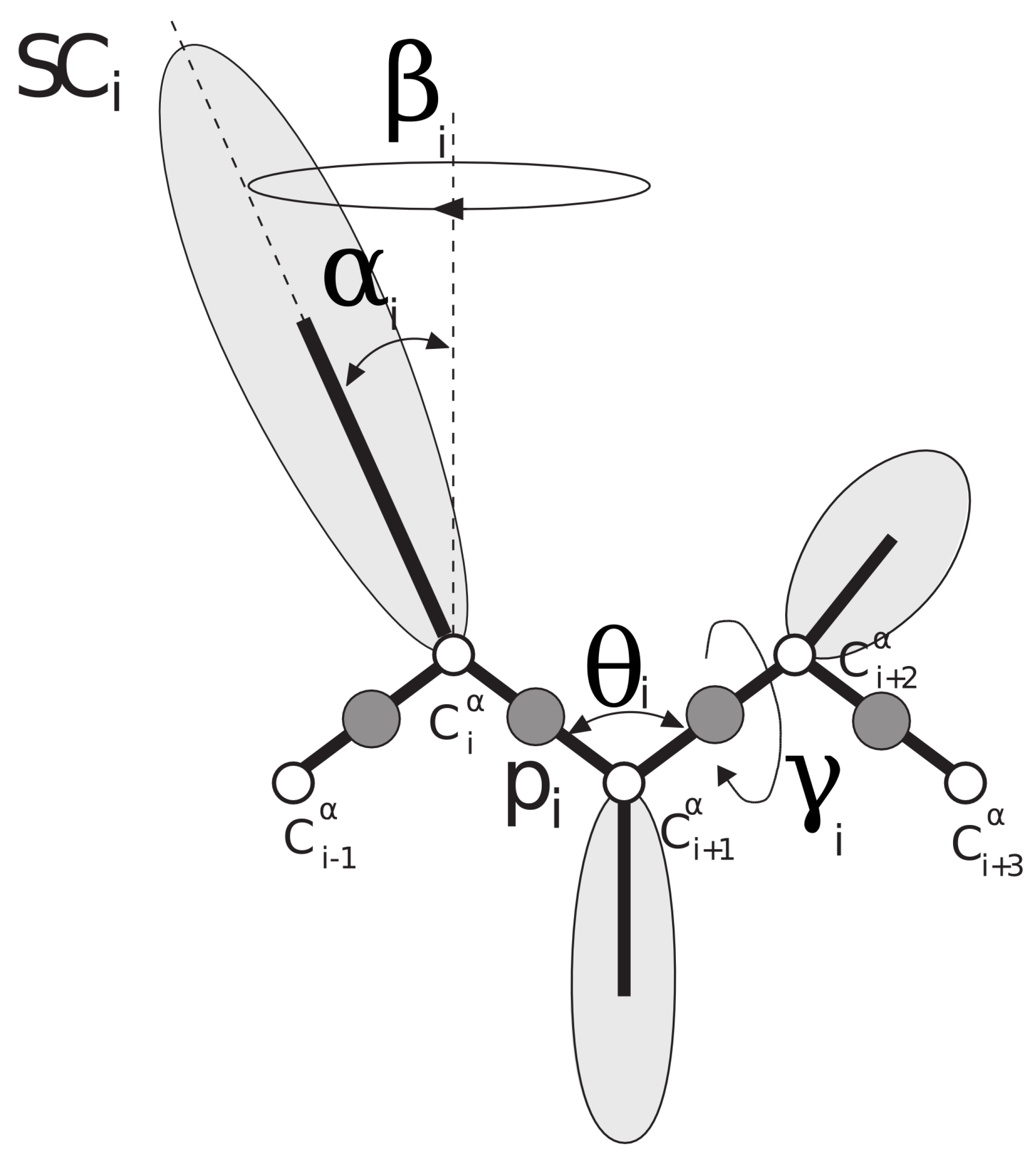
2.1. UNited RESidue (UNRES)
UNRES [
31,
32,
33,
34] is a coarse-grained model for proteins in which each amino-acid residue is reduced to two interaction sites: the united peptide group (p), located halfway between two consecutive C
atoms (which are not interaction sites and are only used to define the geometry of the chain), and the united side chain (SC) (
Figure 1). Due to the model’s reduced number of interaction sites and the exclusion of averaging out the degrees of freedom (the secondary degrees of freedom), the UNRES force field provides a speed of at least 3 order of magnitude higher compared with the all-atom simulations [
35].
