De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. PacBio Library Preparation and Sequencing
2.3. De Novo Assembly of SMRT Sequencing Reads
2.4. Generation of BioNano Optical Maps and Hybrid Assembly
2.5. The hg38 Reference Genome
2.6. Quality Control and Alignment of the Two Swedish De Novo Assemblies
2.7. Detection of Structural Variation in PacBio Data
2.8. Detection of Novel Sequences
2.9. Repeat Analysis and BLAST Comparison of Novel Sequences
2.10. Anchoring Novel Sequences on Human Chromosomes
2.11. Construction of an Extended Reference Based on Swedish Novel Sequences
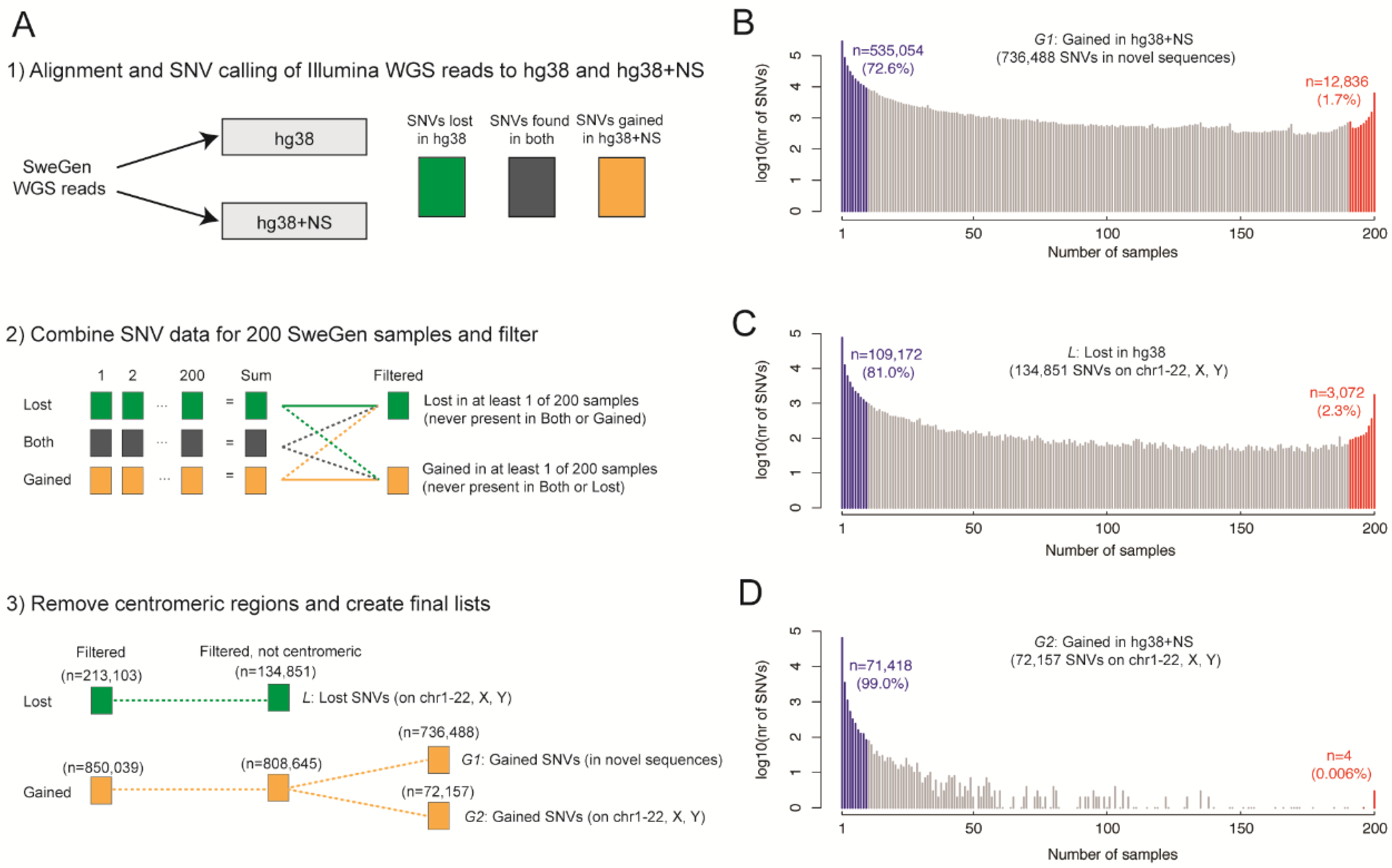
2.12. Re-Alignment of SweGen Illumina Data to hg38 and hg38+NS
2.13. Analysis and Annotation of SNVs in SweGen Re-Alignments
3. Results
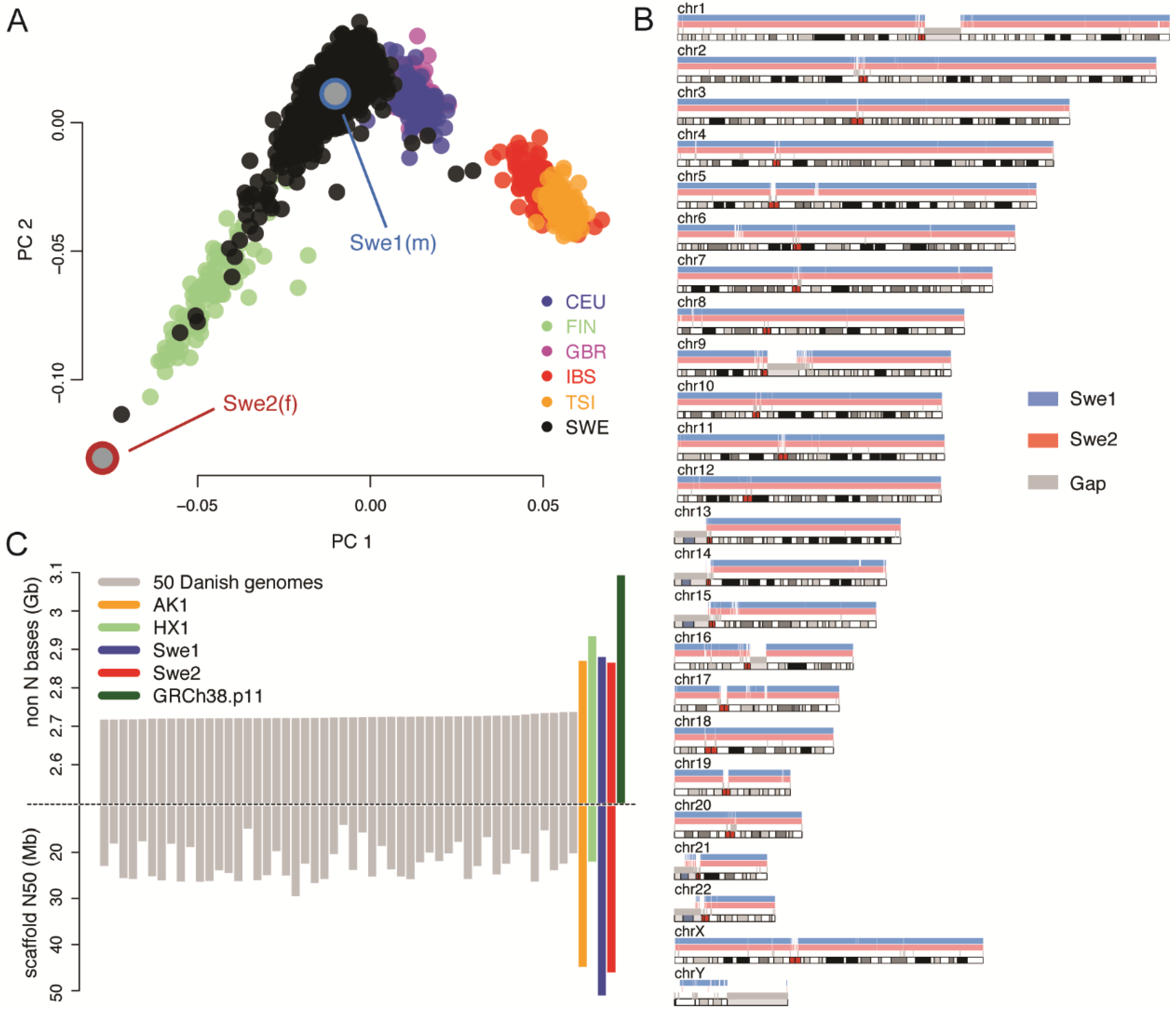
3.1. De Novo Assembly of Two Swedish Individuals
3.2. Evaluating the Quality of the De Novo Assemblies
3.3. Structural Variation in Swedish Genomes
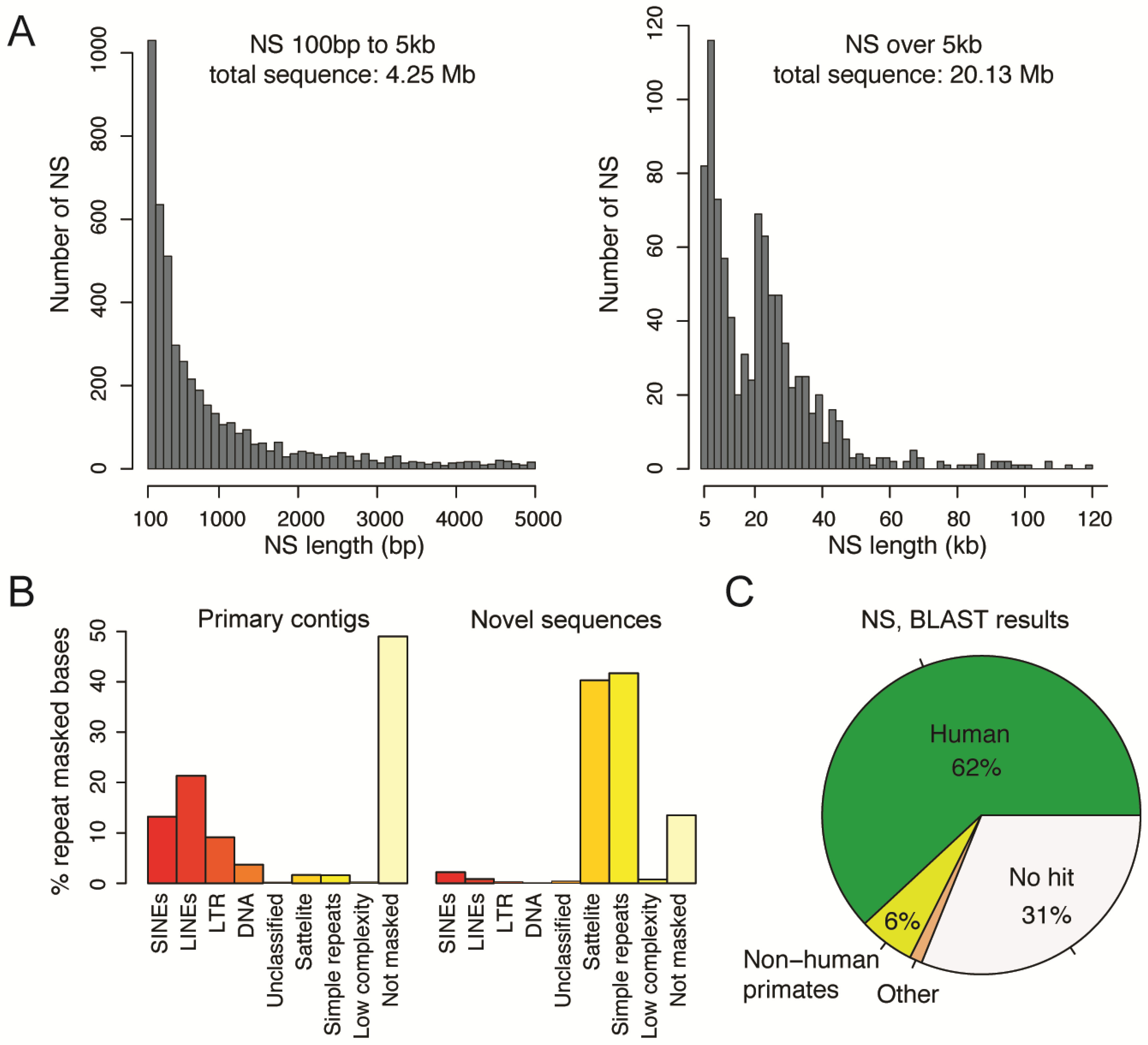
3.4. Detection of Novel Sequences Not Present in the Human Reference
3.5. Origin of the Novel Sequences
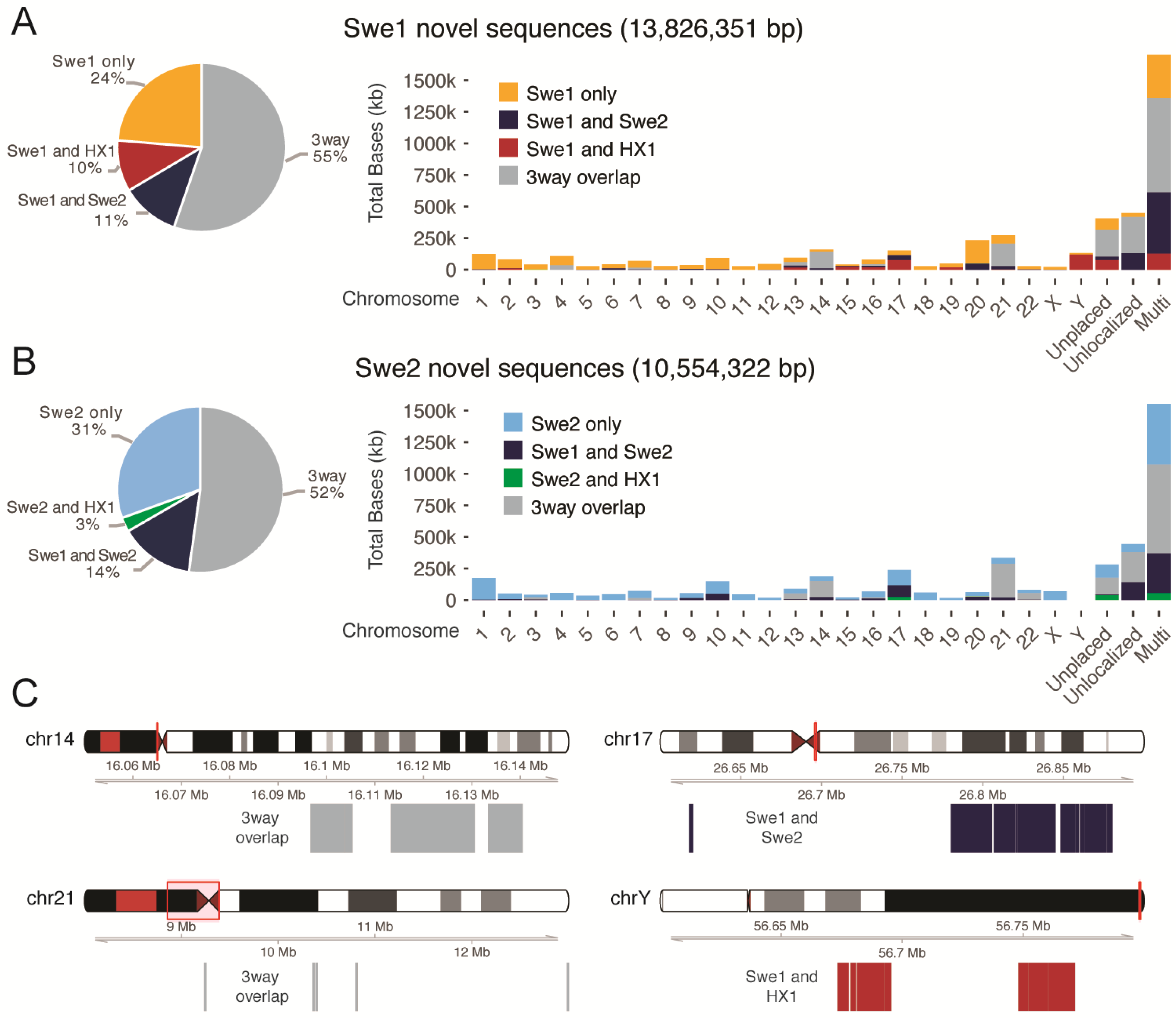
3.6. Comparing Novel Sequences between Swedish Individuals and the Chinese HX1
3.7. Anchoring Novel Sequences on Human Chromosomes
