renal in vitro toxicology [51] through incubation of cell lines with airborne substances present in the sampled places. According to the defined threshold levels for cytotoxicity, airborne substances with a potential medium cytotoxic effect were recovered from three EDC samples from locals with some of the highest bacterial (TSA counts) and/or fungal (MEA) contamination levels: “canteen” and “core lab”. However, no cytotoxic effect was observed in the “office”, where the highest microbial contamination was found. Statistical analysis revealed no relation between fungal contamination and cytotoxicity and contradictory results, regarding the role of bacterial contamination on cytotoxicity, depending on the sampling method. While bacterial contamination (TSA) on EDC seems to negatively affect A549 cell viability, the contrary was observed with swab sampling. These results suggest that other factors besides microbial contamination may be associated with in vitro cytotoxicity, namely, the presence of specific toxigenic species and/or particulate matter and/or other irritant agents in the collected samples. It is noteworthy that toxigenic Aspergillus species were detected in EDC samples collected in canteen (Aspergillus sections Fumigati and Versicolores) and in core lab (Aspergillus section Versicolores). However, such a statement may not provide grounds for a definite conclusion about the compared species of fungi that display a more cytotoxic effect than others.
The comparison between sampling sites regarding microbial contamination allowed us to prioritize intervention regarding which biocidals (for fungi/bacteria elimination) should be applied in each sampling site. However, it was found among the bioburden analyzed that a positive correlation between both sampling methods was applied. Thus, an intervention aiming at air cleaning with ultraviolet germicidal irradiation, such as ultraviolet with wavelength between 200 nm and 280 nm (UVC) [30] should be ensured, covering both fungi and bacteria elimination in the Clinical Pathology Service. Additionally, HVAC systems (including ventilation grids) should be maintained and properly preserved, since their efficacy in reducing microbial contamination has already been demonstrated [52,53].
5. Conclusions
Overall, it was possible to conclude that the two sampling methods used in this study—swabs and EDCs—unveiled a more complete characterization of the bioburden, permitting us to obtain information regarding workers and patient’s potential exposure. Additionally, culture-based methods and molecular tools used in parallel should be used in order to perform an accurate characterization of the fungal contamination. This study also reinforces the need of analyzing the azole resistance profile for fungal species that might represent some health risk. EDC sampling allowed a preliminary risk characterization regarding the cytotoxicity present in the sampled places. Still, further studies are needed for a definite conclusion about the fungal species that display more cytotoxic effects when compared to others.
An intervention including the use of a UVC-emitting device in the Clinical Pathology Service and an increased maintenance and cleaning of the HVAC systems should be ensured to promote the reduction of microbial contamination.
Author Contributions
Conceptualization, C.V.; data curation, C.V.; formal analysis, C.V., M.T., R.L., M.D., B.A., L.A.C., E.C., A.Q.G., R.K., and E.S.; funding acquisition, C.V. and M.T.; investigation, C.V.; methodology, C.V., L.A.C., and S.V.; project administration, C.V.; resources, C.V.; supervision, C.V., A.Q.G., and S.V.; validation, C.V., M.T., A.Q.G., and S.V.; writing—original draft, C.V.; writing—review and editing, C.V., L.A.C., and S.V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by FCT—Fundação para Ciência e Tecnologia for funding the project EXPOsE—establishing protocols to assess occupational exposure to microbiota in clinical settings (02/SAICT/2016—Project No. 23222) and to the Polish Minister of Science and Higher Education, under the program “Regional Initiative of Excellence” in 2019–2022 (Grant No. 008/RID/2018/19).