Overlooked Diversity of Ultramicrobacterial Minorities at the Air-Sea Interface
Abstract
:1. Introduction
2. Experiments
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | %LNA | %HNA |
|---|---|---|
| ULW_21st_April_2016_set1 | 55.0 | 45.0 |
| SML_21st_April_2016_set1 | 76.9 | 23.1 |
| Foam_21st_April_2016_set1 | 42.5 | 57.5 |
| ULW_21st_April_2016_set2 | 53.7 | 46.3 |
| SML_21st_April_2016_set2 | 55.8 | 44.2 |
| Foam_21st_April_2016_set2 | 52.0 | 48.0 |
| ULW_19th_May_2016_set1 | 63.1 | 36.9 |
| SML_19th_May_2016_set1 | 62.5 | 37.5 |
| Foam_19th_May_2016_set1 | 32.5 | 67.5 |
| ULW_19th_May_2016_set2 | 65.0 | 35.0 |
| SML_19th_May_2016_set2 | 60.0 | 40.0 |
| Foam_19th_May_2016_set2 | 24.9 | 75.1 |
| ULW_19th_July_2016_set1 | 50.3 | 49.7 |
| SML_19th_July_2016_set1 | 49.7 | 50.3 |
| Foam_19th_July_2016_set1 | 39.8 | 60.2 |
| ULW_19th_July_2016_set2 | 51.6 | 48.4 |
| SML_19th_July_2016_set2 | 52.8 | 47.2 |
| Foam_19th_July_2016_set2 | 39.2 | 60.8 |
| Foam | SML | 1-m depth | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cDNA | DNA | cDNA | DNA | cDNA | DNA | |||||||||
| FL | PA | FL | PA | FL | PA | FL | PA | FL | PA | FL | PA | |||
| Foam | cDNA | FL | 1.00 | 0.90 | 0.03 | 1.00 | 1.00 | 0.04 | 0.89 | 1.00 | 1.00 | 5.43 × 10−4 | 0.97 | |
| PA | 0.73 | 0.56 | 3.34 × 10−3 | 1.00 | 1.00 | 0.01 | 0.48 | 1.00 | 1.00 | 4.19 × 10−5 | 0.70 | |||
| DNA | FL | 2.26 | 3.11 | 0.78 | 0.81 | 0.76 | 0.74 | 1.00 | 0.93 | 0.91 | 0.06 | 1.00 | ||
| PA | 5.10 | 6.24 | 2.63 | 0.02 | 0.01 | 1.00 | 0.56 | 0.07 | 0.02 | 0.77 | 0.34 | |||
| SML | cDNA | FL | 0.29 | 0.43 | 2.55 | 5.42 | 1.00 | 0.02 | 0.78 | 1.00 | 1.00 | 2.83 × 10−4 | 0.92 | |
| PA | 0.28 | 0.47 | 2.66 | 5.75 | 0.02 | 0.01 | 0.71 | 1.00 | 1.00 | 1.19 × 10−4 | 0.89 | |||
| DNA | FL | 4.97 | 5.97 | 2.71 | 0.35 | 5.26 | 5.52 | 0.55 | 0.08 | 0.03 | 0.94 | 0.35 | ||
| PA | 2.31 | 3.27 | 0.16 | 3.12 | 2.63 | 2.78 | 3.13 | 0.93 | 0.89 | 0.02 | 1.00 | |||
| 1-m depth | cDNA | FL | 0.04 | 0.63 | 2.13 | 4.70 | 0.23 | 0.22 | 4.64 | 2.16 | 1.00 | 1.60 × 10−3 | 0.98 | |
| PA | 0.14 | 0.93 | 2.24 | 5.28 | 0.45 | 0.45 | 5.10 | 2.31 | 0.18 | 3.16 × 10−4 | 0.98 | |||
| DNA | FL | 7.07 | 8.18 | 4.81 | 2.64 | 7.36 | 7.73 | 2.10 | 5.43 | 6.58 | 7.31 | 0.01 | ||
| PA | 1.87 | 2.80 | 0.61 | 3.61 | 2.18 | 2.30 | 3.58 | 0.50 | 1.75 | 1.83 | 5.87 |

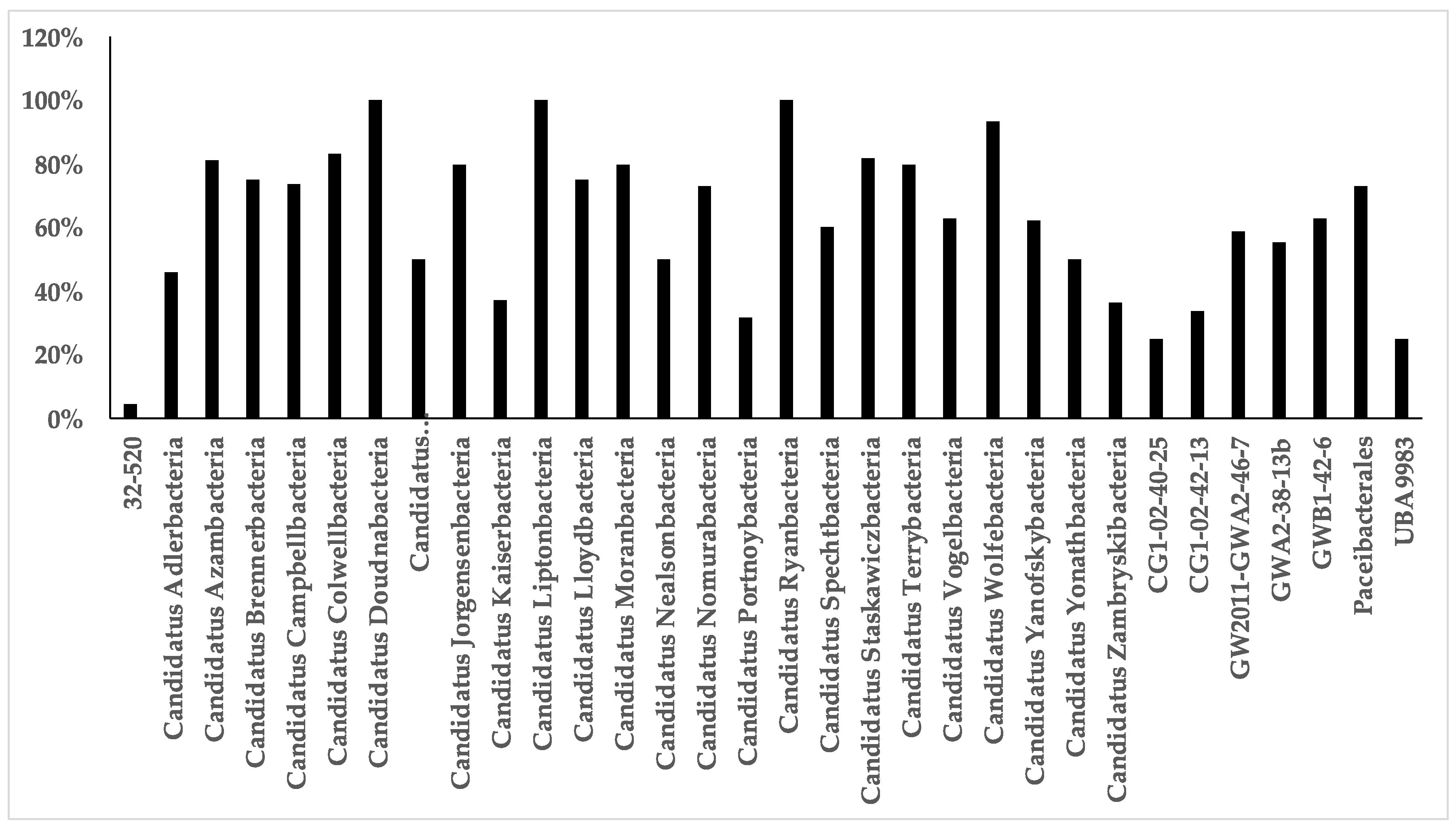

References
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Miller, C.S.; Castelle, C.J.; VerBerkmoes, N.C.; Wilkins, M.J.; Hettich, R.L.; Lipton, M.S.; Williams, K.H.; et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 2012, 337, 1661–1665. [Google Scholar] [CrossRef] [Green Version]
- He, X.; McLean, J.S.; Edlund, A.; Yooseph, S.; Hall, A.P.; Liu, S.Y.; Dorrestein, P.C.; Esquenazi, E.; Hunter, R.C.; Cheng, G.; et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2015, 112, 244–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemos, L.N.; Medeiros, J.D.; Dini-Andreote, F.; Fernandes, G.R.; Varani, A.M.; Oliveira, G.; Pylro, V.S. Genomic signatures and co-occurrence patterns of the ultra-small Saccharimonadia (phylum CPR/Patescibacteria) suggest a symbiotic lifestyle. Mol. Ecol. 2019, 28, 4259–4271. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Paul, B.G.; Castelle, C.J.; Hu, P.; Tringe, S.G.; Valentine, D.L.; Andersen, G.L.; Banfield, J.F. Unusual metabolism and hypervariation in the genome of a Gracilibacteria (BD1-5) from an oil-degrading community. mBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Castelle, C.J.; Banfield, J.F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 2018, 172, 1181–1197. [Google Scholar] [CrossRef] [Green Version]
- Castelle, C.J.; Brown, C.T.; Anantharaman, K.; Probst, A.J.; Huang, R.H.; Banfield, J.F. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol. 2018, 16, 629–645. [Google Scholar] [CrossRef]
