Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains
Abstract
1. Introduction
2. Materials and Methods
2.1. Tetraploid Wheat Mapping Populations
2.2. Growing Conditions and Experimental Design
2.3. QTL Analysis
2.4. Genome-Based Meta- Quantitative Trait Loci Analysis
2.5. Identification of Wheat Orthologs to Yield-Related Genes in Rice
2.6. GRF4-A Single Nucleotide Polymorphism Marker Development
2.7. Development and Evaluation of Introgression Line IL-21.1
2.8. Allelic Variation Studies
3. Results
3.1. Phenotypic Characterization of Grain Parameters in Parental Lines
3.2. QTL Analysis
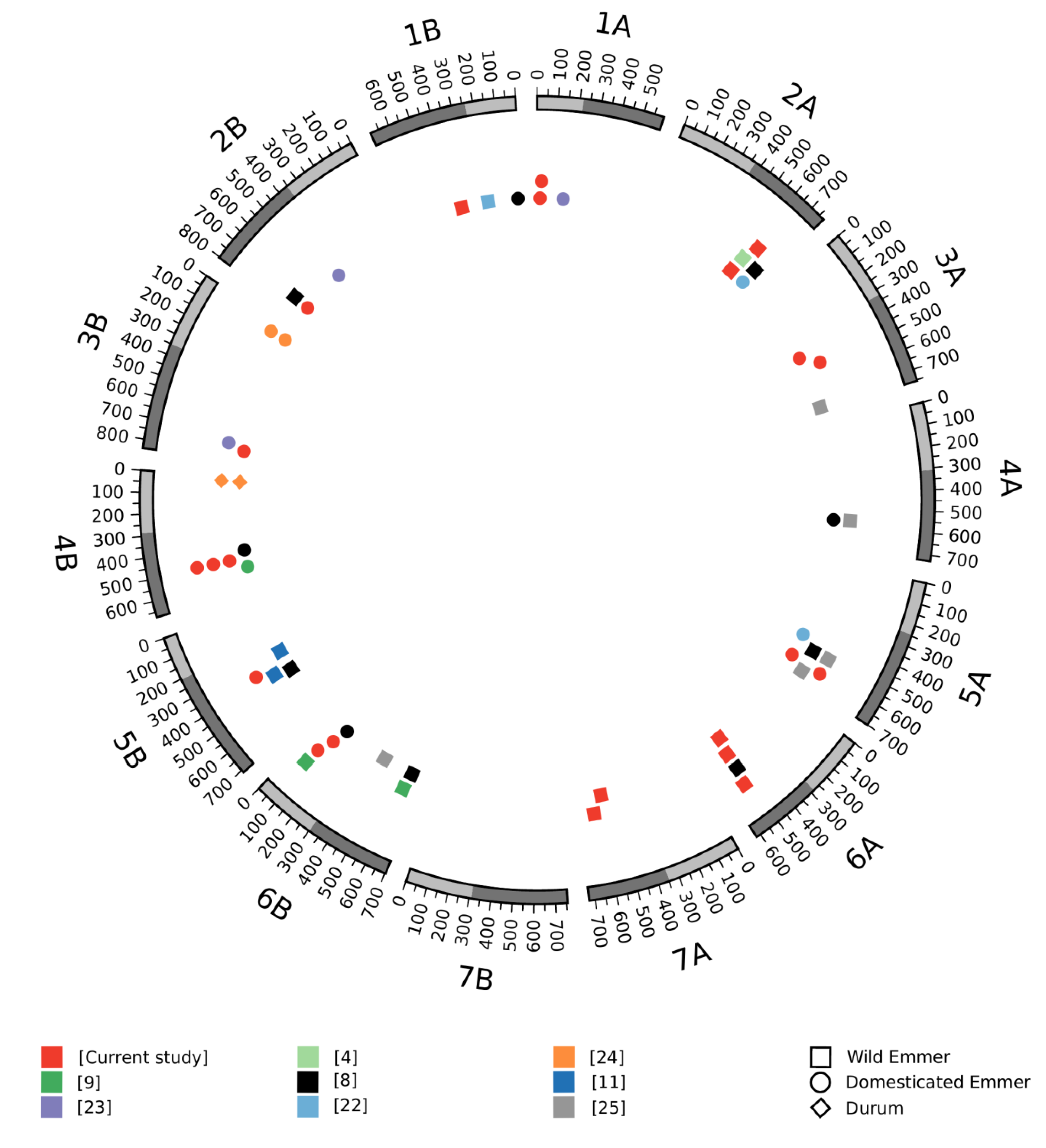
3.3. Meta-QTL Analysis
3.4. Validation of mQTL-GW-6A Using Sv × Zv Introgression Lines
3.5. Wheat Orthologs to Yield-Related Genes in Rice
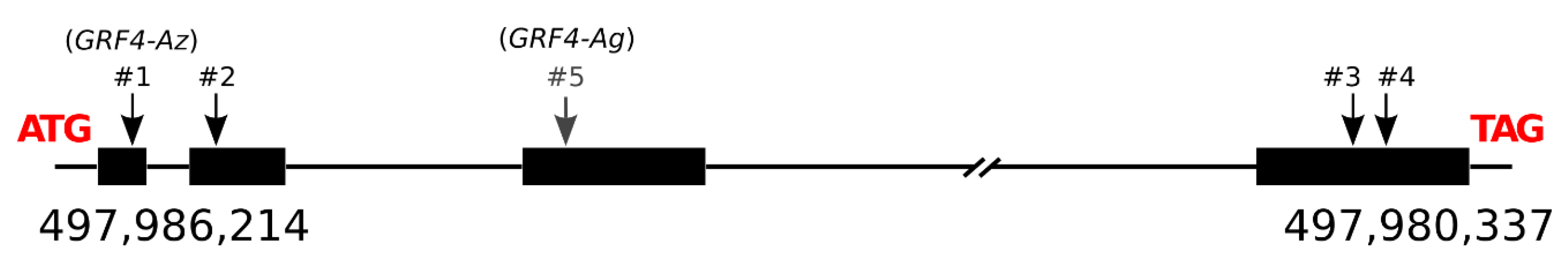
3.6. GRF4-A Polymorphisms
3.7. Allelic Diversity Study of GRF4-A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Campbell, K.G.; Bergman, C.J.; Gualberto, D.G.; Anderson, J.A.; Giroux, M.J.; Hareland, G.; Fulcher, R.G.; Sorrells, M.E.; Finney, P.L. Quantitative trait loci associated with kernel traits in a soft x hard wheat cross. Crop Sci. 1999, 39, 1184–1195. [Google Scholar] [CrossRef]
- Gegas, V.C.; Nazari, A.; Griffiths, S.; Simmonds, J.; Fish, L.; Orford, S.; Sayers, L.; Doonan, J.H.; Snape, J.W. A genetic framework for grain size and shape variation in wheat. Plant Cell 2010, 22, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Pinhasi van-Oss, R.; Gopher, A.; Saranga, Y.; Ofner, I.; Peleg, Z. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef]
- Golan, G.; Oksenberg, A.; Peleg, Z. Genetic evidence for differential selection of grain and embryo weight during wheat evolution under domestication. J. Exp. Bot. 2015, 66, 5703–5711. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, H.; Brandolini, A.; Pozzi, C.; Effgen, S.; Wunder, J.; Salamini, F. A reconsideration of the domestication geography of tetraploid wheats. Theor. Appl. Genet. 2005, 110, 1052–1060. [Google Scholar] [CrossRef]
- Poyarkova, H.; Gerechter-Amitai, Z.K. Two variants of wild emmer (Triticum dicoccoides) native to Israel: Morphology and distribution. Can. J. Bot. 1991, 69, 2772–2789. [Google Scholar] [CrossRef]