The effective energy function in the UNRES model is defined as the restricted free energy (RFE) or the potential of mean force (PMF), and it is given by Equation (
1). A detailed description of UNRES is provided elsewhere [
36].
with
where each potential is multiplied by an appropriate weight,
, and these weights are optimized. In Equation (
1),
and
are side-chain–side-chain and side-chain–peptide-group interaction potentials, respectively. The peptide-group–peptide-group interaction potential is split into the Lennard-Jones term
and the electrostatic term (
). The local properties of the polypeptide chain are described by
,
,
,
, and
potentials, which are torsional, double torsional, bending, rotameric, and virtual-bond-deformation terms, respectively.
and
are higher-order correlation terms that are necessary for the correct reproduction of secondary-structure elements [
37],
is a disulfide bond potential, and
is a recently implemented potential that couples the local positions of the backbone and side chains, which improves the predictive capacities of the UNRES force field [
38,
39]. Additionally, because the UNRES energy function originates from the PMF of polypeptide chains in water, in which the fine-grained degrees of freedom have been averaged out, it is temperature-dependent. The factors
arise from multiplying the terms of the respective order in the cluster-cumulant expansion of the PMF [
37]. Because the current implementation of UNRES involves scoring the decoys, which correspond to folded structures and not folding simulations, we set the temperature at
K, assuming that all proteins considered are folded at this temperature. The UNRES model uses an anisotropic potential for the interactions between side chains, which are represented by the Gay–Berne model [
40]. This model allows for a more accurate approximation of the side-chain interactions than simpler spherical models.
The energy-term weights in the initial version of the UNRES force field were optimized using only one
-helical protein (PDB code: 1GAB) [
27]. We shall term this version of UNRES ‘GB’. In later versions, the force field was re-parameterized using two training mini-proteins: the
-helical tryptophan cage (PDB code: 1L2Y) and tryptophan zipper (PDB code: 1LE1) [
28]. The latter force field was recently extended by the addition of the local torsional potentials [
39] with very limited manual optimization of the weights of the torsional terms (
Table 1). We term this version of UNRES ‘EL’. In the current force field, all the energy terms are physics-based except for side-chain–side-chain interaction terms, which were obtained by an analysis of the PDB [
41]. Recently, a new approach to efficient force field optimization was developed [
29] based on the maximum likelihood method [
42]. However, even with the use of this method, only a very limited number of training proteins can be included in the optimization due to the high computational cost of the iterative procedure based on extensive folding simulations.
For the fold-recognition application reported in this paper, the side-chain–side-chain interaction (
), torsional (
), and side-chain-correlation (
) terms of Equation (
1) are the most important because they account for sequence-specific long- and short-range interactions. Therefore, in addition to the common weights for these terms, we introduced residue-pair-type-specific weights (a total of 400 for each of the three kinds of potentials). Residue-type-specific weights of the excluded-volume contributions (
) were introduced, because these potentials control the size of the proteins and depend on a single residue type. Likewise, residue-type-specific weights were introduced for the contributions to the virtual-bond-angle (
), side-chain-rotamer (
), and double-torsional (
) potentials, the type being that of the central residue. Because, for the decoys taken from the PDB database [
41], the regular secondary structure is already present, the electrostatic (
) and correlation (
and
) terms, which determine the regular secondary structure in free simulations, matter only as much as they contribute to the energy of the “bulk” of the secondary structure of different types. Therefore, only the weights corresponding to total
,
,
,
,
, and
were optimized (one weight per each kind of term).
Multiple types of calculations can be performed with UNRES, from single-point energy calculations and energy minimization in internal and external coordinates to Monte Carlo and Molecular Dynamics calculations of various variants and modifications. Including serial (sequential) and parallel runs with scaling up to 70% on 16K cores [
43]. Example of UNRES usage include: Conformational Space Annealing (CSA) [
44], Hybrid Monte Carlo (HMC) [
45], Replica Exchange Molecular Dynamics (REMD) [
46], and Multiplexed Replica Exchange Molecular Dynamics (MREMD) [
47]. UNRES has been successfully used for studies of protein folding pathways, thermodynamics, and kinetics [
48,
49,
50]; in studies of multimeric systems [
51,
52] with the use of periodic boundary conditions [
53]; and in systems with nonstandard amino acids [
54] and links [
55]. More detail on UNRES can be found elsewhere [
36].
2.2. Reoptimization
As stated in the introduction, the current version of UNRES was optimized to run free simulations in which the potential clashes are removed; however, in general, this is not the case for scoring fixed decoys. Therefore, first, we modified the potential to limit its repulsive components. Specifically, we imposed a cutoff on the repulsive parts of the potential to limit the maximum repulsion to 3 kcal/mol for a given interaction type between each pair of interaction centers. Only with such an approach can the UNRES force field be used without the prior energy minimization of a system because, otherwise, even slight overlapping of the interaction centers can outweigh all other energy components. Another possibility would be to use soft potentials, such as the 8–6 Lennard-Jones potential, but, even then, short energy minimization is needed [
56]. We then reoptimized the force field to accommodate this change and to customize it to the task of decoy scoring.