- Danczak, R.E.; Johnston, M.D.; Kenah, C.; Slattery, M.; Wrighton, K.C.; Wilkins, M.J. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 2017, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Wegner, C.E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazina, T.N.; Babich, T.L.; Kostryukova, N.K.; Sokolova, D.S.; Abdullin, R.R.; Tourova, T.P.; Poltaraus, A.B.; Kalmykov, S.N.; Zakharova, E.V.; Myasoedov, B.F.; et al. Microbial diversity and possible activity in nitrate- and radionuclide-contaminated groundwater. In Behavior of Radionuclides in the Environment I; Springer: Singapore, 2020; pp. 35–66. [Google Scholar] [CrossRef]
- Probst, A.J.; Ladd, B.; Jarett, J.K.; Geller-McGrath, D.E.; Sieber, C.M.K.; Emerson, J.B.; Anantharaman, K.; Thomas, B.C.; Malmstrom, R.R.; Stieglmeier, M.; et al. Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat. Microbiol. 2018, 3, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.O.; Quince, C.; Shaiber, A.; Esen, O.C.; Lee, S.T.; Rappé, M.S.; McLellan, S.L.; Lücker, S.; Eren, A.M. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 2018, 3, 804–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigneron, A.; Cruaud, P.; Langlois, V.; Lovejoy, C.; Culley, A.I.; Vincent, W.F. Ultra-small and abundant: Candidate phyla radiation bacteria are potential catalysts of carbon transformation in a thermokarst lake ecosystem. Limnol. Oceanogr. Lett. 2019, 5, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Cabello-Yeves, P.J.; Zemskaya, T.I.; Zakharenko, A.S.; Sakirko, M.V.; Ivanov, V.G.; Ghai, R.; Rodriguez-Valera, F. Microbiome of the deep Lake Baikal, a unique oxic bathypelagic habitat. Limnol. Oceanogr. 2019, 65, 1471–1488. [Google Scholar] [CrossRef] [Green Version]
- Borrel, G.; Lehours, A.C.; Bardot, C.; Bailly, X.; Fonty, G. Members of candidate divisions OP11, OD1 and SR1 are widespread along the water column of the meromictic Lake Pavin (France). Arch. Microbiol. 2010, 192, 559–567. [Google Scholar] [CrossRef]
- Castelle, C.J.; Brown, C.T.; Thomas, B.C.; Williams, K.H.; Banfield, J.F. Unusual respiratory capacity and nitrogen metabolism in a Parcubacterium (OD1) of the Candidate Phyla Radiation. Sci. Rep. 2017, 7, 40101. [Google Scholar] [CrossRef]
- Nelson, W.C.; Stegen, J.C. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 2015, 6, 713. [Google Scholar] [CrossRef] [Green Version]
- Lannes, R.; Olsson-Francis, K.; Lopez, P.; Bapteste, E. Carbon fixation by marine ultrasmall prokaryotes. Genome Biol. Evol. 2019, 11, 1166–1177. [Google Scholar] [CrossRef] [Green Version]
- Mustaffa, N.I.H.; Ribas-Ribas, M.; Banko-Kubis, H.M.; Wurl, O. Global reduction of in situ CO2 transfer velocity by natural surfactants in the sea-surface microlayer. Proc. Math. Phys. Eng. Sci. 2020, 476, 20190763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Wurl, O.; Stolle, C.; Van Thuoc, C.; Thu, P.T.; Mari, X. Biofilm-like properties of the sea surface and predicted effects on air-sea CO2 exchange. Prog. Oceanogr. 2016, 144, 15–24. [Google Scholar] [CrossRef]
- Cunliffe, M.; Engel, A.; Frka, S.; Gašparović, B.; Guitart, C.; Murrell, J.C.; Salter, M.; Stolle, C.; Upstill-Goddard, R.; Wurl, O. Sea surface microlayers: A unified physicochemical and biological perspective of the air–ocean interface. Progr. Oceanogr. 2013, 109, 104–116. [Google Scholar] [CrossRef]
- Parks, G.; Dean, C.W.; Kluge, J.A.; Soloviev, A.V.; Shivji, M.; Tartar, A.; Howe, K.L.; Lehner, S.; Schwarz, E.; Shen, H.; et al. Analysis of surfactant-associated bacteria in the sea surface microlayer using deoxyribonucleic acid sequencing and synthetic aperture radar. Int. J. Remote Sens. 2020, 41, 3886–3901. [Google Scholar] [CrossRef]
