A Systematic Study on DNA Barcoding of Medicinally Important Genus Epimedium L. (Berberidaceae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Isolation, Amplification, and Sequencing
2.3. Statistical Analysis
3. Results
3.1. Efficiency of PCR Amplification and Sequencing
3.2. Genetic Divergence Determination
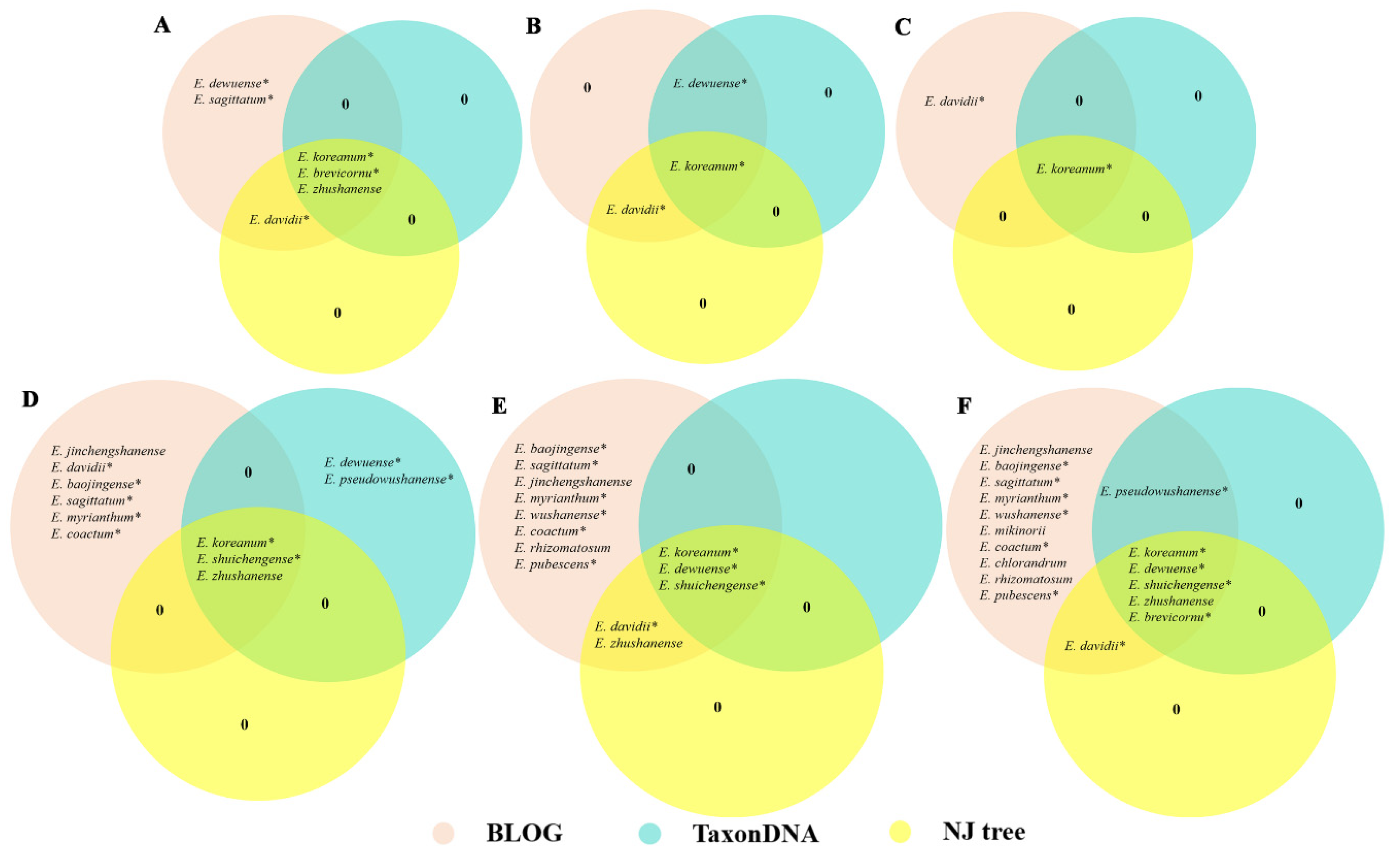
3.3. Evaluation of the Identification Efficiency of the DNA Barcodes
4. Discussion
4.1. Development of DNA Barcode Resources for Genus Epimedium
4.2. Chloroplast Genome-Based Super Barcode Has the Potential to Solve the Problem of Species Discrimination and Classification in Genus Epimedium
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reginster, J.Y.; Burlet, N. Osteoporosis: A still increasing prevalence. Bone 2006, 38, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Burge, R.; Dawson-Hughes, B.; Solomon, D.H.; Wong, J.B.; King, A.; Tosteson, A. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J. Bone Miner. Res. 2007, 22, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.P.; Sheu, S.Y.; Sun, J.S.; Chen, M.H.; Liu, M.H. Icariin isolated from Epimedium pubescens regulates osteoblasts anabolism through BMP-2, SMAD4, and Cbfa1 expression. Phytomedicine 2010, 17, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Zhang, X.; Lu, L.; Wu, X.; Li, S.; Meng, J. Regulation of Cbfa1 expression by total flavonoids of Herba epimedii. Endocr. J. 2006, 53, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lien, E.J.; Lien, L.L. Chemical and pharmacological investigations of Epimedium species: A survey. Prog. Drug Res. 2003, 60, 1–57. [Google Scholar]
- Ma, H.; He, X.; Yang, Y.; Li, M.; Hao, D.; Jia, Z. The genus Epimedium: An ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2011, 134, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; Chemical Industry Press: Beijing, China, 2015; Volume I. [Google Scholar]
- Cho, W.-K.; Kim, H.; Choi, Y.J.; Yim, N.H.; Yang, H.J.; Ma, J.Y. Epimedium koreanum Nakai Water Extract Exhibits Antiviral Activity against Porcine Epidermic Diarrhea Virus In Vitro and In Vivo. Evid.-Based Complement. Altern. 2012, 2012, 1–10. [Google Scholar]
- Yuan, D.; Wang, H.; He, H.; Jia, L.; He, Y.; Wang, T.; Zeng, X.; Li, Y.; Li, S.; Zhang, C. Protective effects of total flavonoids from Epimedium on the male mouse reproductive system against cyclophosphamide-induced oxidative injury by up-regulating the expressions of SOD3 and GPX1. Phytother. Res. 2014, 28, 88–97. [Google Scholar] [CrossRef]
- Jiang, J.; Song, J.; Jia, X.B. Phytochemistry and ethnopharmacology of Epimedium L. species. Chin. Herb. Med. 2015, 7, 204–222. [Google Scholar] [CrossRef]