We employed the neural network technique for optimization. In the implementation of the UNRES model developed in this work, although energy is a linear function of the parameters, the error function to minimize is expressed as:
where index
p runs over the training proteins, index
i runs over the decoys corresponding to a given training protein (including the native structure, which has an index of 0),
is the total number of training proteins,
is the total number of decoys for protein
p (including the native structure of this protein),
and
denote the predicted TM-scores and those calculated from the respective decoy and native structures, and the input features are
, the set of UNRES energy components (Equation
1) calculated for decoy
i of protein
p. As described later, other input features will be used alongside
. In this work, we used the back-propagation neural network method to approximate the values of
that will minimize Equation (
3). The neural network we used is a nonlinear function from the input features (
) to the output feature (
). We started with a random set of neural network weights and passed the input features through the neural network weights to calculate
. This part of the process is known as feed-forward. We then calculated the error associated with said prediction and, by the steepest descent method, modified the neural network weights to reduce this error. This process is known as back-propagation. It was carried out repeatedly until an overfit-protection test was violated. In this case, for the overfit protection, we left out part of the data from training and chose weights that gave the best results for this left-out set.
From UNRES, we first extracted information characterizing the state of the protein as an initial step toward UNRES reoptimization. Besides giving the overall UNRES energy value and the values for each of the components in Equation (
1), we also split these components into their residue-type-specific contributions. For example, for paired interactions, we calculated separate values for the contributions from interactions between one type of residue and another. In total, we have the following input features from UNRES: one overall energy, nine single-value components, four 20-valued components that are residue-type-dependent, and three 400-value components that are dependent on the type of a residue pair. See
Table 2 for a list and description of these labels. Additionally, the weights of each kind of UNRES energy component and that of the total UNRES energy were also optimized. In total, we have