- Rahlff, J.; Stolle, C.; Giebel, H.-A.; Mustaffa, N.I.H.; Wurl, O.; Herlemann, D. Marine foams represent compressed sea-surface microlayer with distinctive bacterial communities. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Schilling, K.; Zessner, M. Foam in the aquatic environment. Water Res. 2011, 45, 4355–4366. [Google Scholar] [CrossRef]
- Carlson, D.J. Surface microlayer phenolic enrichments indicate sea surface slicks. Nature 1982, 296, 426–429. [Google Scholar] [CrossRef]
- Hugoni, M.; Vellet, A.; Debroas, D. Unique and highly variable bacterial communities inhabiting the surface microlayer of an oligotrophic lake. Aquat. Microb. Ecol. 2017, 79, 115–125. [Google Scholar] [CrossRef]
- Uetake, J.; Hill, T.C.J.; Moore, K.A.; DeMott, P.J.; Protat, A.; Kreidenweis, S.M. Airborne bacteria confirm the pristine nature of the Southern Ocean boundary layer. Proc. Natl. Acad. Sci. USA 2020, 117, 13275–13282. [Google Scholar] [CrossRef]
- Harvey, G.W.; Burzell, L.A. A simple microlayer method for small samples. Limnol. Oceanogr. 1972, 17, 156–157. [Google Scholar] [CrossRef]
- Giebel, H.A.; Wolterink, M.; Brinkhoff, T.; Simon, M. Complementary energy acquisition via aerobic anoxygenic photosynthesis and carbon monoxide oxidation by Planktomarina temperata of the Roseobacter group. FEMS Microbiol. Ecol. 2019, 95, fiz050. [Google Scholar] [CrossRef] [PubMed]
- Proctor, C.R.; Besmer, M.D.; Langenegger, T.; Beck, K.; Walser, J.C.; Ackermann, M.; Burgmann, H.; Hammes, F. Phylogenetic clustering of small low nucleic acid-content bacteria across diverse freshwater ecosystems. ISME J. 2018, 12, 1344–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvier, T.; Del Giorgio, P.A.; Gasol, J.M. A comparative study of the cytometric characteristics of high and low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 2007, 9, 2050–2066. [Google Scholar] [CrossRef] [PubMed]
- Schlitzer, R. Ocean Data View. Available online: https://odv.awi.de (accessed on 9 November 2020).
- Rahlff, J.; Stolle, C.; Giebel, H.A.; Brinkhoff, T.; Ribas-Ribas, M.; Hodapp, D.; Wurl, O. High wind speeds prevent formation of a distinct bacterioneuston community in the sea-surface microlayer. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Franklin, M.P.; McDonald, I.R.; Bourne, D.G.; Owens, N.J.; Upstill-Goddard, R.C.; Murrell, J.C. Bacterial diversity in the bacterioneuston (sea surface microlayer): The bacterioneuston through the looking glass. Environ. Microbiol. 2005, 7, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Druzhkov, N.V.; Makarevich, P.R.; Bardan, S.I. Sea foam as an object of sea-surface film studies. Polar Res. 1997, 16, 117–121. [Google Scholar] [CrossRef]
- Maynard, N.G. Aquatic foams as an ecological habitat. Z. Allg. Mikrobiol. 1968, 8, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Stolle, C.; Labrenz, M.; Meeske, C.; Jürgens, K. Bacterioneuston community structure in the southern Baltic sea and its dependence on meteorological conditions. Appl. Environ. Microbiol. 2011, 77, 3726–3733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Nakai, R. Size Matters: Ultra-small and filterable microorganisms in the environment. Microbes Environ. 2020, 35, ME20025. [Google Scholar] [CrossRef]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.Y.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015, 6, 6372. [Google Scholar] [CrossRef]
- Bor, B.; Poweleit, N.; Bois, J.S.; Cen, L.; Bedree, J.K.; Zhou, Z.H.; Gunsalus, R.P.; Lux, R.; McLean, J.S.; He, X.; et al. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont Actinomyces. Microb. Ecol. 2016, 71, 243–255. [Google Scholar] [CrossRef]