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kaoutar, B.; Joly, C.; L’Hériteau, F.; Barbut, F.; Robert, J.; Denis, M.; Espinasse, F.; Merrer, J.; Doit, C.; Costa, Y.; et al. Nosocomial infections and hospital mortality: A multicentre epidemiological study. J. Hosp. Infect. 2004, 58, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, W. Hospital Airborne Infection Control; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Gupta, J.K.; Lin, C.H.; Chen, Q. Flow dynamics and characterization of a cough. Indoor Air 2009, 19, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.K.; Lin, C.H.; Chen, Q. Characterizing exhaled airflow from breathing and talking. Indoor Air 2010, 20, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.W.; Nicolle, A.; Pantelic, J.; Koh, G.C.; De Wang, L.; Amin, M.; Klettner, C.A.; Cheong, D.K.W.; Sekhar, C.; Tham, K.W. Airflow Dynamics of Coughing in Healthy Human Volunteers by Shadowgraph Imaging: An Aid to Aerosol Infection Control. PLoS ONE 2012, 7, e34818. [Google Scholar] [CrossRef]
- Tang, J.W.; Nicolle, A.D.; Klettner, C.A.; Pantelic, J.; Wang, L.; Bin Suhaimi, A.; Tan, A.Y.L.; Ong, G.W.X.; Su, R.; Sekhar, C.; et al. Airflow Dynamics of Human Jets: Sneezing and Breathing—Potential Sources of Infectious Aerosols. PLoS ONE 2013, 8, e59970. [Google Scholar] [CrossRef]
- Ferdyn-Grygierek, J. Monitoring of indoor air parameters in large museum exhibition halls with and without air-conditioning systems. Build. Environ. 2016, 107, 113–126. [Google Scholar] [CrossRef]
- Zahar, J.-R.; Jolivet, S.; Adam, H.; Dananché, C.; Lizon, J.; Alfandari, S.; Boulestreau, H.; Baghdadi, N.; Bay, J.-O.; Bénéteau, A.-M.; et al. Quelles mesures pour maîtriser le risque infectieux chez les patients immunodéprimés? Recommandations formalisées d’experts. J. Mycol. Med. 2017, 27, 449–456. [Google Scholar] [CrossRef]
- Roussel, S.; Reboux, G.; Bellanger, A.-P.; Sornin, S.; Grenouillet, F.; Dalphin, J.-C.; Piarroux, R.; Millon, L. Characteristics of dwellings contaminated by moulds. J. Environ. Monit. 2008, 10, 724. [Google Scholar] [CrossRef]
- Würtz Jürgensen, C.; Mette Madsen, A. Influence of everyday activities and presence of people in common indoor environments on exposure to airborne fungi. AIMS Environ. Sci. 2016, 3, 77–95. [Google Scholar] [CrossRef]
- Mensah-Attipoe, J.; Reponen, T.; Veijalainen, A.-M.; Rintala, H.; Täubel, M.; Rantakokko, P.; Ying, J.; Hyvärinen, A.; Pasanen, P. Comparison of methods for assessing temporal variation of growth of fungi on building materials. Microbiology 2016, 162, 1895–1903. [Google Scholar] [CrossRef]
- Viegas, C.; Faria, T.; Caetano, L.A.; Carolino, E.; Gomes, A.Q.; Viegas, S. Aspergillus spp. prevalence in different Portuguese occupational environments: What is the real scenario in high load settings? J. Occup. Environ. Hyg. 2017, 14, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Viegas, C.; Almeida, B.; Monteiro, A.; Caetano, L.A.; Carolino, E.; Gomes, A.Q.; Twarużek, M.; Kosicki, R.; Marchand, G.; Viegas, S. Bioburden in health care centers: Is the compliance with Portuguese legislation enough to prevent and control infection? Build. Environ. 2019, 160, 106226. [Google Scholar] [CrossRef]
- Viegas, C.; Santos, P.; Almeida, B.; Monteiro, A.; Carolino, E.; Gomes, A.Q.; Viegas, S. Electrostatic dust collector: A passive screening method to assess occupational exposure to organic dust in primary health care centers. Air Qual. Atmos. Heal. 2019, 12, 573–583. [Google Scholar] [CrossRef]
- Snelders, E.; Melchers, W.J.; Verweij, P.E. Azole resistance in Aspergillus fumigatus: A new challenge in the management of invasive aspergillosis? Future Microbiol. 2011, 6, 335–347. [Google Scholar] [CrossRef]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.G.; Meis, J.F. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef]
- Alberti, C.; Bouakline, A.; Ribaud, P.; Lacroix, C.; Rousselot, P.; Leblanc, T.; Derouin, F. Relationship between environmental fungal contamination and the incidence of invasive aspergillosis in haematology patients. J. Hosp. Infect. 2001, 48, 198–206. [Google Scholar] [CrossRef]