- Aaronsohn, A. Agricultural and botanical explorations in Palestine. Bur. Plant Ind. Bull. USDA 1910, 180, 1–63. [Google Scholar]
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. Bot. 2011, 62, 5051–5061. [Google Scholar] [CrossRef]
- Elouafi, I.; Nachit, M.M. A genetic linkage map of the Durum x Triticum dicoccoides backcross population based on SSRs and AFLP markers, and QTL analysis for milling traits. Theor. Appl. Genet. 2004, 108, 401–413. [Google Scholar] [CrossRef]
- Nave, M.; Avni, R.; Ben-Zvi, B.; Hale, I.; Distelfeld, A. QTLs for uniform grain dimensions and germination selected during wheat domestication are co-located on chromosome 4B. Theor. Appl. Genet. 2016, 129, 1303–1315. [Google Scholar] [CrossRef]
- Thanh, P.T.; Vladutu, C.I.; Kianian, S.F.; Thanh, P.T.; Ishii, T.; Nitta, M.; Nasuda, S.; Mori, N. Molecular genetic analysis of domestication traits in emmer wheat. I: Map construction and QTL analysis using an F2 population. Agric. Environ. Biotechnol. 2013, 27, 3627–3637. [Google Scholar] [CrossRef]
- Hu, M.J.; Zhang, H.P.; Cao, J.J.; Zhu, X.F.; Wang, S.X.; Jiang, H.; Wu, Z.Y.; Lu, J.; Chang, C.; Sun, G.; et al. Characterization of an IAA-glucose hydrolase gene TaTGW6 associated with grain weight in common wheat (Triticum aestivum L.). Mol. Breed. 2016. [Google Scholar] [CrossRef]
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Y.; Wang, C. Molecular functions of genes related to grain shape in rice. Breed. Sci. 2015, 65, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, M.; Zhang, Q.; Xu, Z.; Xu, Q. The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed. Sci. 2016, 66, 659–667. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2016, 2, 15203. [Google Scholar] [CrossRef]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef]
- Tsukaya, H. Yield increase: GRFs provide the key. Nat. Plants 2015, 2, 15210. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Eilam, T.; Sela, H.; Alekperov, C.; Peleg, Z.; Dvorak, J.; Korol, A.; Distelfeld, A. Ultra-dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol. Breed. 2014, 34, 1549–1562. [Google Scholar] [CrossRef]