- Mary, I.; Heywood, J.L.; Fuchs, B.M.; Amann, R.; Tarran, G.A.; Burkill, P.H.; Zubkov, M.V. SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic meridional transect. Aquat. Microb. Ecol. 2006, 45, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Eloe-Fadrosh, E.A.; Ivanova, N.N.; Woyke, T.; Kyrpides, N.C. Metagenomics uncovers gaps in amplicon-based detection of microbial diversity. Nat. Microbiol. 2016, 1, 15032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peura, S.; Eiler, A.; Bertilsson, S.; Nykanen, H.; Tiirola, M.; Jones, R.I. Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J. 2012, 6, 1640–1652. [Google Scholar] [CrossRef] [PubMed]
- Gies, E.A.; Konwar, K.M.; Beatty, J.T.; Hallam, S.J. Illuminating microbial dark matter in meromictic Sakinaw Lake. Appl. Environ. Microbiol. 2014, 80, 6807–6818. [Google Scholar] [CrossRef] [Green Version]
- Kadnikov, V.V.; Savvichev, A.S.; Mardanov, A.V.; Beletsky, A.V.; Merkel, A.Y.; Ravin, N.V.; Pimenov, N.V. Microbial communities involved in the methane cycle in the near-bottom water layer and sediments of the meromictic subarctic Lake Svetloe. Antonie Leeuwenhoek 2019, 112, 1801–1814. [Google Scholar] [CrossRef]
- Galand, P.E.; Bourrain, M.; De Maistre, E.; Catala, P.; Desdevises, Y.; Elifantz, H.; Kirchman, D.L.; Lebaron, P. Phylogenetic and functional diversity of Bacteria and Archaea in a unique stratified lagoon, the Clipperton atoll (N Pacific). FEMS Microbiol. Ecol. 2012, 79, 203–217. [Google Scholar] [CrossRef] [Green Version]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Grzymski, J.J.; Riesenfeld, C.S.; Williams, T.J.; Dussaq, A.M.; Ducklow, H.; Erickson, M.; Cavicchioli, R.; Murray, A.E. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012, 6, 1901–1915. [Google Scholar] [CrossRef]
- Grueneberg, J.; Engelen, A.H.; Costa, R.; Wichard, T. Macroalgal morphogenesis induced by waterborne compounds and bacteria in coastal seawater. PLoS ONE 2016, 11, e0146307. [Google Scholar] [CrossRef] [Green Version]
- Poretsky, R.S.; Sun, S.; Mou, X.; Moran, M.A. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 2010, 12, 616–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longford, S.R.; Tujula, N.A.; Crocetti, G.R.; Holmes, A.J.; Holmström, C.; Kjelleberg, S.; Steinberg, P.D.; Taylor, M.W. Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat. Microb. Ecol. 2007, 48, 217–229. [Google Scholar] [CrossRef]
- Liu, M.; Xiao, T.; Sun, J.; Wei, H.; Wu, Y.; Zhao, Y.; Zhang, W. Bacterial community structures associated with a natural spring phytoplankton bloom in the Yellow Sea, China. Deep Sea Res. Part II Top. Stud. Oceanogr. 2013, 97, 85–92. [Google Scholar] [CrossRef]
- Bik, E.M.; Costello, E.K.; Switzer, A.D.; Callahan, B.J.; Holmes, S.P.; Wells, R.S.; Carlin, K.P.; Jensen, E.D.; Venn-Watson, S.; Relman, D.A. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 2016, 7, 10516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Li, L.; Liu, J.; Zhang, M. Microbial structure and chemical components of aerosols caused by rotating brushes in a wastewater treatment plant. Environ. Sci. Pollut. Res. Int. 2012, 19, 4097–4108. [Google Scholar] [CrossRef]
- Lee, I.; Jang, G.I.; Cho, Y.; Yoon, S.J.; Pham, H.M.; Nguyen, A.V.; Lee, Y.M.; Park, H.; Rhee, T.S.; Kim, S.H.; et al. Sandaracinobacter neustonicus sp. nov., isolated from the sea surface microlayer in the Southwestern Pacific Ocean, and emended description of the genus Sandaracinobacter. Int. J. Syst. Evol. Microbiol. 2020, 70, 4698–4703. [Google Scholar] [CrossRef]
- Wurl, O.; Miller, L.; Vagle, S. Production and fate of transparent exopolymer particles in the ocean. J. Geophys Res. Oceans 2011, 116, C00H13. [Google Scholar] [CrossRef] [Green Version]
- Hunter, K. Processes affecting particulate trace metals in the sea surface microlayer. Mar. Chem. 1980, 9, 49–70. [Google Scholar] [CrossRef]
- Alldredge, A.L.; Cohen, Y. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science 1987, 235, 689–691. [Google Scholar] [CrossRef]
