“Out of the Can”: A Draft Genome Assembly, Liver Transcriptome, and Nutrigenomics of the European Sardine, Sardina pilchardus
Abstract
1. Introduction
2. Methods, Results and Discussion
2.1. Sampling, DNA Extraction, Library Preparation and Genome Sequencing
2.2. RNA Extraction, Library Preparation and Sequencing
2.3. RNA-Seq Raw Data Clean-Up and De Novo Assembly Transcriptome
2.4. DNA Raw Data Clean-Up and Genome Size Estimation
2.5. Assembly and Assessment of Sardine Genome
2.6. Genome Annotation
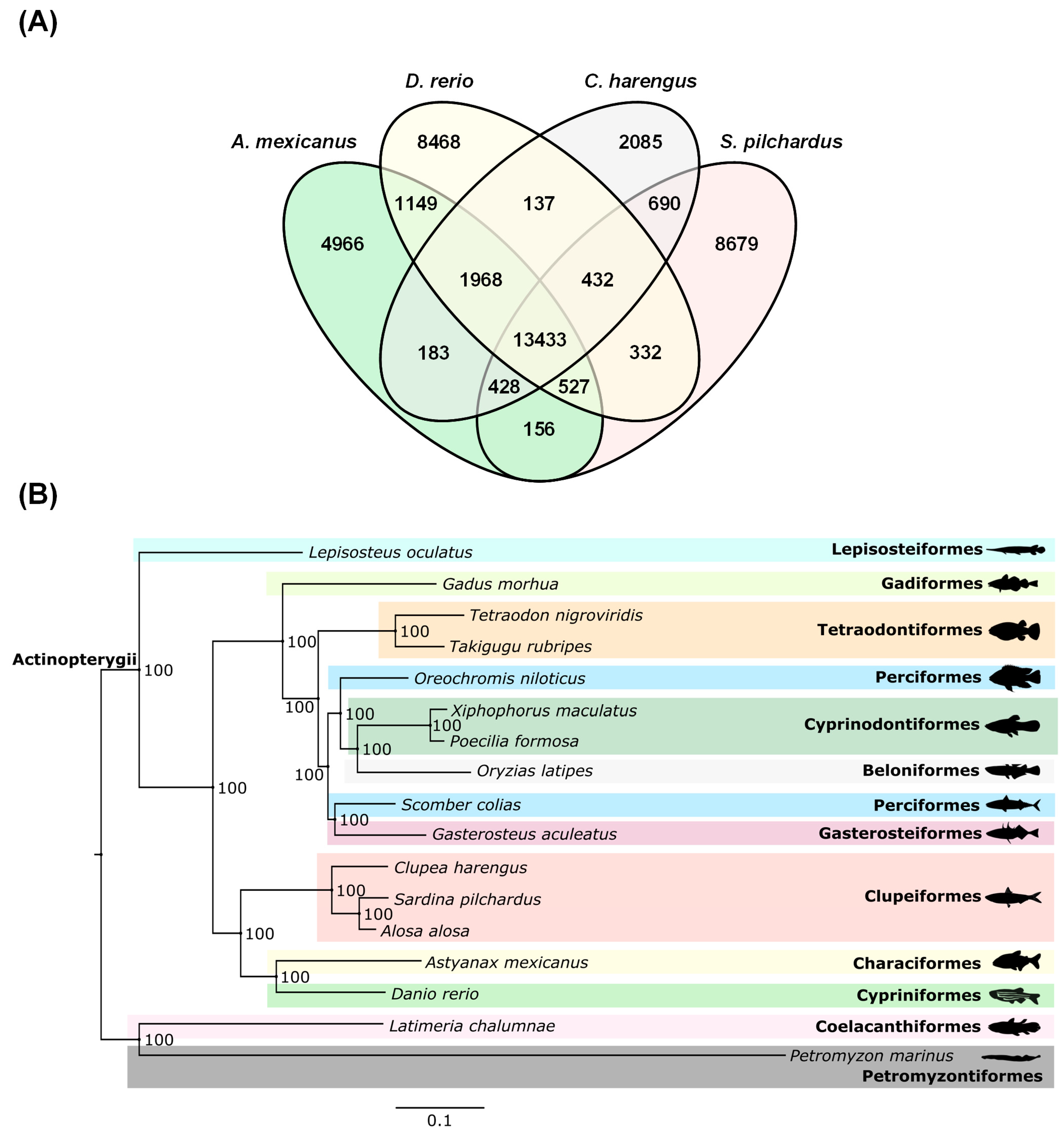
2.7. Sardine Phylogenomics
2.8. Mitochondrial Genome
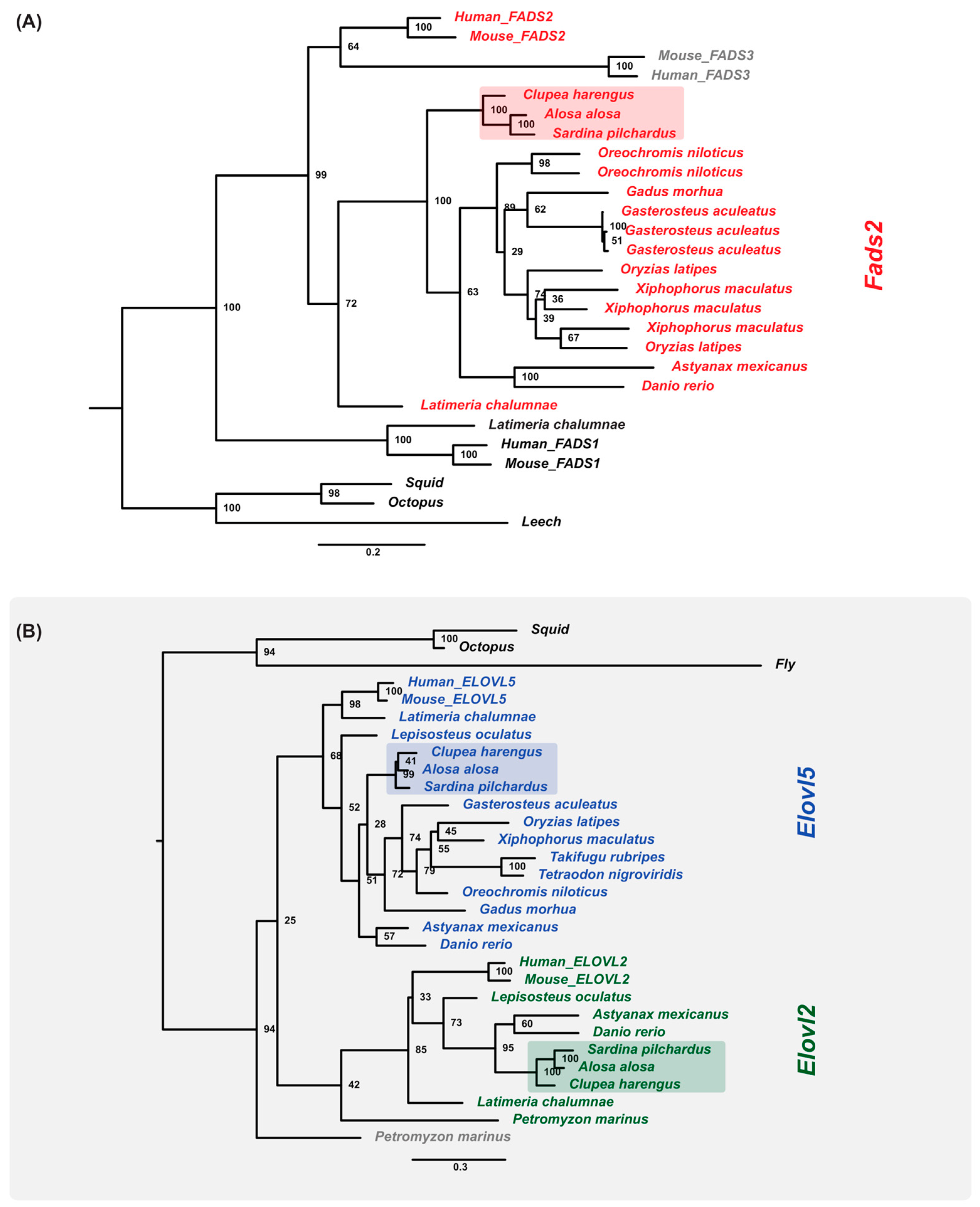
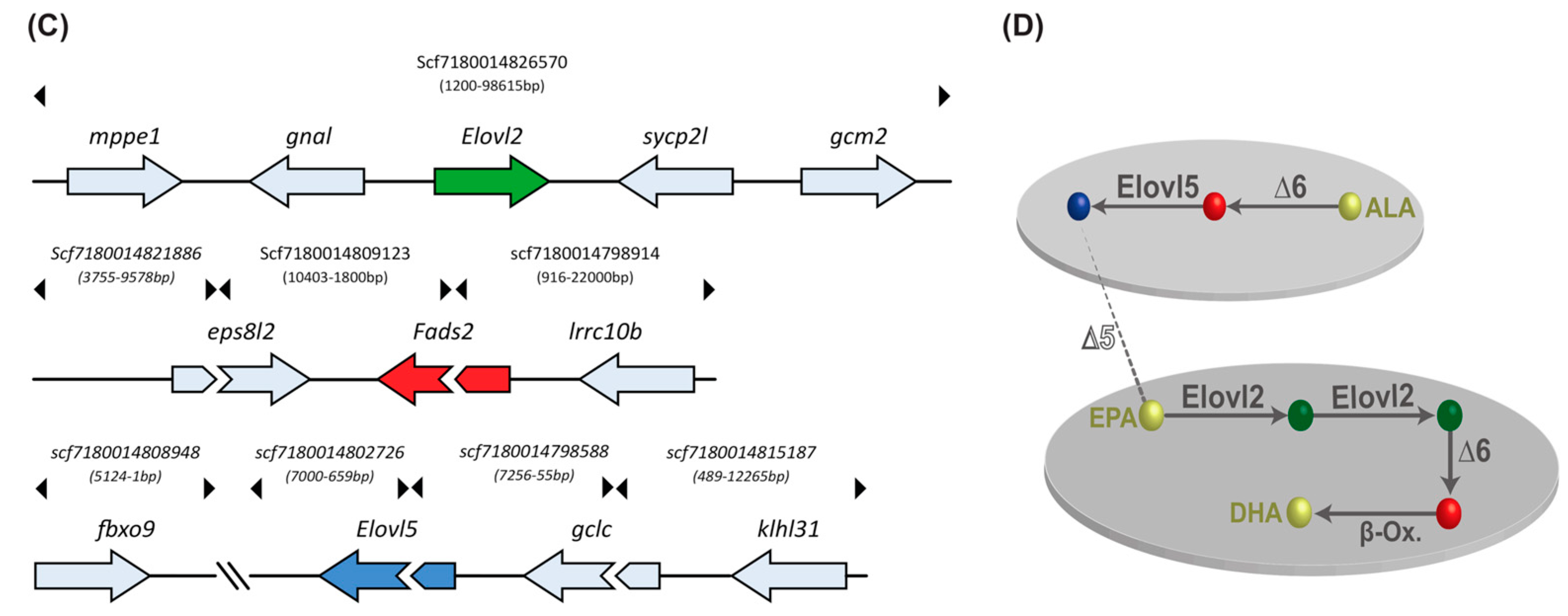
2.9. Gene Orthologs of LC-PUFA Desaturation and Elongation Are Present in the Sardine Genome and Transcriptome
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ravi, V.; Venkatesh, B. The divergent genomes of teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.C.; Ortí, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L.; et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.F.C.; Tocher, D.R.; Monroig, O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016, 62, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Ghasemifard, S.; Turchini, G.M.; Sinclair, A.J. Omega-3 long chain fatty acid “bioavailability”: A review of evidence and methodological considerations. Prog. Lipid Res. 2014, 56, 92–108. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. The State of World Fisheries And Aquaculture (SOFIA)—Meeting the Sustainable Development Goals, 1st ed.; FAO: Rome, Italy, 2018. [Google Scholar]
- Instituto Nacional de Estatística (INE). Estatísticas da Pesca—2016, 1st ed.; Instituto Nacional de Estatística: Lisboa, Portugal, 2017. [Google Scholar]
- Silva, A.; Moreno, A.; Riveiro, I.; Santos, B.; Pita, C.; Rodrigues, J.G.; Villasante, S.; Pavlowski, L.; Duhamel, E. Sardine Fisheries: Resource Assessment and Social and Economic Situation; European Parliament: Brussels, Belgium, 2015; ISBN 978-92-823-8384-1. [Google Scholar]