- Peng, J.; Ronin, Y.; Fahima, T.; Röder, M.S.; Li, Y.; Nevo, E.; Korol, A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 2003, 100, 2489–2494. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Q.; Chao, S.; Zhang, Z.; Xu, S.S. Analysis of agronomic and domestication traits in a durum × cultivated emmer wheat population using a high-density single nucleotide polymorphism-based linkage map. Theor. Appl. Genet. 2014, 127, 2333–2348. [Google Scholar] [CrossRef]
- Russo, M.A.; Ficco, D.B.M.; Laido, G.; Marone, D.; Papa, R.; Blanco, A.; Gadaleta, A.; De Vita, P.; Mastrangelo, M.A. Dense durum wheat × T. dicoccum linkage map based on SNP markers for the study of seed morphology. Mol. Breed. 2014, 34, 1579–1597. [Google Scholar] [CrossRef]
- Tzarfati, R.; Barak, V.; Krugman, T.; Fahima, T.; Abbo, S.; Saranga, Y.; Korol, A.B. Novel quantitative trait loci underlying major domestication traits in tetraploid wheat. Mol. Breed. 2014, 34, 1613–1628. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Avni, R.; Moran Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 97, 93–97. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, W.; Wang, Y.; He, Q.; Shu, F.; Liu, H.; Wang, J.; Wang, J.; Yuan, L.; Deng, H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef]
- Arcade, A.; Labourdette, A.; Falque, M.; Mangin, B.; Chardon, F.; Charcosset, A.; Joets, J. BioMercator: Integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 2004, 20, 2324–2326. [Google Scholar] [CrossRef]
- Goffinet, B.; Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 2000, 155, 463–473. [Google Scholar]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef]
- Mester, D.; Ronin, Y.; Schnable, P.; Aluru, S.; Korol, A. Fast and accurate construction of ultra-dense consensus genetic maps using evolution strategy optimization. PLoS ONE 2015, 10, e0122485. [Google Scholar] [CrossRef]
- Bezant, J.H.; Laurie, D.A.; Pratchett, N.; Chojecki, J.; Kearsey, M.J. Mapping of QTL controlling NIR predicted hot water extract and grain nitrogen content in a spring barley cross using marker-regression. Plant Breed. 1997, 116, 141–145. [Google Scholar] [CrossRef]