- Ploug, H.; Kühl, M.; Buchholz-Cleven, B.; Jørgensen, B.B. Anoxic aggregates—An ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 1997, 13, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Harold, E.; Schlichting, J.R. A preliminary study of the algae and protozoa in seafoam. Bot. Mar. 1971, 14, 24–28. [Google Scholar] [CrossRef]
- Roveillo, Q.; Dervaux, J.; Wang, Y.; Rouyer, F.; Zanchi, D.; Seuront, L.; Elias, F. Trapping of swimming microalgae in foam. J. R. Soc. Interface 2020, 17, 20200077. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Qing, Y.; Guo, X.; Warren, A. “Candidatus Sonnebornia yantaiensis”, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea). Syst. Appl. Microbiol. 2014, 37, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.J.; Elling, F.J.; Castelle, C.J.; Zhu, Q.; Elvert, M.; Birarda, G.; Holman, H.-Y.N.; Lane, K.R.; Ladd, B.; Ryan, M.C.; et al. Lipid analysis of CO2-rich subsurface aquifers suggests an autotrophy-based deep biosphere with lysolipids enriched in CPR bacteria. ISME J. 2020, 14, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Starr, E.P.; Shi, S.; Blazewicz, S.J.; Probst, A.J.; Herman, D.J.; Firestone, M.K.; Banfield, J.F. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 2018, 6, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bor, B.; Collins, A.J.; Murugkar, P.P.; Balasubramanian, S.; To, T.T.; Hendrickson, E.L.; Bedree, J.K.; Bidlack, F.B.; Johnston, C.D.; Shi, W.; et al. Insights obtained by culturing Saccharibacteria with their bacterial hosts. J. Dent. Res. 2020, 99, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Cross, K.L.; Campbell, J.H.; Balachandran, M.; Campbell, A.G.; Cooper, S.J.; Griffen, A.; Heaton, M.; Joshi, S.; Klingeman, D.; Leys, E.; et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 2019, 37, 1314–1321. [Google Scholar] [CrossRef]
- Kindaichi, T.; Yamaoka, S.; Uehara, R.; Ozaki, N.; Ohashi, A.; Albertsen, M.; Nielsen, P.H.; Nielsen, J.L. Phylogenetic diversity and ecophysiology of Candidate phylum Saccharibacteria in activated sludge. FEMS Microbiol. Ecol. 2016, 92, fiw078. [Google Scholar] [CrossRef] [Green Version]
- Bärlocher, F.; Gordon, J.; Ireland, R.J. Organic composition of seafoam and its digestion by Corophium volutator (Pallas). J. Exp. Mar. Biol. Ecol. 1988, 115, 179–186. [Google Scholar] [CrossRef]
- Wegner, C.; Hamburger, M. Occurrence of stable foam in the upper Rhine River caused by plant-derived surfactants. Environ. Sci. Technol. 2002, 36, 3250–3256. [Google Scholar] [CrossRef]
- Velimirov, B. Sugar and lipid components in sea foam near kelp beds. Mar. Ecol. 1982, 3, 97–107. [Google Scholar] [CrossRef]
- Ram, A.S.P.; Mari, X.; Brune, J.; Torreton, J.P.; Chu, V.T.; Raimbault, P.; Niggemann, J.; Sime-Ngando, T. Bacterial-viral interactions in the sea surface microlayer of a black carbon-dominated tropical coastal ecosystem (Halong Bay, Vietnam). Elementa Sci. Anthrop. 2018, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Jeffrey, W.H.; Kase, J.P.; Wilhelm, S.W. UV Radiation effects on heterotrophic bacterioplankton and viruses in marine ecosystems. In The effects of UV Radiation in the Marine Environment; Cambridge University Press: Cambridge, UK, 2000; Volume 10, pp. 206–236. [Google Scholar] [CrossRef]
- Agogué, H.; Joux, F.; Obernosterer, I.; Lebaron, P. Resistance of marine bacterioneuston to solar radiation. Appl. Environ. Microbiol. 2005, 71, 5282–5289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tranvik, L.; Olofsson, H.; Bertilsson, S. Photochemical effects on bacterial degradation of dissolved organic matter in lake water. In Proceedings of the Microbial Biosystems: New Frontiers, the 8th International Symposium on Microbial Ecology, Halifax, NS, Canada, 9–14 August 1999. [Google Scholar]