- Bandarra, N.M.; Marçalo, A.; Cordeiro, A.R.; Pousão-Ferreira, P. Sardine (Sardina pilchardus) lipid composition: Does it change after one year in captivity? Food Chem. 2018, 244, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Olmedo, M.; Iglesias, J.; Peleteiro, J.; Forés, R.; Miranda, A. Acclimatization and induced spawning of sardine Sardina pilchardus Walbaum in captivity. J. Exp. Mar. Biol. Ecol. 1990, 140, 61–67. [Google Scholar] [CrossRef]
- Fernandez-Silva, I.; Henderson, J.B.; Rocha, L.A.; Simison, W.B. Whole-genome assembly of the coral reef Pearlscale Pygmy Angelfish (Centropyge vrolikii). Sci. Rep. 2018, 8, 1498. [Google Scholar] [CrossRef] [PubMed]
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Star, B.; Snipen, L.G.; Hansen, T.F.; Baalsrud, H.T.; Nederbragt, A.J.; Hanel, R.; Salzburger, W.; et al. Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 2016, 48, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Mori, K.; Saitoh, K.; Oshima, K.; Mekuchi, M.; Sugaya, T.; Shigenobu, Y.; Ojima, N.; Muta, S.; Fujiwara, A.; et al. Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. Proc. Natl. Acad. Sci. USA 2013, 110, 11061–11066. [Google Scholar] [CrossRef] [PubMed]
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Jakobsen, K.S.; Jentoft, S. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci. Data 2017, 4, 160132. [Google Scholar] [CrossRef] [PubMed]
- Martinez Barrio, A.; Lamichhaney, S.; Fan, G.; Rafati, N.; Pettersson, M.; Zhang, H.; Dainat, J.; Ekman, D.; Höppner, M.; Jern, P.; et al. The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. Elife 2016, 5, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Monroig, O.; Tocher, D.R.; Castro, L.F.C. Polyunsaturated Fatty Acid Biosynthesis and Metabolism in Fish. In Polyunsaturated Fatty Acid Metabolism, 1st ed.; Burdge, G., Ed.; AOCS Press: Urbana, IL, USA, 2018. [Google Scholar]