- Telang, N.T.; Li, G.; Katdare, M.; Sepkovic, D.W.; Bradlow, H.L.; Wong, G.Y.C. The nutritional herb Epimedium grandiflorum inhibits the growth in a model for the Luminal A molecular subtype of breast cancer. Oncol. Lett. 2017, 13, 2477–2482. [Google Scholar] [CrossRef]
- Stearn, W.T. The genus Epimedium and other herbaceous Berberidaceae; Timber Press: Portland, OR, USA, 2002. [Google Scholar]
- Zhang, Y.; Du, L.; Liu, A.; Chen, J.; Wu, L.; Hu, W.; Zhang, W.; Kim, K.; Lee, S.; Yang, T.; et al. The Complete Chloroplast Genome Sequences of Five Epimedium Species: Lights into Phylogenetic and Taxonomic Analyses. Front. Plant Sci. 2016, 7, 306. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Xu, W.F.; He, S.Z. Epimedium muhuangense (Berberidaceae), a new species from China. Phytotaxa 2017, 319, 277–282. [Google Scholar] [CrossRef]
- Wei, N.; Zhang, Y.J.; Xu, Z.; Kamande, E.M.; Ngumbau, V.M.; Hu, G.W. Epimedium zhaotongense (Berberidaceae), a New Species from Yunnan Province, China. Phytotaxa 2017, 296, 88. [Google Scholar] [CrossRef]
- Takahashi, C. Karyomorphological studies on speciation of Epimedium and its allied Vancouveria with special reference to C-bands. J. Sci. Hiroshima Univ. B 1989, 2, 159–269. [Google Scholar]
- Ying, J. Petal evolution and distribution patterns of Epimedium L. (Berberidaceae). Acta Phytotaxon. Sin. 2002, 40, 481–489. [Google Scholar]
- De Smet, Y.; Goetghebeur, P.; Wanke, S.; Asselman, P.; Samain, M.S. Additional evidence for recent divergence of Chinese Epimedium (Berberidaceae) derived from AFLP, chloroplast and nuclear data supplemented with characterisation of leaflet pubescence. Plant Ecol. Evol. 2012, 145, 73–87. [Google Scholar] [CrossRef]
- Guo, B.L.; Pei, L.K.; Xiao, P.G. Further research on taxonomic significance of flavonoids in Epimedium (Berberidaceae). J. Syst. Evol. 2008, 46, 874–885. [Google Scholar]
- Ying, T.S.; Boufford, D.E.; Brach, A.R. Epimedium L. In Flora of China; Science Press: St. Louis, MO, USA; Botanical Garden Press: Beijing, China, 2011; Volume 19, pp. 787–799. [Google Scholar]
- Govindaraghavan, S.; Hennell, J.R.; Sucher, N.J. From classical taxonomy to genome and metabolome: Towards comprehensive quality standards for medicinal herb raw materials and extracts. Fitoterapia 2012, 83, 979–988. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Cai, W.Z.; Hu, S.F.; Huang, X.H.; Ge, F.; Wang, Y. Morphological variation of non-glandular hairs in cultivated Epimedium sagittatum (Berberidaceae) populations and implications for taxonomy. Biodivers. Sci. 2013, 21, 185–196. [Google Scholar]
- He, S.Z. The Genus Epimedium of China in Colour; Guizhou Publishing Group: Guizhou, China, 2014. [Google Scholar]
