Virus-Like Particle Production in Atmospheric Eubacteria Isolates
Abstract
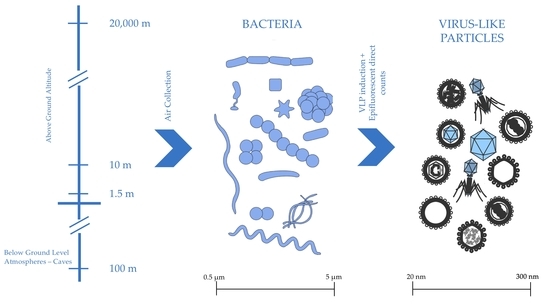
1. Introduction
2. Experiments
2.1. Origin of Isolates
2.2. Isolate Collection Assay
2.3. Mitomycin C Induction Plate Setup
2.4. Epifluorescent Direct Counts
2.5. Statistics
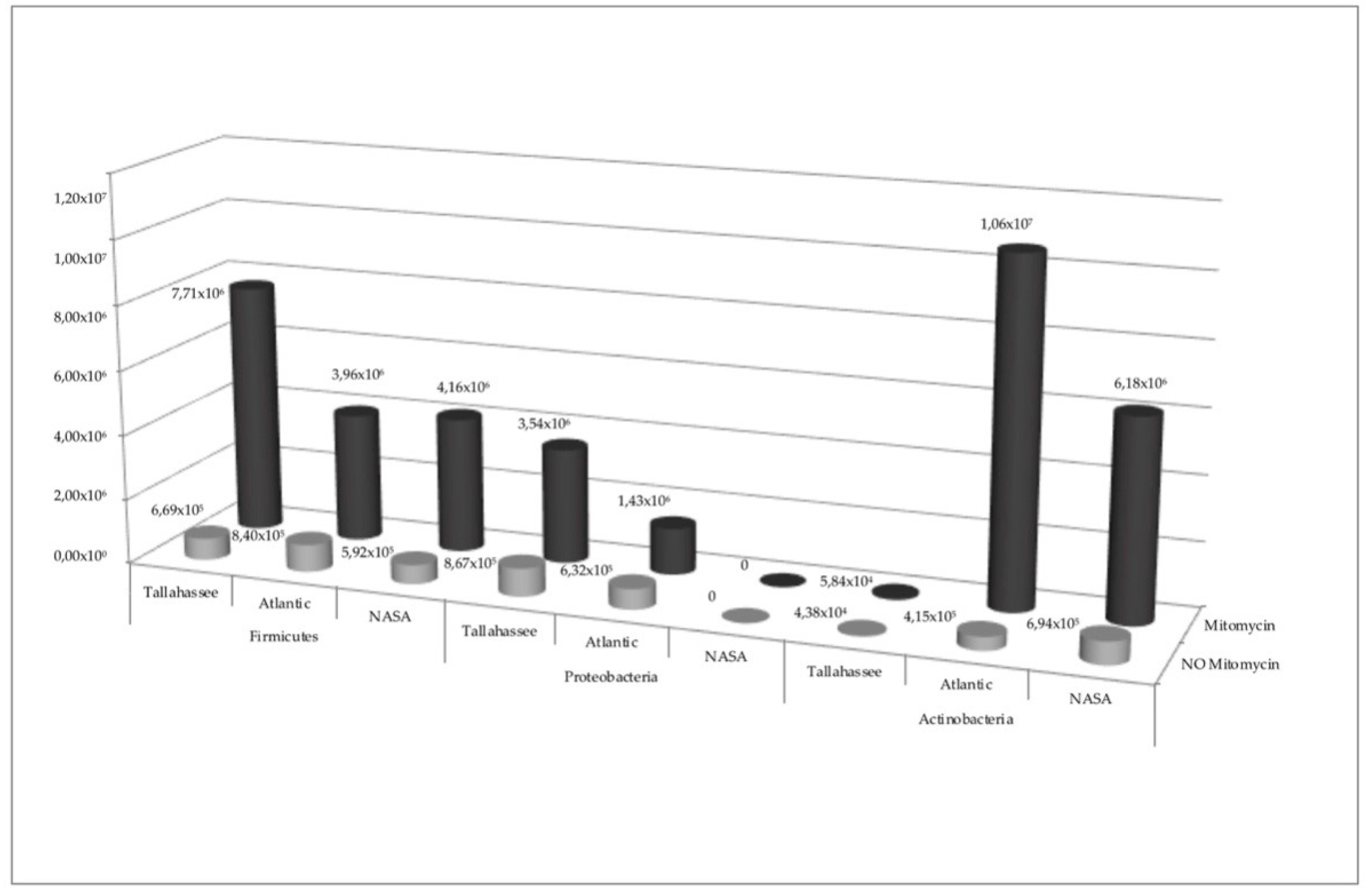
3. Results
Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Bergh, O.; Borsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Ashelford, K.E.; Day, M.J.; Bailey, M.J.; Lilley, A.K.; Fry, J.C. In Situ Population Dynamics of Bacterial Viruses in a Terrestrial Environment. Appl. Environ. Microbiol. 1999, 65, 169–174. [Google Scholar] [PubMed]
- Gonzalez-Martin, C.; Teigell-Perez, N.; Lyles, M.; Valladares, B.; Griffin, D.W.D.W. Epifluorescent direct counts of bacteria and viruses from topsoil of various desert dust storm regions. Res. Microbiol. 2013, 164, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Srinivasiah, S.; Bhavsar, J.; Thapar, K.; Liles, M.; Schoenfeld, T.; Wommack, K.E. Phages across the biosphere: Contrasts of viruses in soil and aquatic environments. Res. Microbiol. 2008, 159, 349–357. [Google Scholar] [CrossRef]
- Williamson, K.E.; Radosevich, M.; Smith, D.W.; Wommack, K.E. Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 2007, 9, 2563–2574. [Google Scholar] [CrossRef]
- Breitbart, M.; Wegley, L.; Leeds, S.; Schoenfeld, T.; Rohwer, F. Phage community dynamics in hot springs. Appl. Environ. Microbiol. 2004, 70, 1633–1640. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef]
- Forterre, P.; Soler, N.; Krupovic, M.; Marguet, E.; Ackermann, H.-W. Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 2013, 21, 1–5. [Google Scholar] [CrossRef]
- Biers, E.J.; Wang, K.; Pennington, C.; Belas, R.; Chen, F.; Moran, M.A. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl. Environ. Microbiol. 2008, 74, 2933–2939. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, L.D.; Young, E.; Delaney, J.; Ruhnau, F.; Ritchie, K.B.; Paul, J.H. High frequency of horizontal gene transfer in the oceans. Science 2010, 330, 50. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Paul, J.H. Significance of Lysogeny in the Marine Environment: Studies with Isolates and a Model of Lysogenic Phage Production. Microb. Ecol. 1998, 35, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Muhling, M.; Fuller, N.J.; Millard, A.; Somerfield, P.J.; Marie, D.; Wilson, W.H.; Scanlan, D.J.; Post, A.F.; Joint, I.; Mann, N.H. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: Evidence for viral control of phytoplankton. Environ. Microbiol. 