values characterizing the UNRES energy function in this study. It should be noted that the parameters of the pairwise side-chain–side-chain interaction energies are not symmetric. The reason for this is the directionality of the protein chain (from the N to the C terminus). For example, a parameter for an ‘AC’ pair of side chains means that alanine precedes cysteine in sequence. Such non-symmetry of interactions is quite commonly used in fold-recognition studies [
1,
2,
3,
4,
5,
6] and partially accounts for the “through sequence” long-range interactions.
The same approach we used in the Seder1 scoring function [
18] was used here for scoring a model of a given sequence based on its similarity to the native PDB [
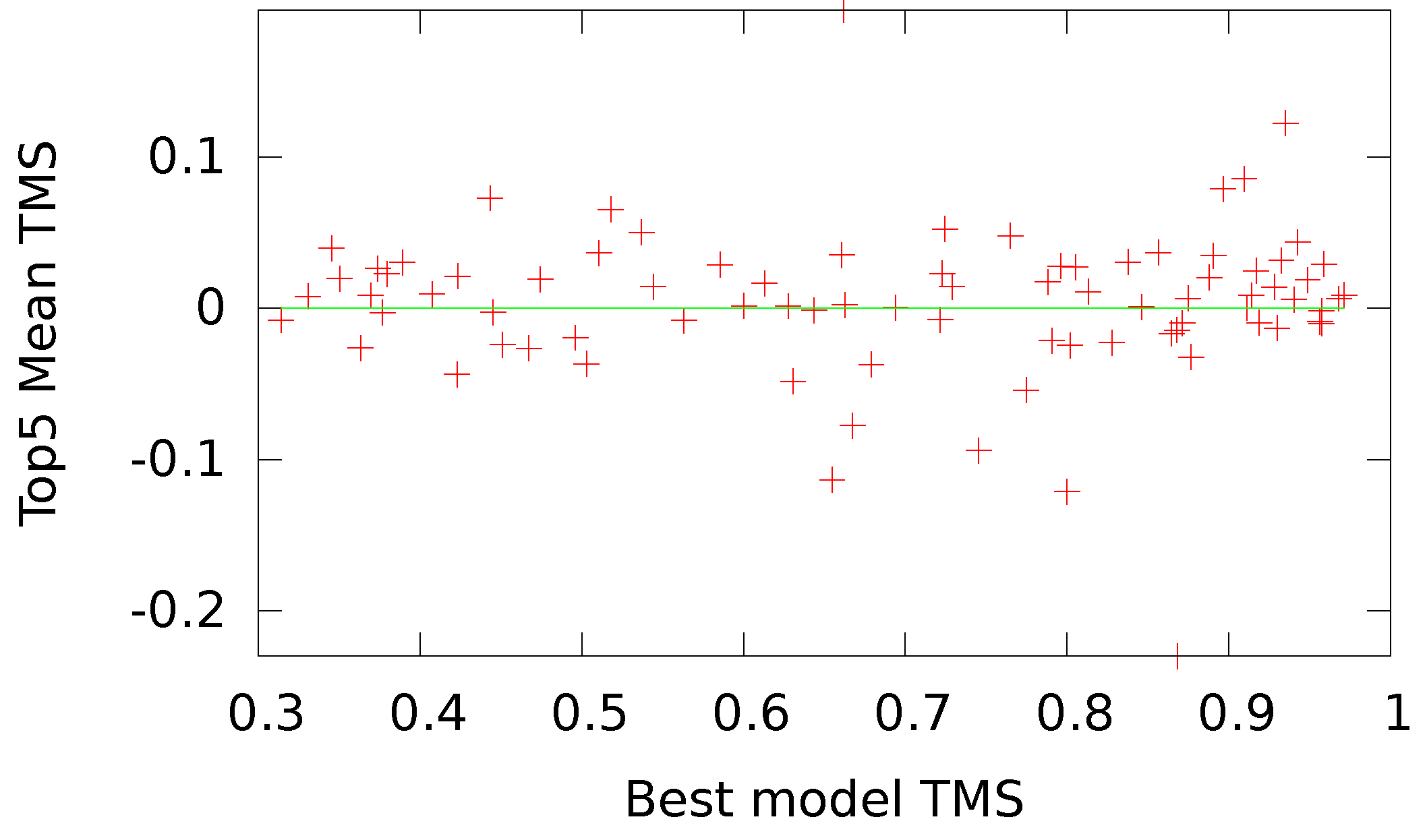
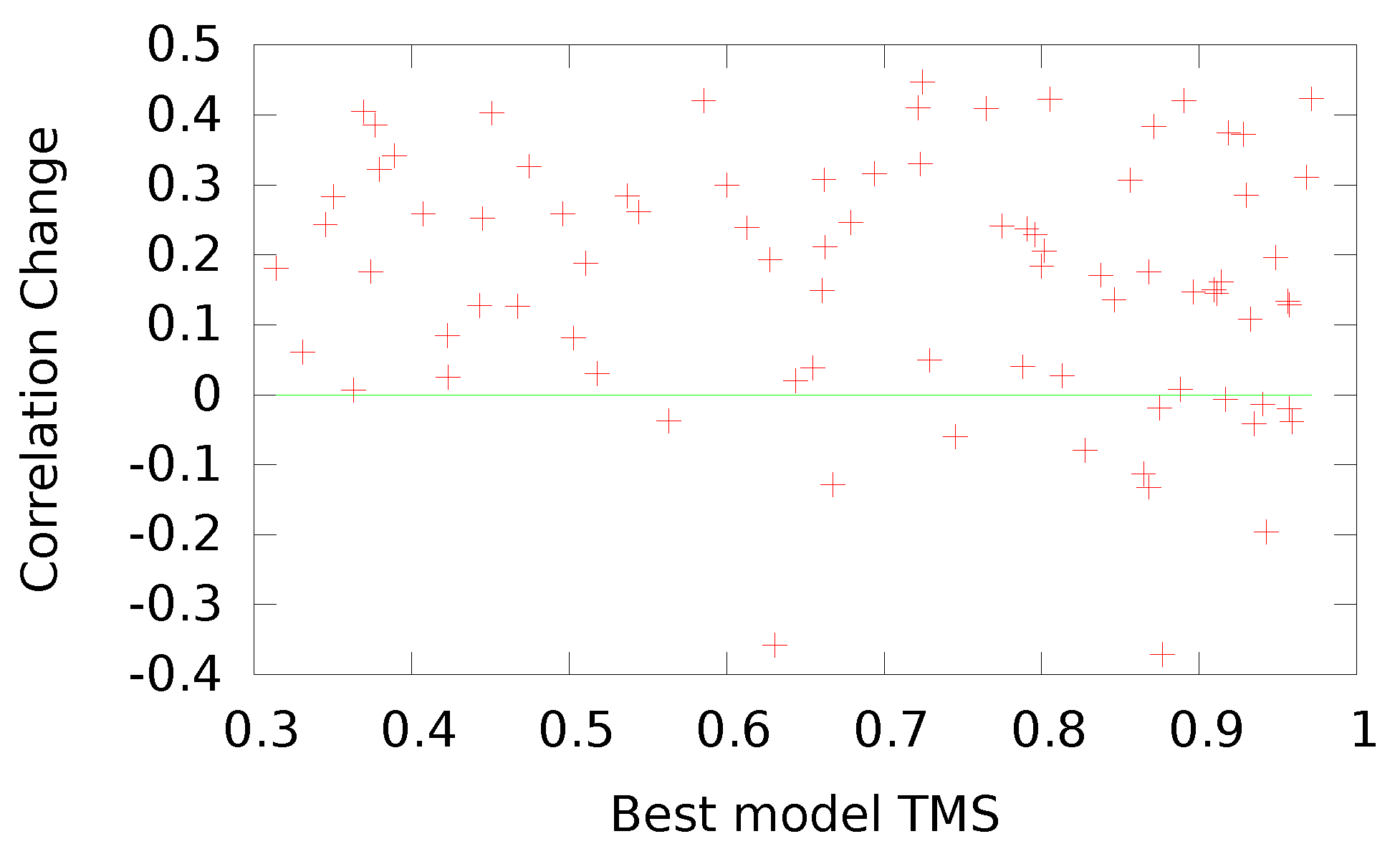
41] structure of the sequence, as measured by the TMS, which provides a normalized value for training our networks. In addition, we established a formula for transforming the TM-score,
, to achieve a distribution of values more fitting to our bipolar selection of neural networks and to align the directionality between our score and the energy values. With our transformation, a native structure scores a ‘−1’.
We used a two-layer feed-forward neural network with momentum, recently described in detail [
30]. A diagram of the neural network architecture is given in
Figure 2. HL1 and HL2 refer to the first and second hidden layers, respectively, and W1, W2, and W3 refer to the weights connecting the different layers. We used an all-connected network in which weights connect all the nodes of one layer to all the nodes of the next layer. The number of weights for a given network will depend on the number of inputs, as this will determine the number of weights in W1. For an all-connected network, the number of weights connecting layer L1 with
nodes (plus a bias node) to layer L2 with
nodes is
. At each node, the weighted sum of the previous layer is passed through an activation function to give the value of that node. We used a bipolar hyperbolic tangent activation function. Momentum refers to the contribution of the gradient calculated at the previous time step to the correction of the weights.
We used the steepest-descent back-propagation algorithm to optimize the neural network weights. We started the analyses with a randomly selected training set of 22,805 protein chain models from the full training set of 296,381. The use of a small training set enabled us to optimize the architecture. After that initial optimization, deviations from the optimized values were tested for the full set. The final optimized values are given in