- Bor, B.; McLean, J.S.; Foster, K.R.; Cen, L.; To, T.T.; Serrato-Guillen, A.; Dewhirst, F.E.; Shi, W.; He, X. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl. Acad. Sci. USA 2018, 115, 12277–12282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utter, D.R.; He, X.; Cavanaugh, C.M.; McLean, J.S.; Bor, B. The saccharibacterium TM7x elicits differential responses across its host range. ISME J. 2020, 14, 3054–3067. [Google Scholar] [CrossRef] [PubMed]
- Eisenreich, S.J.; Elzerman, A.W.; Armstrong, D.E. Enrichment of micronutrients, heavy metals, and chlorinated hydrocarbons in wind-generated lake foam. Environ. Sci. Technol. 1978, 12, 413–417. [Google Scholar] [CrossRef]
- Kuznetsova, M.; Lee, C. Dissolved free and combined amino acids in nearshore seawater, sea surface microlayers and foams: Influence of extracellular hydrolysis. Aquat. Sci. 2002, 64, 252–268. [Google Scholar] [CrossRef]
- Craig, D.; Ireland, R.J.; Bärlocher, F. Seasonal variation in the organic composition of seafoam. J. Exp. Mar. Biol. Ecol. 1989, 130, 71–80. [Google Scholar] [CrossRef]
- Wurl, O.; Obbard, J.P. A review of pollutants in the sea-surface microlayer (SML): A unique habitat for marine organisms. Mar. Pollut. Bull. 2004, 48, 1016–1030. [Google Scholar] [CrossRef]
- Santos, A.L.; Lopes, S.; Baptista, I.; Henriques, I.; Gomes, N.C.; Almeida, A.; Correia, A.; Cunha, A. Diversity in UV sensitivity and recovery potential among bacterioneuston and bacterioplankton isolates. Lett. Appl. Microbiol. 2011, 52, 360–366. [Google Scholar] [CrossRef] [Green Version]
- Robinson, T.B.; Wurl, O.; Bahlmann, E.; Jürgens, K.; Stolle, C. Rising bubbles enhance the gelatinous nature of the air–sea interface. Limnol. Oceanogr. 2019, 64, 2358–2372. [Google Scholar] [CrossRef]
- Hardy, J.T. The sea surface microlayer: Biology, chemistry and anthropogenic enrichment. Prog. Oceanogr. 1982, 11, 307–328. [Google Scholar] [CrossRef]
- Aller, J.Y.; Kuznetsova, M.R.; Jahns, C.J.; Kemp, P.F. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J. Aerosol. Sci. 2005, 36, 801–812. [Google Scholar] [CrossRef]
- Rahlff, J. The virioneuston: A review on viral-bacterial associations at air-water interfaces. Viruses 2019, 11, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaud, J.M.; Thompson, L.R.; Kaul, D.; Espinoza, J.L.; Richter, A.R.; Xu, Z.Z.; Lee, C.; Pham, K.M.; Beall, C.M.; Malfatti, F.; et al. Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat. Commun. 2018, 9, 2017. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, C.; Hagstrom, A.; Nilsson, D.; Zweifel, U.L. Annual variations in the diversity, viability, and origin of airborne bacteria. Appl. Environ. Microbiol. 2010, 76, 3015–3025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rastelli, E.; Corinaldesi, C.; Dell’Anno, A.; Lo Martire, M.; Greco, S.; Cristina Facchini, M.; Rinaldi, M.; O’Dowd, C.; Ceburnis, D.; Danovaro, R. Transfer of labile organic matter and microbes from the ocean surface to the marine aerosol: An experimental approach. Sci. Rep. 2017, 7, 11475. [Google Scholar] [CrossRef]
- Fleck, R.; Gill, R.L.; Pettit, T.; Irga, P.J.; Williams, N.L.R.; Seymour, J.R.; Torpy, F.R. Characterisation of fungal and bacterial dynamics in an active green wall used for indoor air pollutant removal. Build. Environ. 2020, 179, 106987. [Google Scholar] [CrossRef]
- Mortazavi, R.; Attiya, S.; Ariya, P.A. Arctic microbial and next-generation sequencing approach for bacteria in snow and frost flowers: Selected identification, abundance and freezing nucleation. Atmos. Chem. Phys. 2015, 15, 6183–6204. [Google Scholar] [CrossRef] [Green Version]