2005, 7, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.H.; Sullivan, M.B. Marine phage genomics: What have we learned? Curr. Opin. Biotechnol. 2005, 16, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G.; Rassoulzadegan, F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 2004, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sano, E.; Carlson, S.; Wegley, L.; Rohwer, F. Movement of Viruses between Biomes. Appl. Environ. Microbiol. 2004, 70, 5842–5846. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Evolution and Ecology. Adv. Appl. Microbiol. 2009, 67, 1–45. [Google Scholar]
- Ackermann, H.; DuBow, M.S. Viruses of Prokaryotes. Vol. 1, General Properties of Bacteriophages; CRC: Boca Raton, FL, USA, 1987. [Google Scholar]
- Lamont, I.; Brumby, A.M.; Egan, J.B. UV induction of coliphage 186: Prophage induction as an SOS function. Proc. Natl. Acad. Sci. USA 1989, 86, 5492–5496. [Google Scholar] [CrossRef]
- Boyd, E.F.; Waldor, M.K. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: Generalized transduction of CTXPhi by bacteriophage CP-T1. Infect. Immun. 1999, 67, 5898–5905. [Google Scholar]
- Mlynarczyk, G.; Mlynarczyk, A.; Zabicka, D.; Jeljaszewicz, J. Lysogenic conversion as a factor influencing the vancomycin tolerance phenomenon in Staphylococcus aureus. J. Antimicrob. Chemother. 1997, 40, 136–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roossinck, M.J. The good viruses: Viral mutualistic symbioses. Nat. Rev. Microbiol. 2011, 9, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Yaron, S.; Kolling, G.L.; Simon, L.; Matthews, K.R. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420. [Google Scholar] [CrossRef] [PubMed]
- Schooling, S.R.; Hubley, A.; Beveridge, T.J. Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 2009, 191, 4097–4102. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A.; Suttle, C.A. Viruses in Marine Planktonic Systems. Oceanography 1993, 6, 51–63. [Google Scholar] [CrossRef]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brussow, H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Paul, J.H. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar. Ecol. Prog. Ser. 1994, 104, 163–172. [Google Scholar] [CrossRef]
- Curtis, T.P.; Sloan, W.T.; Scannell, J.W. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 2002, 99, 10494–10499. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Suttle, C.A. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the gulf of Mexico. Appl. Environ. Microbiol. 1996, 62, 4374–4380. [Google Scholar]
- Williamson, S.J.; Houchin, L.A.; Mcdaniel, L.; Paul, J.H. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 2002, 68, 4307–4314. [Google Scholar] [CrossRef]