- Cooper, M.; Woodruff, D.R.; Eisemann, R.L.; Brennan, P.S.; DeLacy, I.H. A selection strategy to accommodate genotype-by-environment interaction for grain yield of wheat: Managed-environments for selection among genotypes. Theor. Appl. Genet. 1995, 90, 492–502. [Google Scholar] [CrossRef]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; del Blanco, A.; Dubcovsky, J.; Uauy, C. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef]
- Distelfeld, A.; Uauy, C.; Olmos, S.; Schlatter, A.R.; Dubcovsky, J.; Fahima, T. Microcolinearity between a 2-cM region encompassing the grain protein content locus Gpc-6B1 on wheat chromosome 6B and a 350-kb region on rice chromosome 2. Funct. Integr. Genom. 2004, 4, 59–66. [Google Scholar] [CrossRef]
- Lu, L.; Shao, D.; Qiu, X.; Sun, L.; Yan, W.; Zhou, X.; Yang, L.; He, Y.; Yu, S.; Xing, Y. Natural variation and artificial selection in four genes determine grain shape in rice. New Phytol. 2013, 200, 1269–1280. [Google Scholar] [CrossRef]
- Ozkan, H.; Willcox, G.; Graner, A.; Salamini, F.; Kilian, B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 2011, 58, 11–53. [Google Scholar] [CrossRef]
- Sela, H.; Ezrati, S.; Ben-Yehuda, P.; Manisterski, J.; Akhunov, E.; Dvorak, J.; Breiman, A.; Korol, A. Linkage disequilibrium and association analysis of stripe rust resistance in wild emmer wheat (Triticum turgidum ssp. dicoccoides) population in Israel. Theor. Appl. Genet. 2014, 127, 2453–2463. [Google Scholar] [CrossRef]
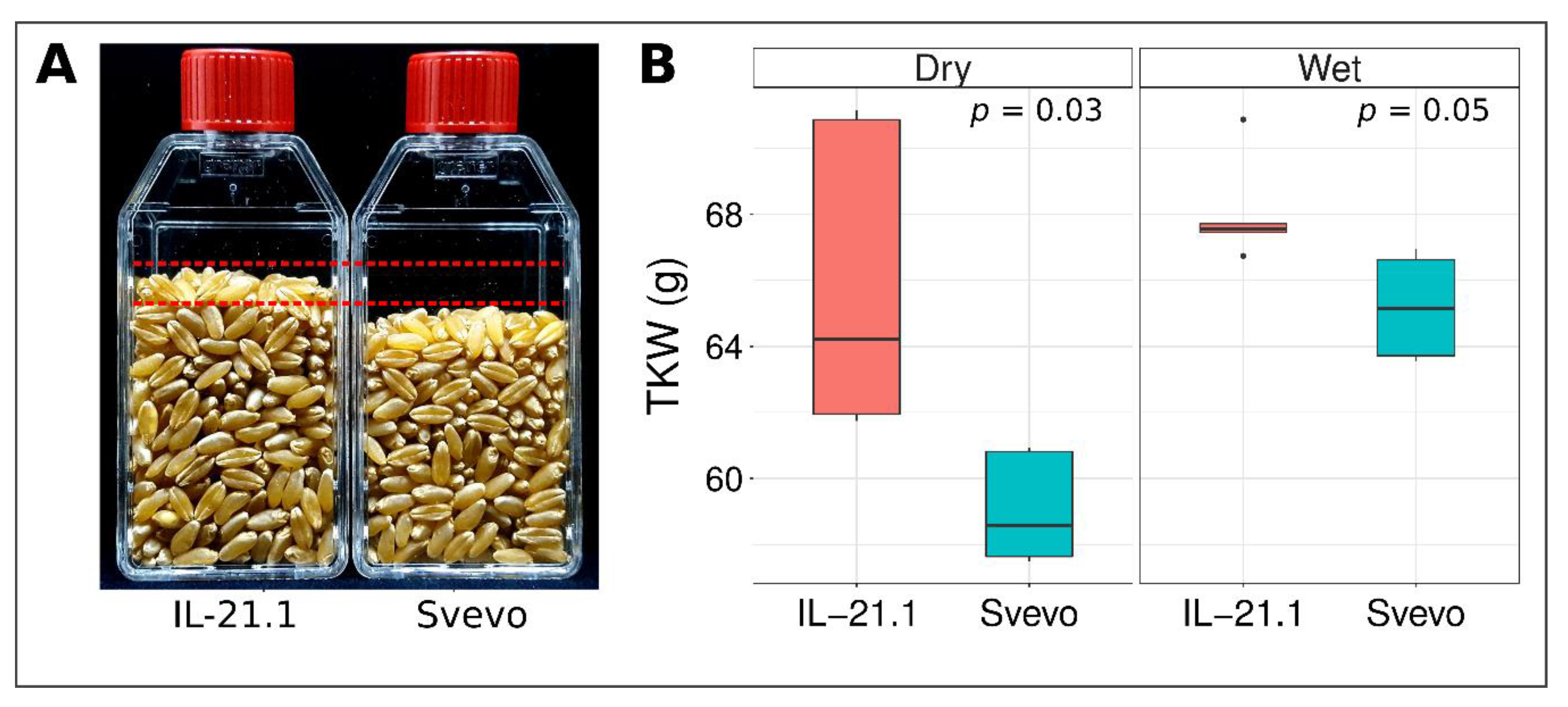
| Trait | Environment | Sv | Zv |
|---|---|---|---|
| Mean spike weight (g) | 2014R | 3.9 ± 0.1 | 2.5 ± 0.1 |
| 2015A | 4.0 ± 0.1 | 1.6 ± 0.8 | |
| TKW (g) | 2014R | 61.9 ± 0.2 | 45.8 ± 0.1 |
| 2015A | 55.7 ± 0.1 | 29.1 ± 0.1 | |
| Grain area (mm2) | 2014R | 21.6 ± 0.2 | 23.3 ± 0.3 |
| 2015A | 23.0 ± 0.3 | 18.6 ± 0.4 | |
| Grain width (mm) | 2014R | 3.2 ± 0.03 | 2.8 ± 0.0 |
| 2015A | 3.7 ± 0.03 | 2.5 ± 0.0 | |
| Grain length (mm) | 2014R | 8.1 ± 0.08 | 10.2 ± 0.1 |
| 2015A | 8.5 ± 0.05 | 10.1 ± 0.1 |
| Rice Gene | Rice Gene Function | Source | Wheat Chr. | Wheat Alignment Start | Wheat Alignment End | Wheat Gene Function | WEW Gene ID |
|---|---|---|---|---|---|---|---|