- Castro, L.F.C.; Monroig, Ó.; Leaver, M.J.; Wilson, J.; Cunha, I.; Tocher, D.R. Functional desaturase fads1 (Δ5) and fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates. PLoS ONE 2012, 7, e31950. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.M.; Felício, M.; Fonseca, E.; da Fonseca, R.R.; Castro, L.F.C. A resource for sustainable management: De novo assembly and annotation of the liver transcriptome of the Atlantic chub mackerel, Scomber colias. Data Brief 2018, 18, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Lafond-Lapalme, J.; Duceppe, M.O.; Wang, S.; Moffett, P.; Mimee, B. A new method for decontamination of de novo transcriptomes using a hierarchical clustering algorithm. Bioinformatics 2017, 33, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef] [PubMed]
- Chikhi, R.; Medvedev, P. Informed and automated k-mer size selection for genome assembly. Bioinformatics 2014, 30, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.H.; Shin, S.C.; Kim, B.M.; Kang, S.; Kim, J.H.; Ahn, I.; Park, J.; Park, H. Draft genome of the Antarctic dragonfish, Parachaenichthys charcoti. Gigascience 2017, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Depristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Mapleson, D.; Accinelli, G.G.; Kettleborough, G.; Wright, J.; Clavijo, B.J. KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 2017, 33, 574–576. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.M.; Tan, M.H.; Harrisson, K.A.; Lee, Y.P.; Croft, L.J.; Sunnucks, P.; Pavlova, A.; Gan, H.M. De novo genome assembly and annotation of Australia’s largest freshwater fish, the Murray cod (Maccullochella peelii), from Illumina and Nanopore sequencing read. Gigascience 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, B.Y.; Han, J.; Jeong, C.B.; Hwang, D.S.; Lee, M.C.; Kang, H.M.; Kim, D.H.; Lee, D.; Kim, J.; et al. The genome of the marine medaka Oryzias melastigma. Mol. Ecol. Resour. 2018, 18, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; Austin, C.M.; Hammer, M.P.; Lee, Y.P.; Croft, L.J.; Gan, H.M. Finding Nemo: Hybrid assembly with Oxford Nanopore and Illumina reads greatly improves the clownfish (Amphiprion ocellaris) genome assembly. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Marcionetti, A.; Rossier, V.; Bertrand, J.A.M.; Litsios, G.; Salamin, N. First draft genome of an iconic clownfish species (Amphiprion frenatus). Mol. Ecol. Resour. 2018, 18, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.S.; Holt, C.; Moore, B.; Yandell, M. Genome Annotation and Curation Using MAKER and MAKER-P. Curr. Protoc. Bioinform. 2014, 2014, 4.11.1–4.11.39. [Google Scholar] [CrossRef]
- Tørresen, O.K.; Star, B.; Jentoft, S.; Reinar, W.B.; Grove, H.; Miller, J.R.; Walenz, B.P.; Knight, J.; Ekholm, J.M.; Peluso, P.; et al. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genom. 2017, 18, 95. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Gremme, G.; Steinbiss, S.; Kurtz, S. Genome tools: A comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 645–656. [Google Scholar] [CrossRef] [PubMed]
- TransposonPSI: An Application of PSI-Blast to Mine (Retro-)Transposon ORF Homologies. Available online: http://transposonpsi.sourceforge.net/ (accessed on 18 April 2018).
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [PubMed]
- Hoff, K.J.; Lange, S.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER1: Unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 2015, 32, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Mapleson, D.; Venturini, L.; Kaithakottil, G.; Swarbreck, D. Efficient and accurate detection of splice junctions from RNAseq with Portcullis. bioRxiv 2017, 217620. [Google Scholar] [CrossRef]
- Venturini, L.; Caim, S.; Kaithakottil, G.G.; Mapleson, D.L.; Swarbreck, D. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience 2018, 7, giy093. [Google Scholar] [CrossRef] [PubMed]
- Grenon, P.; Smith, B. SNAP and SPAN: Towards dynamic spatial ontology. Spat. Cognit. Comput. 2004, 4, 69–104. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Coleman-Derr, D.; Chen, G.; Gu, Y.Q. OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015, 43, W78–W84. [Google Scholar] [CrossRef] [PubMed]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Aberer, A.J.; Stamatakis, A. ExaML version 3: A tool for phylogenomic analyses on supercomputers. Bioinformatics 2015, 31, 2577–2579. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Monroig, Ó.; Lopes-Marques, M.; Navarro, J.C.; Hontoria, F.; Ruivo, R.; Santos, M.M.; Venkatesh, B.; Tocher, D.R.C.; Castro, L.F. Evolutionary functional elaboration of the Elovl2/5 gene family in chordates. Sci. Rep. 2016, 6, 20510. [Google Scholar] [CrossRef] [PubMed]
- Morais, S.; Monroig, O.; Zheng, X.; Leaver, M.J.; Tocher, D.R. Highly unsaturated fatty acid synthesis in Atlantic salmon: Characterization of ELOVL5- and ELOVL2-like elongases. Mar. Biotechnol. 2009, 11, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Oboh, A.; Betancor, M.B.; Tocher, D.R.; Monroig, O. Biosynthesis of long-chain polyunsaturated fatty acids in the African catfish Clarias gariepinus: Molecular cloning and functional characterisation of fatty acyl desaturase (fads2) and elongase (elovl2) cDNAs. Aquaculture 2016, 462, 70–79. [Google Scholar] [CrossRef]
- Kabeya, N.; Yevzelman, S.; Oboh, A.; Tocher, D.R.; Monroig, O. Essential fatty acid metabolism and requirements of the cleaner fish, ballan wrasse Labrus bergylta: Defining pathways of long-chain polyunsaturated fatty acid biosynthesis. Aquaculture 2018, 488, 199–206. [Google Scholar] [CrossRef]

| Features | Genome # | Liver Transcriptome # |
|---|---|---|
| Raw Data | ||
| Raw sequencing reads | 456,775,568 | 122,806,922 |
| Clean reads | 412,914,751 | 111,524,231 |
| Contig statistics | ||
| Number of contigs | 90,290 | 245,053 |
| Total contig size, Mb | 640.1 | 278.5 |
| Contig N50 size, bp | 10,878 | 1760 |
| Longest contig, bp | 87,474 | 15,773 |
| GC/AT/N, % | 44.45 | 48.10 |
| Scaffold statistics | ||
| Number of scaffolds | 45,321 | - |
| Total scaffold size, Mb | 641.5 | - |
| Scaffold N50 size, bp | 25,577 | - |
| Longest scaffold, bp | 285,113 | - |
| Genome coverage, × | 59 | - |
| BUSCO Completeness (Met/Ver/Actino) | ||
| Complete, % | 82.7/70.5/68.8 | 99.1/80.6/72 |
| Complete and single copy, % | 78.8/68.4/66.3 | 41.5/31.2/29.1 |