- Prestel, E.; Salamitou, S.; DuBow, M.S. An examination of the bacteriophages and bacteria of the Namib desert. J. Microbiol. 2008, 46, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Sawstrom, C.; Lisle, J.; Anesio, A.M.; Priscu, J.C.; Laybourn-Parry, J. Bacteriophage in polar inland waters. Extremophiles 2008, 12, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Prigent, M.; Leroy, M.; Confalonieri, F.; Dutertre, M.; DuBow, M.S. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles 2005, 9, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Paul, J. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 1996, 142, 27–38. [Google Scholar] [CrossRef]
- Griffin, D.W.D.W. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 2007, 20, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Vettori, C.; Gallori, E.; Stotzky, G. Clay minerals protect bacteriophage PBS1 of Bacillus subtilis against inactivation and loss of transducing ability by UV radiation. Can. J. Microbiol. 2000, 46, 770–773. [Google Scholar] [CrossRef]
- Griffin, D.W.D.W.; Garrison, V.H.V.H.; Herman, J.R.; Shinn, E.A. African desert dust in the Caribbean atmosphere: Microbiology and public health. Aerobiologia (Bologna) 2001, 17, 203–213. [Google Scholar] [CrossRef]
- De Deckker, P.; Abed, R.M.M.; de Beer, D.; Hinrichs, K.U.; O’Loingsigh, T.; Schefufl, E.; Stuut, J.B.W.; Tapper, N.J.; van der Kaars, S. Geochemical and microbiological fingerprinting of airborne dust that fell in Canberra, Australia, in October 2002. Geochem. Geophys. Geosyst. 2008, 9, Q12Q10. [Google Scholar] [CrossRef]
- Favet, J.; Lapanje, A.; Giongo, A.; Kennedy, S.; Aung, Y.-Y.; Cattaneo, A.; Davis-Richardson, A.G.; Brown, C.T.; Kort, R.; Brumsack, H.-J.; et al. Microbial hitchhikers on intercontinental dust: Catching a lift in Chad. ISME J. 2013, 7, 850–867. [Google Scholar] [CrossRef]
- Gorbushina, A.A.; Kort, R.; Schulte, A.; Lazarus, D.; Schnetger, B.; Brumsack, H.-J.; Broughton, W.J.; Favet, J. Life in Darwin’s dust: Intercontinental transport and survival of microbes in the nineteenth century. Environ. Microbiol. 2007, 9, 2911–2922. [Google Scholar] [CrossRef]
- Smith, D.J.; Jaffe, D.A.; Birmele, M.N.; Griffin, D.W.; Schuerger, A.C.; Hee, J.; Roberts, M.S. Free tropospheric transport of microorganisms from Asia to North America. Microb. Ecol. 2012, 64, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Timonen, H.J.; Jaffe, D.A.; Griffin, D.W.; Birmele, M.N.; Perry, K.D.; Ward, P.D.; Roberts, M.S. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl. Environ. Microbiol. 2013, 79, 1134–1139. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Ravichandar, J.D.; Jain, S.; Griffin, D.W.; Yu, H.; Tan, Q.; Thissen, J.; Lusby, T.; Nicoll, P.; Shedler, S.; et al. Airborne bacteria in earth’s lower stratosphere resemble taxa detected in the troposphere: Results from a new NASA Aircraft Bioaerosol Collector (ABC). Front. Microbiol. 2018, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.W.; Gonzalez, C.; Teigell, N.; Petrosky, T.; Northup, D.E.; Lyles, M. Observations on the use of membrane filtration and liquid impingement to collect airborne microorganisms in various atmospheric environments. Aerobiologia (Bologna) 2011, 27, 25–35. [Google Scholar] [CrossRef]
- Griffin, D.W.; Westphal, D.L.; Gray, M.A. Airborne microorganisms in the African desert dust corridor over the mid-Atlantic ridge, Ocean Drilling Program, Leg 209. Aerobiologia 2006, 22, 211–226. [Google Scholar] [CrossRef]
- Griffin, D.W. Non-spore forming eubacteria isolated at an altitude of 20,000 m in Earth’s atmosphere: Extended incubation periods needed for culture-based assays. Aerobiologia (Bologna) 2008, 24, 19–25. [Google Scholar] [CrossRef]
- Griffin, D.W. Terrestrial microorganisms at an altitude of 20,000 m in Earth ’ s atmosphere. Aerobiologia (Bologna) 2004, 20, 135–140. [Google Scholar] [CrossRef]
- Smith, D.J.; Griffin, D.W.; Schuerger, A.C. Stratospheric microbiology at 20 km over the Pacific Ocean. Aerobiologia (Bologna) 2010, 26, 35–46. [Google Scholar] [CrossRef]
- Griffin, D.W.; Gray, M.A.; Lyles, M.B.; Northup, D.E. The Transport of Nonindigenous Microorganisms Into Caves by Human The Transport of Nonindigenous Microorganisms Into Caves by Human Visitation: A Case Study at Carlsbad Caverns National Park. Geomicrobiol. J. 2014, 31, 175–185. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef]
- NCBI (National Center for Biotechnology Information). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 8 August 2010).
- Noble, R.T.; Fuhrman, J.A. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 1997, 63, 77–83. [Google Scholar] [PubMed]
- Mercanti, D.J.; Carminati, D.; Reinheimer, J.A.; Quiberoni, A. Widely distributed lysogeny in probiotic lactobacilli represents a potentially high risk for the fermentative dairy industry. Int. J. Food Microbiol. 2011, 144, 503–510. [Google Scholar] [CrossRef] [PubMed]
| Isolate (Date Collected-ID) | Location | Altitude (m) | Taxonomy (GenBank Closest Neighbor) | GenBank Accession Number |
|---|---|---|---|---|
| 07/06/10-A | Tallahassee | 1.5 | Bacillus gibsonii | HE604,338 |
| 07/06/10-B | Tallahassee | 1.5 | Bacillus megaterium | HE604,339 |
| 07/06/10-C | Tallahassee | 1.5 | Bacillus sp. | HE604,340 |
| 07/12/10-A | Tallahassee | 1.5 | Bacillus sp. | HE604,341 |