| D11/DWARF11 | Cytochrome P450 (CYP724B1) enzyme | Reviewed by [14] | 2A | 561795447 | 561798557 | Cytochrome P450 superfamily protein | TRIDC2AG048380 |
| 2B | 496938005 | 496941148 | Cytochrome P450 superfamily protein | TRIDC2BG050840 | |||
| D2 | Cytochrome P450 (CYP90D) enzyme | Reviewed by [14] | 2A | 4843464 | 5209969 | Cytochrome P450 superfamily protein | TRIDC2AG001470 |
| 2B | 5686865 | 5987817 | Cytochrome P450 superfamily protein | TRIDC2BG001370 | |||
| D61 | BR insensitive (BRI)-like leucine-rich repeat (LRR) receptor kinase | Reviewed by [14] | 3A | 465976238 | 465979780 | Leucine-rich receptor-like protein kinase family protein | TRIDC3AG036670 |
| 3B | 453931439 | 453935096 | receptor-like protein kinase 2 | TRIDC3BG041310 | |||
| GIF1 | Cell wall invertase | Reviewed by [14] | 2A | 503854205 | 503855081 | Beta-fructofuranosidase, insoluble isoenzyme 2 (Cell wall invertase 2) | TRIDC2AG042730 |
| 2B | 447195335 | 447196211 | Beta-fructofuranosidase, insoluble isoenzyme 2 (Cell wall invertase 2) | TRIDC2BG045820 | |||
| GRF4/GS2 | Growth-Regulating Factor 4 (OsGRF4) | [18,29] | 2A | 680343644 | 680346735 | Growth-regulating factor 3 | TRIDC2AG062550 |
| 2B | 649416512 | 649417723 | Growth-regulating factor 3 | TRIDC2BG066890 | |||
| 6A | 497985063 | 497985958 | Growth-regulating factor 4 | TRIDC6AG041360 | |||
| 6B | 517412993 | 517416246 | Growth-regulating factor 4 | TRIDC6BG048340 | |||
| GS3 | Membrane protein with multiple domains | Reviewed by [14] | 4A | 714924235 | 714925670 | Grain length protein | TRIDC4AG069340 |
| 7A | 5283743 | 5283958 | Grain length protein | TRIDC7AG001510 | |||
| GS5 | Serine carboxypeptidase | Reviewed by [14] | 3A | 182355936 | 182359086 | serine carboxypeptidase-like 33 | TRIDC3AG023140 |
| 3B | 212372375 | 212373474 | Carboxypeptidase Y homolog A | TRIDC3BG026960 | |||
| GW2 | RING-type E3 ubiquitin ligase | Reviewed by [14] | 6A | 230789449 | 230809149 | Protein SIP5 (*TaGW2) | TRIDC6AG027660 |
| 6B | 294434000 | 294448424 | Protein SIP5 | TRIDC6BG033820 | |||
| GW5 | Arginine-rich protein of 144 amino acids | Reviewed by [14] | 1A | 142379896 | 142381359 | IQ-domain 26 | TRIDC1AG017640 |