3.8. Application of Novel Sequences for Population Scale WGS Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ameur, A.; Dahlberg, J.; Olason, P.; Vezzi, F.; Karlsson, R.; Martin, M.; Viklund, J.; Kahari, A.K.; Lundin, P.; Che, H.; et al. SweGen: A whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur. J. Hum. Genet. 2017, 25, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, D.I.; Wijmenga, C.; Slagboom, E.P.; Swertz, M.A.; Karssen, L.C.; Abdellaoui, A.; Ye, K.; Guryev, V.; Vermaat, M.; van Dijk, F.; et al. The Genome of the Netherlands: Design, and project goals. Eur. J. Hum. Genet. 2014, 22, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Fakhro, K.A.; Staudt, M.R.; Ramstetter, M.D.; Robay, A.; Malek, J.A.; Badii, R.; Al-Marri, A.A.; Abi Khalil, C.; Al-Shakaki, A.; Chidiac, O.; et al. The Qatar genome: A population-specific tool for precision medicine in the Middle East. Hum. Genome Var. 2016, 3, 16016. [Google Scholar] [CrossRef] [PubMed]
- Gudbjartsson, D.F.; Helgason, H.; Gudjonsson, S.A.; Zink, F.; Oddson, A.; Gylfason, A.; Besenbacher, S.; Magnusson, G.; Halldorsson, B.V.; Hjartarson, E.; et al. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015, 47, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, N.; Moorjani, P.; Rai, N.; Sarkar, B.; Tandon, A.; Patterson, N.; Bhavani, G.S.; Girisha, K.M.; Mustak, M.S.; Srinivasan, S.; et al. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017, 49, 1403. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.P.; Ong, R.T.; Poh, W.T.; Liu, X.; Chen, P.; Li, R.; Lam, K.K.; Pillai, N.E.; Sim, K.S.; Xu, H.; et al. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 2013, 92, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U.K.; Walter, K.; Min, J.L.; Huang, J.; Crooks, L.; Memari, Y.; McCarthy, S.; Perry, J.R.; Xu, C.; Futema, M.; et al. The UK10K project identifies rare variants in health and disease. Nature 2015, 526, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Pierce, L.C.; Biggs, W.H.; di Iulio, J.; Wong, E.H.; Fabani, M.M.; Kirkness, E.F.; Moustafa, A.; Shah, N.; Xie, C.; et al. Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. USA 2016, 113, 11901–11906. [Google Scholar] [CrossRef] [PubMed]
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Maretty, L.; Jensen, J.M.; Petersen, B.; Sibbesen, J.A.; Liu, S.; Villesen, P.; Skov, L.; Belling, K.; Theil Have, C.; Izarzugaza, J.M.; et al. Sequencing and de novo assembly of 150 genomes from Denmark as a population reference. Nature 2017, 548, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.G.; Russ, C.; Costello, M.; Hollinger, A.; Lennon, N.J.; Hegarty, R.; Nusbaum, C.; Jaffe, D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013, 14, R51. [Google Scholar] [CrossRef] [PubMed]