- Gangneux, J.-P.; Bousseau, A.; Cornillet, A.; Kauffmann-Lacroix, C. Maîtrise du risque fongique environnemental dans les établissements de santé. J. Mycol. Med. 2006, 16, 204–211. [Google Scholar] [CrossRef]
- Piecková, E. Adverse Health Effects of Indoor Moulds/Štetni Zdravstveni Učinci Plijesni Unutarnjih Prostora. Arch. Ind. Hyg. Toxicol. 2012, 63, 545–549. [Google Scholar] [CrossRef]
- Rylander, R. Organic dust induced pulmonary disease—The role of mould derived beta-glucan. Ann. Agric. Environ. Med. 2010, 17, 9–13. [Google Scholar]
- Liebers, V.; van Kampen, V.; Bünger, J.; Düser, M.; Stubel, H.; Brüning, T.; Raulf-Heimsoth, M. Assessment of Airborne Exposure to Endotoxin and Pyrogenic Active Dust Using Electrostatic Dustfall Collectors (EDCs). J. Toxicol. Environ. Heal. Part A 2012, 75, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, E.; Campbell, A.W.; Jones, J.; Ehiri, J.E.; Akpan, A.I. The Neurological Significance of Abnormal Natural Killer Cell Activity in Chronic Toxigenic Mold Exposures. Sci. World J. 2003, 3, 1128–1137. [Google Scholar] [CrossRef]
- Gniadek, A.; Krzyściak, P.; Twarużek, M.; Macura, A. Occurrence of fungi and cytotoxicity of the species: Aspergillus ochraceus, Aspergillus niger and Aspergillus flavus isolated from the air of hospital wards. Int. J. Occup. Med. Environ. Health 2017. [Google Scholar] [CrossRef] [PubMed]
- Viegas, S.; Viegas, C.; Oppliger, O. Occupational Exposure to Mycotoxins: Current Knowledge and Prospects. Ann. Work Expos. Health 2018, 62, 923–941. [Google Scholar] [CrossRef] [PubMed]
- National Health Service. Design Considerations: Ventilation in Healthcare Premises, Health Technical Memorandum 2025; National Health Service Estates: London, UK, 1994. [Google Scholar]
- AIA. Guidelines for de Sign and Construction of Hospital and Health Care Facilities; AIA Press: New York, NY, USA, 2001. [Google Scholar]
- American Society of Heating; Refrigerating, and Air Conditioning Engineers. HVAC Design Manual for Hospitals and Clinics; ASHRAE: Atlanta, GA, USA, 2003.
- Chinn, R.Y.W.; Sehulster, L. Guidelines for Environmental Infection Control in Health-Care Facilities; Recommendations of CDC and Healthcare Infection Control Practices Advisory Committee (HICPAC); CDC: Atlanta, GA, USA, 2003.
- Ethington, T.; Newsome, S.; Waugh, J.; Lee, L.D. Cleaning the air with ultraviolet germicidal irradiation lessened contact infections in a long-term acute care hospital. Am. J. Infect. Control. 2018, 46, 482–486. [Google Scholar] [CrossRef]
- Anderson, D.J.; Chen, L.F.; Weber, D.J.; Moehring, R.W.; Lewis, S.S.; Triplett, P.F.; Blocker, M.; Becherer, P.; Schwab, J.C.; Knelson, L.P.; et al. Enhanced terminal room disinfection and acquisition and infection caused by multidrug-resistant organisms and Clostridium difficile (the Benefits of Enhanced Terminal Room Disinfection study): A cluster-randomised, multicentre, crossover study. Lancet 2017, 389, 805–814. [Google Scholar] [CrossRef]
- Rutala, W.A.; Gergen, M.F.; Weber, D.J. Room decontamination with UV radiation. Infect. Control. Hosp. Epidemiol. 2010, 31, 1025–1029. [Google Scholar] [CrossRef]
- Health Quality Ontario. Portable Ultraviolet Light Surface-Disinfecting Devices for Prevention of Hospital-Acquired Infections: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 2018, 18, 1–73. [Google Scholar]
- Mayer, Z.; Bagnara, A.; Färber, P.; Geisen, R. Quantification of the copy number of nor-1, a gene of the aflatoxin biosynthetic pathway by real-time PCR, and its correlation to the cfu of Aspergillus flavus in foods. Int. J. Food Microbiol. 2003, 82, 143–151. [Google Scholar] [CrossRef]
- Cruz-Perez, P.; Buttner, M.P.; Stetzenbach, L.D. Detection and quantitation of Aspergillus fumigatus in pure culture using polymerase chain reaction. Mol. Cell. Probes. 2001, 15, 81–88. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency (EPA), About the National Exposure Research Laboratory (NERL), (n.d.). Available online: http://www.epa.gov/nerlcwww/moldtech.html (accessed on 8 January 2020).