| 1B | 185320338 | 185321816 | IQ-domain 26 | TRIDC1BG021520 | |||
| 3A | 69160021 | 69161092 | IQ-domain 26 | TRIDC3AG013280 | |||
| 3B | 111226601 | 111227636 | IQ-domain 26 | TRIDC3BG017740 | |||
| GW8/SPL16 | SQUAMOSA promoter-binding protein-like 16 | Reviewed by [14] | 7A | 251030195 | 251034936 | undescribed protein | TRIDC7AG033770 |
| 7B | 230000953 | 230005263 | Squamosa promoter-binding-like protein 16 | TRIDC7BG025060 | |||
| qGL3 | Ser/Thr phosphatase of the protein phosphatase kelch-like (PPKL) family | Reviewed by [15] | 5A | 683802818 | 683803388 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | TRIDC5AG075900 |
| SRS3 | Kinesin 13 protein | Reviewed by [14] | 1A | 131830406 | 131835083 | Kinesin-related protein 6 | TRIDC1AG016970 |
| 1B | 142745967 | 142751957 | Kinesin-related protein 6 | TRIDC1BG017960 | |||
| 3A | 274848912 | 274857469 | Kinesin-related protein 6 | TRIDC3AG027550 | |||
| 3B | 295258442 | 295261011 | Kinesin-related protein 6 | TRIDC3BG032430 | |||
| DEP1 | G protein γ subunit | [16] | 5A | 422466437 | 422469555 | Guanine nucleotide-binding protein subunit gamma 3 | TRIDC5AG033880 |
| 5B | 391766206 | 391769237 | undescribed protein | TRIDC5BG035790 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avni, R.; Oren, L.; Shabtay, G.; Assili, S.; Pozniak, C.; Hale, I.; Ben-David, R.; Peleg, Z.; Distelfeld, A. Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains. Genes 2018, 9, 636. https://doi.org/10.3390/genes9120636
Avni R, Oren L, Shabtay G, Assili S, Pozniak C, Hale I, Ben-David R, Peleg Z, Distelfeld A. Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains. Genes. 2018; 9(12):636. https://doi.org/10.3390/genes9120636
Chicago/Turabian StyleAvni, Raz, Leah Oren, Gai Shabtay, Siwar Assili, Curtis Pozniak, Iago Hale, Roi Ben-David, Zvi Peleg, and Assaf Distelfeld. 2018. "Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains" Genes 9, no. 12: 636. https://doi.org/10.3390/genes9120636
APA StyleAvni, R., Oren, L., Shabtay, G., Assili, S., Pozniak, C., Hale, I., Ben-David, R., Peleg, Z., & Distelfeld, A. (2018). Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains. Genes, 9(12), 636. https://doi.org/10.3390/genes9120636