| Complete and duplicated, % | 3.9/2.1/2.5 | 57.6/49.4/42.9 |
| Fragmented, % | 9.2/19.0/13.3 | 0.6/10.5/8.6 |
| Missing, % | 8.1/10.5/17.9 | 0.3/8.9/19.4 |
| Total BUSCO found | 91.9/89.5/82.1 | 99.7/91.1/80.6 |
| Annotation | ||
| Number of protein-coding genes | 29,701 | - |
| Number of functionally annotated proteins | 28,783 | - |
| Average CDS length | 1561.42 | - |
| Longest CDS | 49,643 | - |
| Average protein length | 373.45 | - |
| Longest protein | 16,525 | - |
| Average number of exon per gene | 6.59 | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, A.M.; Tørresen, O.K.; Kabeya, N.; Couto, A.; Petersen, B.; Felício, M.; Campos, P.F.; Fonseca, E.; Bandarra, N.; Lopes-Marques, M.; et al. “Out of the Can”: A Draft Genome Assembly, Liver Transcriptome, and Nutrigenomics of the European Sardine, Sardina pilchardus. Genes 2018, 9, 485. https://doi.org/10.3390/genes9100485
Machado AM, Tørresen OK, Kabeya N, Couto A, Petersen B, Felício M, Campos PF, Fonseca E, Bandarra N, Lopes-Marques M, et al. “Out of the Can”: A Draft Genome Assembly, Liver Transcriptome, and Nutrigenomics of the European Sardine, Sardina pilchardus. Genes. 2018; 9(10):485. https://doi.org/10.3390/genes9100485
Chicago/Turabian StyleMachado, André M., Ole K. Tørresen, Naoki Kabeya, Alvarina Couto, Bent Petersen, Mónica Felício, Paula F. Campos, Elza Fonseca, Narcisa Bandarra, Mónica Lopes-Marques, and et al. 2018. "“Out of the Can”: A Draft Genome Assembly, Liver Transcriptome, and Nutrigenomics of the European Sardine, Sardina pilchardus" Genes 9, no. 10: 485. https://doi.org/10.3390/genes9100485
APA StyleMachado, A. M., Tørresen, O. K., Kabeya, N., Couto, A., Petersen, B., Felício, M., Campos, P. F., Fonseca, E., Bandarra, N., Lopes-Marques, M., Ferraz, R., Ruivo, R., Fonseca, M. M., Jentoft, S., Monroig, Ó., Da Fonseca, R. R., & C. Castro, L. F. (2018). “Out of the Can”: A Draft Genome Assembly, Liver Transcriptome, and Nutrigenomics of the European Sardine, Sardina pilchardus. Genes, 9(10), 485. https://doi.org/10.3390/genes9100485