- Xie, P.S.; Yan, Y.Z.; Guo, B.L.; Lam, C.W.K.; Chui, S.H.; Yu, Q.X. Chemical pattern-aided classification to simplify the intricacy of morphological taxonomy of Epimedium species using chromatographic fingerprinting. J. Pharmaceut. Biomed. 2010, 52, 452–460. [Google Scholar] [CrossRef]
- Zhang, H.F.; Yang, X.H. Asian medicine: Protect rare plants. Nature 2012, 482, 35. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fung, K.P.; Leung, P.C.; Shi, D.; Shaw, P.C. Characterization of medicinal Epimedium species by 5S rRNA gene spacer sequencing. Planta Med. 2004, 70, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Xiao, G.; Guo, J.; Fei, Z.; Xu, Y.; Roe, B.A.; Wang, Y. Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) Maxim. BMC Genom. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xu, Y.; Huang, H.; Wang, Y. PRIMER NOTE: Identification of microsatellite loci from Epimedium koreanum and cross-species amplification in four species of Epimedium (Berberidaceae). Mol. Ecol. Notes 2006, 7, 467–470. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Z.; Wang, Y. Fourteen microsatellite loci for the Chinese medicinal plant Epimedium sagittatum and cross-species application in other medicinal species. Mol. Ecol. Resour. 2008, 8, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, Z.; Hu, W.; Zhang, Y.; Zeng, S.; Wang, Y. Systematic validation of medicinally important genus epimedium species based on microsatellite markers. Pak. J. Bot. 2015, 47, 477–484. [Google Scholar]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Li, D.Z.; van der Bank, M.; Twyford, A.D. Telling plant species apart with DNA: From barcodes to genomes. Philos. Trans. R. Soc. B 2016, 371, 20150338. [Google Scholar] [CrossRef]
- Guo, M.; Ren, L.; Pang, X. Inspecting the true identity of herbal materials from Cynanchum using ITS2 barcode. Front. Plant Sci. 2017, 8, 1945. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Sass, C.; Little, D.P.; Stevenson, D.W.; Specht, C.D. DNA barcoding in the cycadales: Testing the potential of proposed barcoding markers for species identification of cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Schleicher, T.; Schultz, J.; Muller, T.; Dandekar, T.; Wolf, M. 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene 2009, 430, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Zhang, G.; Ali, F. The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Syst. Biol. 2008, 57, 809–813. [Google Scholar] [CrossRef]
- Meier, R.; Shiyang, K.; Vaidya, G.; Ng, P.K. DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst. Biol. 2006, 55, 715–728. [Google Scholar] [CrossRef]