- Møller, A.K.; Søborg, D.A.; Abu Al-Soud, W.; Sørensen, S.J.; Kroer, N. Bacterial community structure in High-Arctic snow and freshwater as revealed by pyrosequencing of 16S rRNA genes and cultivation. Polar Res. 2013, 32, 17390. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Amores-Arrocha, H.; Malard, L.A.; Els, N.; Sattler, B.; Pearce, D.A. Characterisation of Arctic bacterial communities in the air above Svalbard. Biology 2017, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.R.L. A beach structure due to wind-driven foam. J. Sediment. Res. 1967, 37, 691–692. [Google Scholar] [CrossRef]
- Jenkinson, I.R.; Laurent, S.; Ding, H.; Elias, F. Biological modification of mechanical properties of the sea surface microlayer, influencing waves, ripples, foam and air-sea fluxes. Elem. Sci. Anth. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
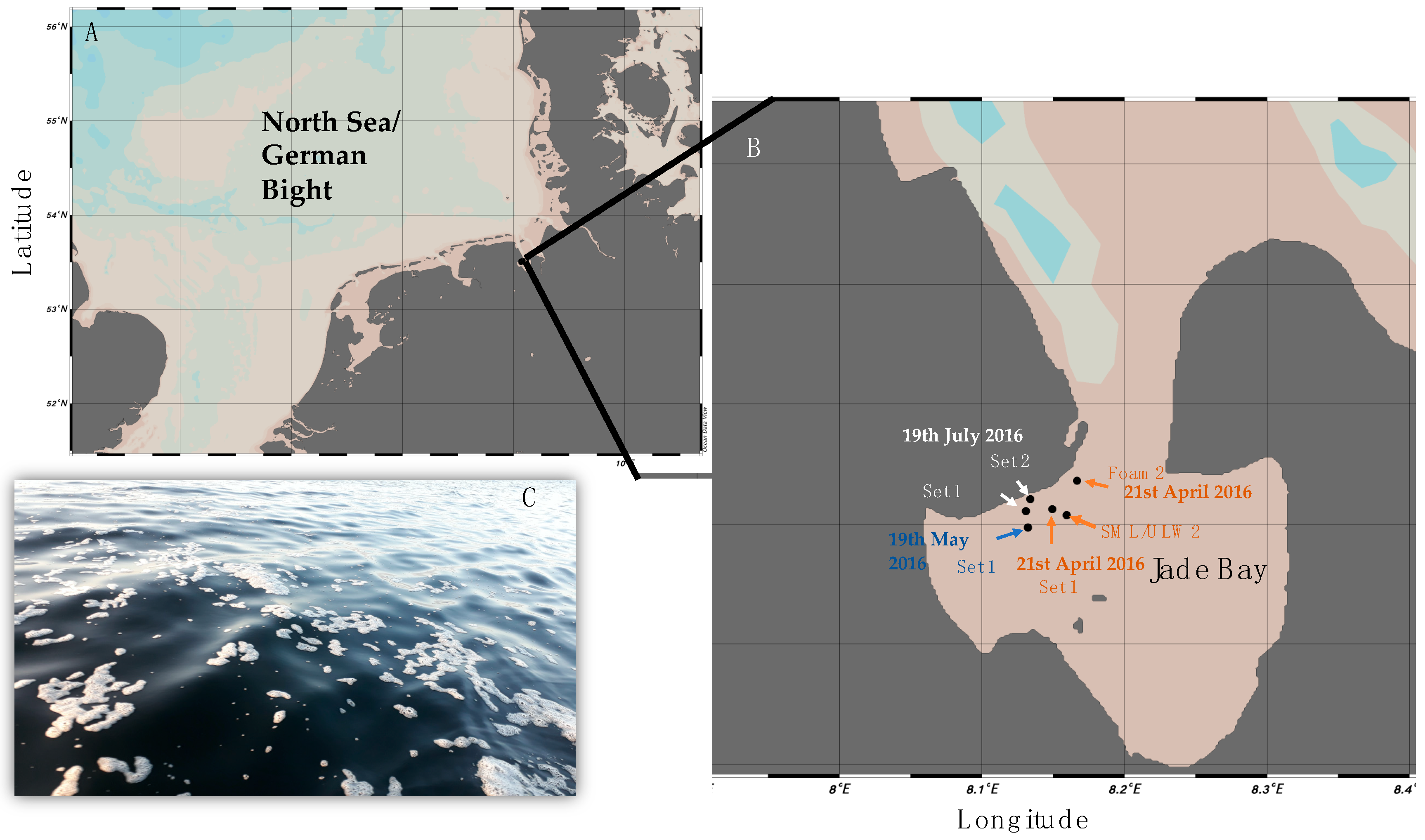
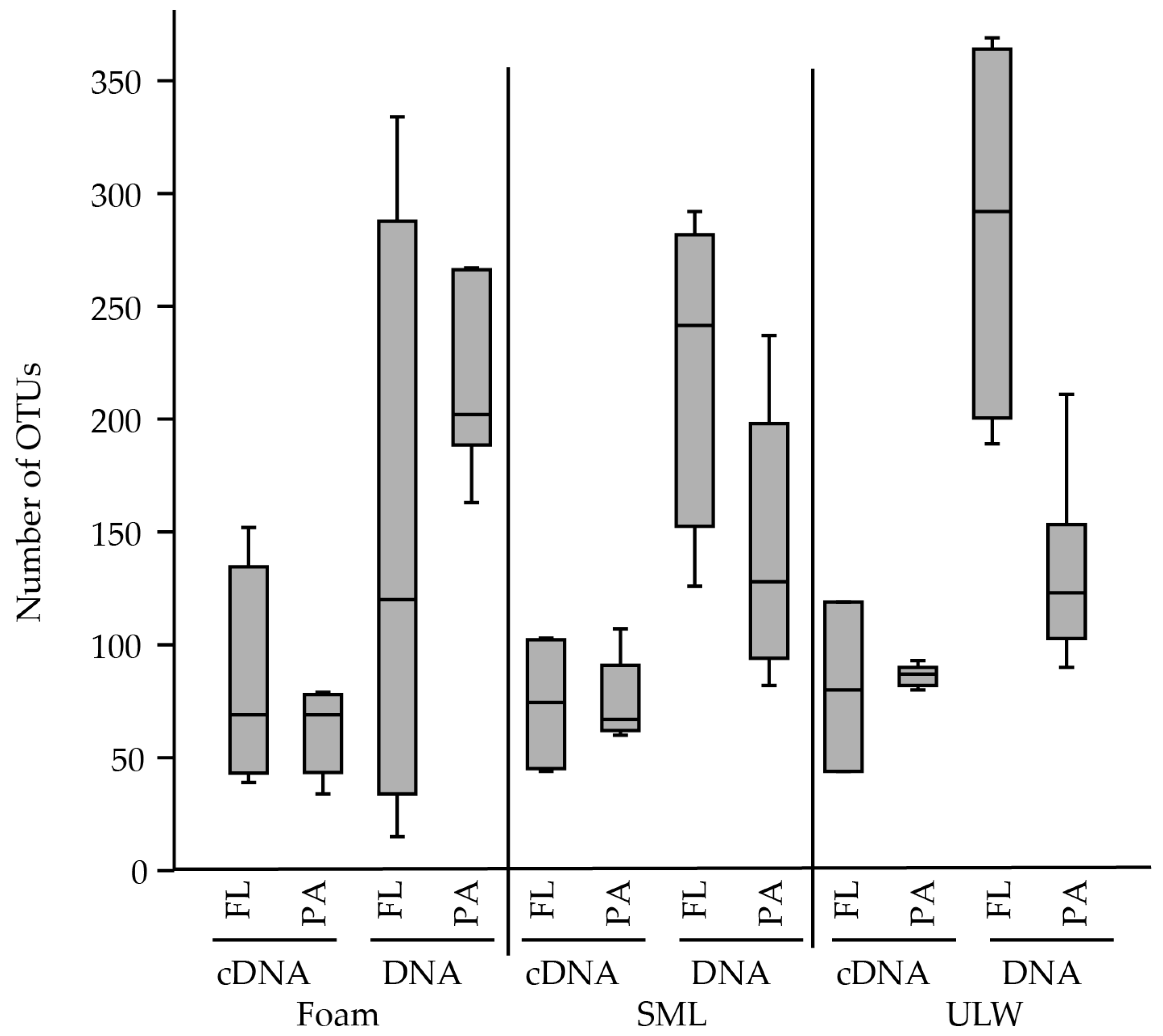
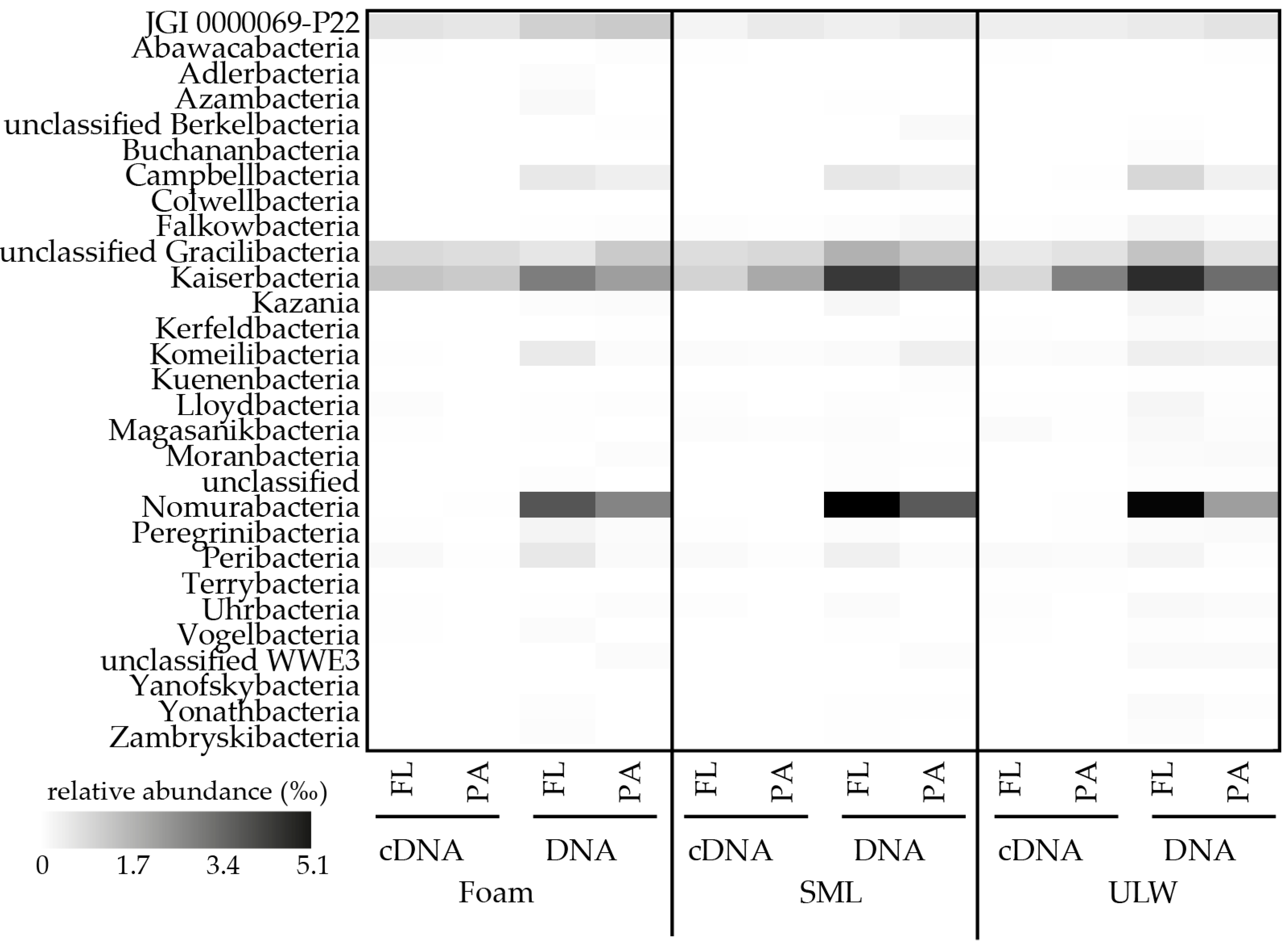

| NCBI Accession # of Sequences from This Study | NCBI Accession # of Phylogenetic Neighbor from Surface Water/Aerosol | Sequence Name | Origin | Phylogenetic Affiliation | Reference |
|---|---|---|---|---|---|
| MW167728, MW167751 | HQ691922 | Uncultured bacterium, 1327, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | JGI 0000069−P22 | [60] |
| MW167732, MW167737, MW167759, MW167763 | HQ691923 | Uncultured bacterium, 1317, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | JGI 0000069−P22 | [60] |
| MW167705, MW167707, MW167722 | HQ691924 | Uncultured bacterium, 1276, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Peregrinibacteria | [60] |
| MW167672, MW167678, MW167694 | HQ691925 | Uncultured bacterium, 1288, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Peribacteria | [60] |
| MW167669, MW167680, MW167690 | HQ691926 | Uncultured bacterium, 1318, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Peribacteria | [60] |
| MW167660, MW167688, MW167703 | HQ691927 | Uncultured bacterium, 1311, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Peribacteria | [60] |
| MW167675, MW167687, MW167717 | HQ691928 | Uncultured bacterium, 1323, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Buchananbacteria | [60] |
| MW167683, MW167684, MW167710, MW167736 | HQ691929 | Uncultured bacterium, 1317, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Moranbacteria | [60] |
| MW167686, MW167713 | HQ691930 | Uncultured bacterium, 1328, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Magasanikbacteria | [60] |
| MW167715, MW167719, MW167721 | HQ691931 | Uncultured bacterium, 1359, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Magasanikbacteria | [60] |
| MW167673, MW167689 | HQ691932 | Uncultured bacterium, 1323, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Komeilibacteria | [60] |
| MW167674, MW167704, MW167711 | HQ691933 | Uncultured bacterium, 1333, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Komeilibacteria | [60] |
| MW167671, MW167699 | HQ691934 | Uncultured bacterium, 1395, stratified lagoon | Clipperton Island atoll, North Pacific Ocean, Meromictic lagoon, marine | Cand. Falkowbacteria | [60] |
| MW167663, MW167666 | AACY023814357 | Marine metagenome, 1412, predominantly from surface water marine samples | Surface water samples, off the coast of Bermuda, marine | Cand. Falkowbacteria | [61] |
| MW167720 | AACY023758110 | Marine metagenome, 1295, predominantly from surface water marine samples | Surface water samples, off the coast of Bermuda, marine | Cand. Kuenenbacteria | [61] |
| MW167725, MW167733, MW167749 | AACY023749576 | Marine metagenome, 1409, predominantly from surface water marine samples | Surface water samples, off the coast of Bermuda, marine | JGI 0000069−P22 | [61] |
| MW167667, MW167676, MW167679, MW167691 | AACY020292957 | Marine metagenome, 1318, predominantly from surface water marine samples | Surface water samples, off the coast of Bermuda, marine | Cand. Kaiserbacteria | [61] |