| 07/12/10-B | Tallahassee | 1.5 | Bacillus sp. | HE604,342 |
| 07/12/10-C | Tallahassee | 1.5 | Enterobacteriaceae | HE604,343 |
| 07/12/10-D | Tallahassee | 1.5 | Enterobacteriaceae | HE604,344 |
| 07/12/10-E | Tallahassee | 1.5 | Bacillus sp. | HE604,342 |
| 07/12/10-F | Tallahassee | 1.5 | Bacillus sp. | HE604,346 |
| 07/13/10-A | Tallahassee | 1.5 | Bacillus sp. | HE604,347 |
| 07/13/10-B | Tallahassee | 1.5 | Bacillus sp. | HE604,348 |
| 07/13/10-C | Tallahassee | 1.5 | Exiguobacterium sp. | HE604,349 |
| 07/13/10-D | Tallahassee | 1.5 | Bacillus sp. | HE604,350 |
| 07/14/10-A | Tallahassee | 1.5 | Bacillus sp. | HE604,345 |
| 07/14/10-B | Tallahassee | 1.5 | Bacillus sp. | HE604,342 |
| 07/15/12-E | Tallahassee | 1.5 | Pseudomonas sp. | HE995,774 |
| 07/15/12-G | Tallahassee | 1.5 | Microccocus sp. | HE995,775 |
| 05/25/03-BY0 | mid-Atlantic | 10 | Actinobacteria | AY857,677 |
| 05/25/03-BY1 | mid-Atlantic | 10 | Frigoribacterium | AY857,767 |
| 05/26/03-BW0 | mid-Atlantic | 10 | Kocuria rosea | AY857,672 |
| 05/27/03-BY0 | mid-Atlantic | 10 | Lentzea sp. | AY857,673 |
| 06/01/03-BY0 | mid-Atlantic | 10 | Novosphingobium subarticum | AY857,675 |
| 06/12/03-BY0 | mid-Atlantic | 10 | Brevibacterium casei | AY857,665 |
| 06/12/03-BY1 | mid-Atlantic | 10 | Staphylococcus epidermis | AY857,685 |
| 06/12/03-BY2 | mid-Atlantic | 10 | Liefsonia sp. | AY857,676 |
| 06/15/03-BY0 | mid-Atlantic | 10 | Bacillus aminovorans | AY857,666 |
| 06/19/03-BC2 | mid-Atlantic | 10 | Bacillus benzoevorans | AY857,668 |
| 06/28/03-BP1 | mid-Atlantic | 10 | Gordonia terrae | AY857,719 |
| 06/28/03-BY0 | mid-Atlantic | 10 | Pseudomonas sp. | AY857,688 |
| 07/01/03-BW1 | mid-Atlantic | 10 | Bacillus aminovorans | AY857,722 |
| 07/01/03-BW0 | mid-Atlantic | 10 | Bacillus sp. | AY857,721 |
| NASA1-1 | Continental USA | 20,000 | Bacillus luciferensis | AY291,461 |
| NASA1-71 | Continental USA | 20,000 | Bacillus sphaericus | AY291,474 |
| 08/13/04-NASA2-8 | Continental USA | 20,000 | Micrococcaceae | EU029,597 |
| 08/13/04-NASA2-25 | Continental USA | 20,000 | Staphylococcus sp. | EU029,614 |
| 08/13/04-NASA2-33 | Continental USA | 20,000 | Micrococcus luteus | EU029,622 |
| 08/13/04-NASA2-34 | Continental USA | 20,000 | Micrococcus thailand | EU029,623 |
| 08/13/04-NASA2-43 | Continental USA | 20,000 | Brevibacterium sp. | EU029,632 |
| 04/28/08-NASA-DS1 | Pacific | 20,000 | Bacillus endophyticus | FJ649,336 |
| 04/28/08-NASA-DS3 | Pacific | 20,000 | Bacillus sp. | FJ649,338 |
| SPG PP1 | Carlsbad Caverns | 100 below surface | Myceligenerans crystallogenes | HE995776 |
| Collection Location | Samples ID | A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|---|---|