Table 3. From the full training set, we selected 30% of the proteins at random for an overfit-protection set. A total of six such random training/overfit sets were used to train different realizations of the neural network. For each of the approaches to optimizing UNRES in
Table 4, the initial weights and the order of the training proteins were randomized for each of the six neural network realizations used for the corresponding approach. We chose to use six realizations based on experience from previous work [
18,
30,
57,
58]. To obtain the final prediction, we averaged the results from the six realizations. This also gives an estimate for the stability of the prediction through the standard deviation. We obtained the full training and overfit-protection sets from three sources (number of models given in parenthesis): server models submitted to CASP4 through CASP10 (123,634) [
59], native models from the PDB (54,084) [
41], and native models from the UCSF database of protein models (118,663) [
60]. The three sources are treated in more detail in our previous publication [
18]. Combining these three sources resulted in 296,381 proteins. From these, we randomly set aside 30% for the overfit-protection set and used the rest for training. We used the published results from CASP11 [
59] as the testing set for the results here. This set contains 83 proteins that were selected by the CASP organizers to represent a variety of protein structures. Our approach here simulates participation in CASP11.
Both the EL and GB versions of UNRES were optimized, resulting in the OUNRES (Optimized UNRES) versions. We also tried to integrate additional external information into UNRES, such as the number of residues, the number of atoms, and the scores from DFire2 [
61] and Seder1 [
18]. All input features were z-scored. We used cutoff values, defined by the values of the top and bottom 1% of the data, to limit the effect of outliers in the UNRES values.
Our testing method consisted of simulating participation in the CASP11 competition [
59]. All of our training instances were restricted to models available before the release of CASP11 targets. We collected a total of 83 surviving CASP11 targets and the top 150 server predictions for them. This resulted in a total testing dataset of 12,240 structures. No information from these structures, such as overfit protection, parameter optimization, or any others, was used in the training of the neural network for the testing reported here. However, the version of OUNRES released with this work (and available from
http://mamiris.com/services.html) used CASP11 information to select the top-performing networks among possible candidates.