- Weitschek, E.; van Velzen, R.; Felici, G.; Bertolazzi, P. BLOG 2.0: A software system for character-based species classification with DNA Barcode sequences. What it does, how to use it. Mol. Ecol. Resour. 2013, 13, 1043–1046. [Google Scholar] [CrossRef]
- China Plant BOL Group. Comparative analysis of a large dataset indicates that ITS should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [PubMed]
- Newmaster, S.G.; Fazekas, A.J.; Ragupathy, S. DNA barcoding in land plants: Evaluation of rbcL in a multigene tiered approach. Botany 2006, 84, 335–341. [Google Scholar] [CrossRef]
- Goldstein, P.Z.; DeSalle, R. Integrating DNA barcode data and taxonomic practice: Determination, discovery, and description. Bioessays 2011, 33, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Dang, H.S.; Wang, Y.; Li, X.W.; Li, J.Q. A taxonomic revision of unifoliolate Chinese Epimedium L. (Berberidaceae). Kew Bull. 2011, 66, 253. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Nguyen, P.A.T.; Kim, J.S.; Kim, J.H. The complete chloroplast genome of colchicine plants (Colchicum autumnale L. and Gloriosa superba L.) and its application for identifying the genus. Planta 2015, 242, 223–237. [Google Scholar] [CrossRef]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef]
- Nock, C.J.; Waters, D.L.E.; Edwards, M.A.; Bowen, S.G.; Rice, N.; Cordeiro, G.M.; Henry, R.J. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 2011, 9, 328–333. [Google Scholar] [CrossRef]
- Kane, N.; Sveinsson, S.; Dempewolf, H.; Yang, J.Y.; Zhang, D.; Engels, J.M.; Cronk, Q. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 2012, 99, 320–329. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 157–166. [Google Scholar] [CrossRef] [PubMed]
| Best Match | Best Close Match (Threshold 3%) | BLOG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Region | Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No Match | Correct | Not Classified | Wrong |
| psbA-trnH | 29.62% | 27.77% | 42.59% | 29.62% | 27.77% | 38.88% | 3.70% | 61.11% | 38.89% | 0.00% |
| ITS | 22.22% | 70.37% | 7.40% | 22.22% | 70.37% | 7.40% | 0.00% | 48.15% | 51.85% | 0.00% |
| ITS2 | 18.51% | 74.07% | 7.40% | 18.51% | 74.07% | 7.40% | 0.00% | 24.07% | 75.93% | 0.00% |
| rbcL | 3.70% | 96.29% | 0.00% | 3.70% | 96.29% | 0.00% | 0.00% | 14.81% | 85.19% | 0.00% |
| psbA-trnH + ITS | 37.03% | 24.07% | 38.88% | 37.03% | 24.07% | 38.88% | 0.00% | 92.59% | 7.41% | 0.00% |
| psbA-trnH + ITS2 | 38.88% | 25.92% | 35.18% | 38.88% | 25.92% | 33.33% | 3.70% | 77.78% | 22.22% | 0.00% |
| psbA-trnH + ITS + rbcL | 37.03% | 18.51% | 44.44% | 37.03% | 18.51% | 44.44% | 0.00% | 92.59% | 7.41% | 0.00% |
| psbA-trnH + ITS2 + rbcL | 37.03% | 24.07% | 38.88% | 37.03% | 24.07% | 38.88% | 0.00% | 81.48% | 18.52% | 0.00% |