| Tallahassee samples | 070,610 A | X | X | X | X | X | X | X | |
| 070,610 B | X | X | X | X | X | X | |||
| 070,610 C | X | X | X | X | |||||
| 071,210 B | X | X | X | X | X | ||||
| 071,210 A | X | X | X | X | X | X | X | ||
| 071,210 C | X | X | X | X | X | X | X | ||
| 071,210 D | X | X | X | X | |||||
| 071,210 E | X | X | X | X | X | X | |||
| 071,210 F | X | X | X | X | X | X | X | ||
| 071,310 A | X | X | X | X | X | X | X | ||
| 071,310 B | X | X | X | X | X | ||||
| 071,310 C | X | X | X | X | X | ||||
| 071,310 D | X | X | X | X | |||||
| 071,410 A | X | X | X | X | X | ||||
| 071,410 B | X | X | X | X | X | X | |||
| 071,512 E | X | X | X | X | X | X | |||
| 071,512 G | X | X | X | X | X | X | |||
| Atlantic samples | 052,503 BY0 | X | X | X | X | X | X | ||
| 052,503 BY1 | X | X | X | X | X | X | |||
| 052,603 BW0 | X | X | X | X | |||||
| 052,703 BY0 | X | X | X | X | X | X | |||
| 060,103 BY0 | X | X | X | X | |||||
| 061,203 BY0 | X | X | X | X | X | X | X | ||
| 061,203 BY1 | X | X | X | X | X | X | X | ||
| 061,203 BY2 | X | X | X | X | X | ||||
| 061,503 BY0 | X | X | X | X | |||||
| 061,903 BC2 | X | X | X | X | |||||
| 062,803 BP1 | X | X | X | X | X | X | X | ||
| 062,803 BY0 | X | X | X | X | |||||
| 070,103 BWO | X | X | X | X | |||||
| 070,103 BW1 | X | X | X | X | X | X | |||
| NASA samples | NASA 1–1 | X | X | X | X | X | |||
| NASA 1–71 | X | X | X | X | |||||
| NASA 2–43 | X | X | X | X | |||||
| NASA 2–25 | X | X | X | X | X | X | X | ||
| NASA 2–8 | X | X | X | X | X | X | X | ||
| NASA 2–33 | X | X | X | X | X | ||||
| NASA 2–34 | X | X | X | X | |||||
| NASA DS1 | X | X | X | X | |||||
| NASA DS3 | X | X | X | X | |||||
| Carlsbad | SPG PP1 | X | X | X | X | X |
| Collection Location | Isolate ID | Wells Cleared a | Mitomycin VLP mL−1 | Control Wells VLP mL−1 | VLP Production |
|---|---|---|---|---|---|
| Tallahassee samples | 070610 A | 1 | 5.60 × 106 | 2.39 × 106 | 3.21 × 106 |
| 070610 B | 2 | 1.31 × 105 | 3.07 × 105 | −1.76 × 105 | |
| 070610 C | 4 | 1.28 × 107 | 0.00 × 101 | 1.28 × 107 | |
| 071210 B | 3 | 1.61 × 105 | 7.30 × 104 | 8.80 × 104 | |
| 071210 A | 0 | 7.30 × 103 | 0.00 × 101 | 7.30 × 103 | |
| 071210 C | 0 | 6.13 × 106 | 6.94 × 105 | 5.44 × 106 | |
| 071210 D | 4 | 4.46 × 106 | 1.90 × 106 | 2.56 × 106 | |
| 071210 E | 2 | 1.99 × 105 | 1.02 × 105 | 9.70 × 104 | |
| 071210 F | 0 | 9.86 × 105 | 1.83 × 105 | 8.03 × 105 | |
| 071310 A | 1 | 4.67 × 105 | 1.31 × 105 | 3.36 × 105 | |
| 071310 B | 3 | 8.98 × 105 | 1.10 × 105 | 7.88 × 105 | |
| 071310 C | 3 | 5.60 × 107 | 4.11 × 106 | 5.19 × 107 | |
| 071310 D | 4 | 2.10 × 107 | 2.04 × 105 | 2.08 × 107 | |