- Donskey, C.J. Beyond high-touch surfaces: Portable equipment and floors as potential sources of transmission of health care–associated pathogens. Am. J. Infect. Control. 2019, 47, A90–A95. [Google Scholar] [CrossRef] [PubMed]
- Hardy, K.J.; Oppenheim, B.A.; Gossain, S.; Gao, F.; Hawkey, P.M. A Study of the Relationship Between Environmental Contamination with Methicillin-Resistant Staphylococcus aureus (MRSA) and Patients’ Acquisition of MRSA. Infect. Control Hosp. Epidemiol. 2006, 27, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Dancer, S.J. Importance of the environment in meticillin-resistant Staphylococcus aureus acquisition: The case for hospital cleaning. Lancet Infect. Dis. 2008, 8, 101–113. [Google Scholar] [CrossRef]
- Gilbert, P.; McBain, A.J. Potential Impact of Increased Use of Biocides in Consumer Products on Prevalence of Antibiotic Resistance. Clin. Microbiol. Rev. 2003, 16, 189–208. [Google Scholar] [CrossRef]
- Russell, A. Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 2003, 3, 794–803. [Google Scholar] [CrossRef]
- Maillard, J.-Y. Antimicrobial biocides in the healthcare environment: Efficacy, usage, policies, and perceived problems. Ther. Clin. Risk Manag. 2005, 1, 307–320. [Google Scholar]
- Maillard, J.-Y. Bacterial resistance to biocides in the healthcare environment: Should it be of genuine concern? J. Hosp. Infect. 2007, 65, 60–72. [Google Scholar] [CrossRef]
- Ghanem, B.; Haddadin, R.N. Multiple drug resistance and biocide resistance in Escherichia coli environmental isolates from hospital and household settings. Antimicrob. Resist. Infect. Control. 2018, 7, 47. [Google Scholar] [CrossRef]
- Degois, J.; Clerc, F.; Simon, X.; Bontemps, C.; Leblond, P.; Duquenne, P. First Metagenomic Survey of the Microbial Diversity in Bioaerosols Emitted in Waste Sorting Plants. Ann. Work Expo. Heal. 2017, 61, 1076–1086. [Google Scholar] [CrossRef]
- Mbareche, H.; Veillette, M.; Bilodeau, G.J.; Duchaine, C. Fungal aerosols at dairy farms using molecular and culture techniques. Sci. Total Environ. 2019, 653, 253–263. [Google Scholar] [CrossRef]
- Segal, B.H. Aspergillosis. N. Engl. J. Med. 2009, 360, 1870–1884. [Google Scholar] [CrossRef] [PubMed]
- Viegas, C.; Almeida, B.; Monteiro, A.; Paciência, I.; Rufo, J.C.; Carolino, E.; Quintal-Gomes, A.; Twarużek, M.; Kosicki, R.; Marchand, G.; et al. Settled dust assessment in clinical environment: Useful for the evaluation of a wider bioburden spectrum. Int. J. Environ. Health Res. 2019, 1–19. [Google Scholar] [CrossRef]
- Foldbjerg, R.; Dang, D.A.; Autrup, H. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch. Toxicol. 2011, 85, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Tabei, Y.; Sonoda, A.; Nakajima, Y.; Biju, V.; Makita, Y.; Yoshida, Y.; Horie, M. In vitro evaluation of the cellular effect of indium tin oxide nanoparticles using the human lung adenocarcinoma A549 cells. Metallomics 2015, 7, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Heussner, A.H.; Dietrich, D.R. Primary porcine proximal tubular cells as an alternative to human primary renal cells in vitro: An initial characterization. BMC Cell Biol. 2013, 14, 55. [Google Scholar] [CrossRef]
- Srikanth, P.; Sudharsanam, S.; Steinberg, R. Bio-aerosols in indoor environment: Composition, health effects and analysis. Indian J. Med. Microbiol. 2008, 26, 302. [Google Scholar] [CrossRef]
- Gołofit-Szymczak, M.; Górny, R.L. Microbiological air quality in office buildings equipped with dventilation systems. Indoor Air 2018, 28, 792–805. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).