- Ameur, A.; Kloosterman, W.P.; Hestand, M.S. Single-molecule sequencing: Towards clinical applications. Trends Biotechnol. 2018. [CrossRef] [PubMed]
- Chaisson, M.J.; Huddleston, J.; Dennis, M.Y.; Sudmant, P.H.; Malig, M.; Hormozdiari, F.; Antonacci, F.; Surti, U.; Sandstrom, R.; Boitano, M.; et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 2015, 517, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Guo, Y.; Dong, C.; Huddleston, J.; Yang, H.; Han, X.; Fu, A.; Li, Q.; Li, N.; Gong, S.; et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 2016, 7, 12065. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Rhie, A.; Kim, J.; Lee, S.; Sohn, M.H.; Kim, C.U.; Hastie, A.; Cao, H.; Yun, J.Y.; Kim, J.; et al. De novo assembly and phasing of a Korean human genome. Nature 2016, 538, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, M.; Sebra, R.; Pang, A.W.; Ummat, A.; Franzen, O.; Rausch, T.; Stutz, A.M.; Stedman, W.; Anantharaman, T.; Hastie, A.; et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 2015, 12, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Mostovoy, Y.; Levy-Sakin, M.; Lam, J.; Lam, E.T.; Hastie, A.R.; Marks, P.; Lee, J.; Chu, C.; Lin, C.; Dzakula, Z.; et