| Species | Formulas | Score | Coverage |
|---|---|---|---|
| E. davidii * | pos1043 = T | 0.136 | 1 |
| E. baojingense * | pos314 = G OR pos1085 = C | 0.174 | 1 |
| E. koreanum * | pos201 = T | 0.136 | 1 |
| E. dewuense * | pos395 = C & pos1220 = G | 0.136 | 1 |
| E. sagittatum * | pos201 = G & pos320 = T & pos395 = T & pos411 = G & pos1052 = T | 0.24 | 1 |
| OR pos91 = C | |||
| OR pos113 = A | |||
| E. jinchengshanense | pos224 = C & pos411 = T & pos1220 = G | 0.136 | 1 |
| OR pos411 = G & pos1052 = A | |||
| E. chlorandrum | pos90 = T & pos150 = G & pos201 = G & pos224 = A & pos395 = T & pos411 = T & pos536 = A & pos1043 = C & pos1052 = T & pos1220 = A & pos1252 = G | 0.174 | 1 |
| OR pos1026 = C & pos1182 = T | |||
| E. pseudowushanense * | pos90 = T & pos91 = A & pos224 = C & pos411 = T & pos788 = C & pos1182 = C & pos1220 = A | 0.136 | 1 |
| E. rhizomatosum | pos411 = T & pos1052 = A | 0.136 | 1 |
| E. pubescens * | pos90 = T & pos150 = G & pos201! = G & pos201! = T & pos395 = T | 0.174 | 1 |
| OR pos90 = G & pos224 = C & pos411 = T & pos1026 = C & pos1182 = C | |||
| OR pos320 = A | |||
| E. shuichengense * | pos201 = A & pos1052 = T | 0.136 | 1 |
| E. myrianthum * | pos90 = G & pos411 = G | 0.208 | 1 |
| OR pos113 = G & pos150 = A & pos314 = A & pos730 = A | |||
| E. wushanense * | pos164 = T & pos224 = A & pos411 = T & pos1085 = T & pos1220 = G | 0.174 | 1 |
| OR pos90 = G & pos201 = G & pos224 = A & pos411 = T & pos1085 = T | |||
| E. brevicornu * | pos1252 = C | 0.136 | 1 |
| E. coactum * | pos505 = C & pos536 = C | 0.136 | 1 |
| OR pos1026 = A | |||
| E. mikinorii | pos788 = T | 0.136 | 1 |
| OR pos90 = T & pos224 = C & pos411 = G & pos1220 = A | |||
| E. zhushanense | pos164 = G | 0.136 | 1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Xu, Y.; Ren, L.; He, S.; Pang, X. A Systematic Study on DNA Barcoding of Medicinally Important Genus Epimedium L. (Berberidaceae). Genes 2018, 9, 637. https://doi.org/10.3390/genes9120637
Guo M, Xu Y, Ren L, He S, Pang X. A Systematic Study on DNA Barcoding of Medicinally Important Genus Epimedium L. (Berberidaceae). Genes. 2018; 9(12):637. https://doi.org/10.3390/genes9120637
Chicago/Turabian StyleGuo, Mengyue, Yanqin Xu, Li Ren, Shunzhi He, and Xiaohui Pang. 2018. "A Systematic Study on DNA Barcoding of Medicinally Important Genus Epimedium L. (Berberidaceae)" Genes 9, no. 12: 637. https://doi.org/10.3390/genes9120637
APA StyleGuo, M., Xu, Y., Ren, L., He, S., & Pang, X. (2018). A Systematic Study on DNA Barcoding of Medicinally Important Genus Epimedium L. (Berberidaceae). Genes, 9(12), 637. https://doi.org/10.3390/genes9120637