| 071410 A | 3 | 8.32 × 105 | 0.00 × 100 | 8.32 × 105 | |
| 071410 B | 3 | 1.18 × 106 | 1.09 × 106 | 9.00 × 104 | |
| 071512 E | 2 | 2.19 × 104 | 7.30 × 103 | 1.46 × 104 | |
| 071512 G | 2 | 5.84 × 104 | 4.38 × 104 | 1.46 × 104 | |
| Atlantic samples | 052503 BY0 | 2 | 4.91 × 106 | 4.02 × 105 | 4.51 × 106 |
| 052503 BY1 | 2 | 1.66 × 106 | 8.03 × 105 | 8.57 × 105 | |
| 052603 BW0 | 4 | 4.46 × 107 | 4.38 × 104 | 4.46 × 107 | |
| 052703 BY0 | 2 | 3.60 × 106 | 3.21 × 105 | 3.28 × 106 | |
| 060103 BY0 | 4 | 1.18 × 106 | 5.40 × 105 | 6.40 × 105 | |
| 061203 BY0 | 1 | 8.07 × 106 | 0.00 × 100 | 8.07 × 106 | |
| 061203 BY1 | 0 | 3.25 × 106 | 2.92 × 105 | 2.96 × 106 | |
| 061203 BY2 | 3 | 7.04 × 106 | 5.84 × 105 | 6.46 × 106 | |
| 061503 BY0 | 4 | 9.24 × 106 | 4.75 × 105 | 8.77 × 106 | |
| 061903 BC2 | 4 | 5.95 × 105 | 1.10 × 106 | −5.05 × 105 | |
| 062803 BP1 | 1 | 1.72 × 106 | 3.37 × 105 | 1.38 × 106 | |
| 062803 BY0 | 4 | 1.68 × 106 | 7.23 × 105 | 9.57 × 105 | |
| 070103 BWO | 4 | 2.59 × 106 | 0.00 × 100 | 2.59 × 106 | |
| 070103 BW1 | 2 | 0.00 × 100 | 3.20 × 106 | −3.20 × 106 | |
| NASA samples | NASA 1-1 | 3 | 6.79 × 106 | 0.00 × 100 | 6.79 × 106 |
| NASA 1-71 | 4 | 4.85 × 106 | 2.23 × 106 | 2.62 × 106 | |
| NASA 2-43 | 4 | 1.41 × 107 | 5.84 × 104 | 1.40 × 107 | |
| NASA 2-25 | 0 | 3.62 × 106 | 4.38 × 104 | 3.58 × 106 | |
| NASA 2-8 | 1 | 1.53 × 106 | 8.18 × 105 | 7.12 × 105 | |
| NASA 2-33 | 3 | 5.50 × 106 | 1.90 × 106 | 3.60 × 106 | |
| NASA 2-34 | 1 | 3.58 × 106 | 0.00 × 100 | 3.58 × 106 | |
| NASA DS1 | 4 | 1.42 × 106 | 1.02 × 105 | 1.32 × 106 | |
| NASA DS3 | 4 | 4.10 × 106 | 5.84 × 105 | 3.52 × 106 | |
| Carlsbad Cavern | SPG PP1 | 3 | 2.40 × 106 | 8.32 × 105 | 1.57 × 106 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teigell-Perez, N.; Gonzalez-Martin, C.; Valladares, B.; Smith, D.J.; Griffin, D.W. Virus-Like Particle Production in Atmospheric Eubacteria Isolates. Atmosphere 2019, 10, 417. https://doi.org/10.3390/atmos10070417
Teigell-Perez N, Gonzalez-Martin C, Valladares B, Smith DJ, Griffin DW. Virus-Like Particle Production in Atmospheric Eubacteria Isolates. Atmosphere. 2019; 10(7):417. https://doi.org/10.3390/atmos10070417
Chicago/Turabian StyleTeigell-Perez, Nuria, Cristina Gonzalez-Martin, Basilio Valladares, David J. Smith, and Dale W. Griffin. 2019. "Virus-Like Particle Production in Atmospheric Eubacteria Isolates" Atmosphere 10, no. 7: 417. https://doi.org/10.3390/atmos10070417
APA StyleTeigell-Perez, N., Gonzalez-Martin, C., Valladares, B., Smith, D. J., & Griffin, D. W. (2019). Virus-Like Particle Production in Atmospheric Eubacteria Isolates. Atmosphere, 10(7), 417. https://doi.org/10.3390/atmos10070417