al. A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods 2016, 13, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Levy-Sakin, M.; Kwok, P.Y. De novo human genome assemblies reveal spectrum of alternative haplotypes in diverse populations. Nat. Commun. 2018, 9, 3040. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Zheng-Bradley, X.; Streeter, I.; Fairley, S.; Richardson, D.; Clarke, L.; Flicek, P. Alignment of 1000 Genomes Project reads to reference assembly GRCh38. Gigascience 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bennett, H.M.; Mok, H.P.; Gkrania-Klotsas, E.; Tsai, I.J.; Stanley, E.J.; Antoun, N.M.; Coghlan, A.; Harsha, B.; Traini, A.; Ribeiro, D.M.; et al. The genome of the sparganosis tapeworm Spirometra erinaceieuropaei isolated from the biopsy of a migrating brain lesion. Genome Biol. 2014, 15, 510. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T.; et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017, 49, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338. [Google Scholar] [CrossRef] [PubMed]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Johnston, H.R.; Zhang, G.; Li, Y.; Hu, Y.J.; Qin, Z.S. One Size Doesn’t Fit All—RefEditor: Building Personalized Diploid Reference Genome to Improve Read Mapping and Genotype Calling in Next Generation Sequencing Studies. PLoS Comput. Biol. 2015, 11, e1004448. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ameur, A.; Che, H.; Martin, M.; Bunikis, I.; Dahlberg, J.; Höijer, I.; Häggqvist, S.; Vezzi, F.; Nordlund, J.; Olason, P.; et al. De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data. Genes 2018, 9, 486. https://doi.org/10.3390/genes9100486
Ameur A, Che H, Martin M, Bunikis I, Dahlberg J, Höijer I, Häggqvist S, Vezzi F, Nordlund J, Olason P, et al. De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data. Genes. 2018; 9(10):486. https://doi.org/10.3390/genes9100486
Chicago/Turabian StyleAmeur, Adam, Huiwen Che, Marcel Martin, Ignas Bunikis, Johan Dahlberg, Ida Höijer, Susana Häggqvist, Francesco Vezzi, Jessica Nordlund, Pall Olason, and et al. 2018. "De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data" Genes 9, no. 10: 486. https://doi.org/10.3390/genes9100486
APA StyleAmeur, A., Che, H., Martin, M., Bunikis, I., Dahlberg, J., Höijer, I., Häggqvist, S., Vezzi, F., Nordlund, J., Olason, P., Feuk, L., & Gyllensten, U. (2018). De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data. Genes, 9(10), 486. https://doi.org/10.3390/genes9100486