| MW167670, MW167692, MW167695, MW167696 | AACY023772748 | Marine metagenome, 1245, predominantly from surface water marine samples | Surface water samples, off the coast of Bermuda, marine | Cand. Gracilibacteria | [61] |
| MW167731, MW167744, MW167765 | GU235593 | Uncultured marine bacterium, 1328, Antarctic sea water collected from 5m | Antarctic sea water collected from 5 m, coastal surface waters at Palmer Station, on the west coast of the Antarctic Peninsula | JGI 0000069−P22 | [62] |
| MW167727, MW167734, MW167735, MW167747, MW167748, MW167756 | GU234860 | Uncultured marine bacterium, 1316, Antarctic sea water collected from 5m | Antarctic sea water collected from 5 m, coastal surface waters at Palmer Station, on the west coast of the Antarctic Peninsula | JGI 0000069−P22 | [62] |
| MW167726, MW167740 | LN681284 | Bacterium RFB D08, 1233, marine alga | Ria Formosa tidal pools close to the Ramalhete Marine Station, marine | JGI 0000069−P22 | [63] |
| MW167668, MW167685, MW167700 | FJ744790 | Uncultured bacterium, 1397, surface water at the UGA Marine Institute | Coastal water samples were collected high tide from Sapelo Island, GA, marine | Cand. Gracilibacteria | [64] |
| MW167677, MW167701, MW167716 | DQ269061 | Uncultured bacterium, 1342, surface of marine macro−alga | One of three sampling sites, Sydney, Australia, marine | Cand. Gracilibacteria | [65] |
| MW167739, MW167743, MW167757 | FJ826108 | Uncultured marine bacterium, 1418, filtered surface sea water | Filtered surface sea water in the decay period after diatom bloom in the Yellow Sea, marine | Cand. Peregrinibacteria | [66] |
| MW167739, MW167743, MW167757 | FJ826198 | Uncultured marine bacterium, 1442, filtered surface sea water | Filtered surface sea water in the decay period after diatom bloom in the Yellow Sea, marine | Cand. Peregrinibacteria | [66] |
| MW167681 | KU578668 | Uncultured bacterium, 1369, ocean water | Marine | Cand. Magasanikbacteria | unpublished |
| MW167681, MW167697, MW167702, MW167712 | FLOH01000114 | Marine metagenome, 1479, water | Marine | Cand. Magasanikbacteria | not specified |
| MW167664, MW167714, MW167718 | CEVN01160041 | Marine metagenome, 1440, saline water (ENVO:00002010), including plankton (ENVO:xxxxxxxx) | Marine | Cand. Kaiserbacteria | not specified |
| MW167661, MW167665, MW167698 | JQ197106 | Uncultured bacterium, 1330, seawater; next to dolphin E | Marine | Cand. Nomurabacteria | [67] |
| MW167693, MW167708, MW167709 | JQ198499 | Uncultured bacterium, 1329, seawater; next to dolphin K | Marine | Cand. Nomurabacteria | [67] |
| MW167745, MW167746, MW167753 | JN981903 | Uncultured beta proteobacterium, 1495, aerosols from orbal oxidation ditch in a municipal WWTP | Aerosols from orbal oxidation ditch in a municipal WWTP | Cand. Gracilibacteria | [68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahlff, J.; Giebel, H.-A.; Stolle, C.; Wurl, O.; Probst, A.J.; Herlemann, D.P.R. Overlooked Diversity of Ultramicrobacterial Minorities at the Air-Sea Interface. Atmosphere 2020, 11, 1214. https://doi.org/10.3390/atmos11111214
Rahlff J, Giebel H-A, Stolle C, Wurl O, Probst AJ, Herlemann DPR. Overlooked Diversity of Ultramicrobacterial Minorities at the Air-Sea Interface. Atmosphere. 2020; 11(11):1214. https://doi.org/10.3390/atmos11111214
Chicago/Turabian StyleRahlff, Janina, Helge-Ansgar Giebel, Christian Stolle, Oliver Wurl, Alexander J. Probst, and Daniel P. R. Herlemann. 2020. "Overlooked Diversity of Ultramicrobacterial Minorities at the Air-Sea Interface" Atmosphere 11, no. 11: 1214. https://doi.org/10.3390/atmos11111214
APA StyleRahlff, J., Giebel, H.-A., Stolle, C., Wurl, O., Probst, A. J., & Herlemann, D. P. R. (2020). Overlooked Diversity of Ultramicrobacterial Minorities at the Air-Sea Interface. Atmosphere, 11(11), 1214. https://doi.org/10.3390/atmos11111214






