Molecular Genotyping (SSR) and Agronomic Phenotyping for Utilization of Durum Wheat (Triticum durum Desf.) Ex Situ Collection from Southern Italy: A Combined Approach Including Pedigreed Varieties
Abstract
1. Introduction
2. Methods
2.1. Plant Material
2.2. Molecular Characterization
2.3. Genomic DNA Extraction and Genotyping
2.4. Morphological Characterization
2.5. Statistical Analysis
3. Results
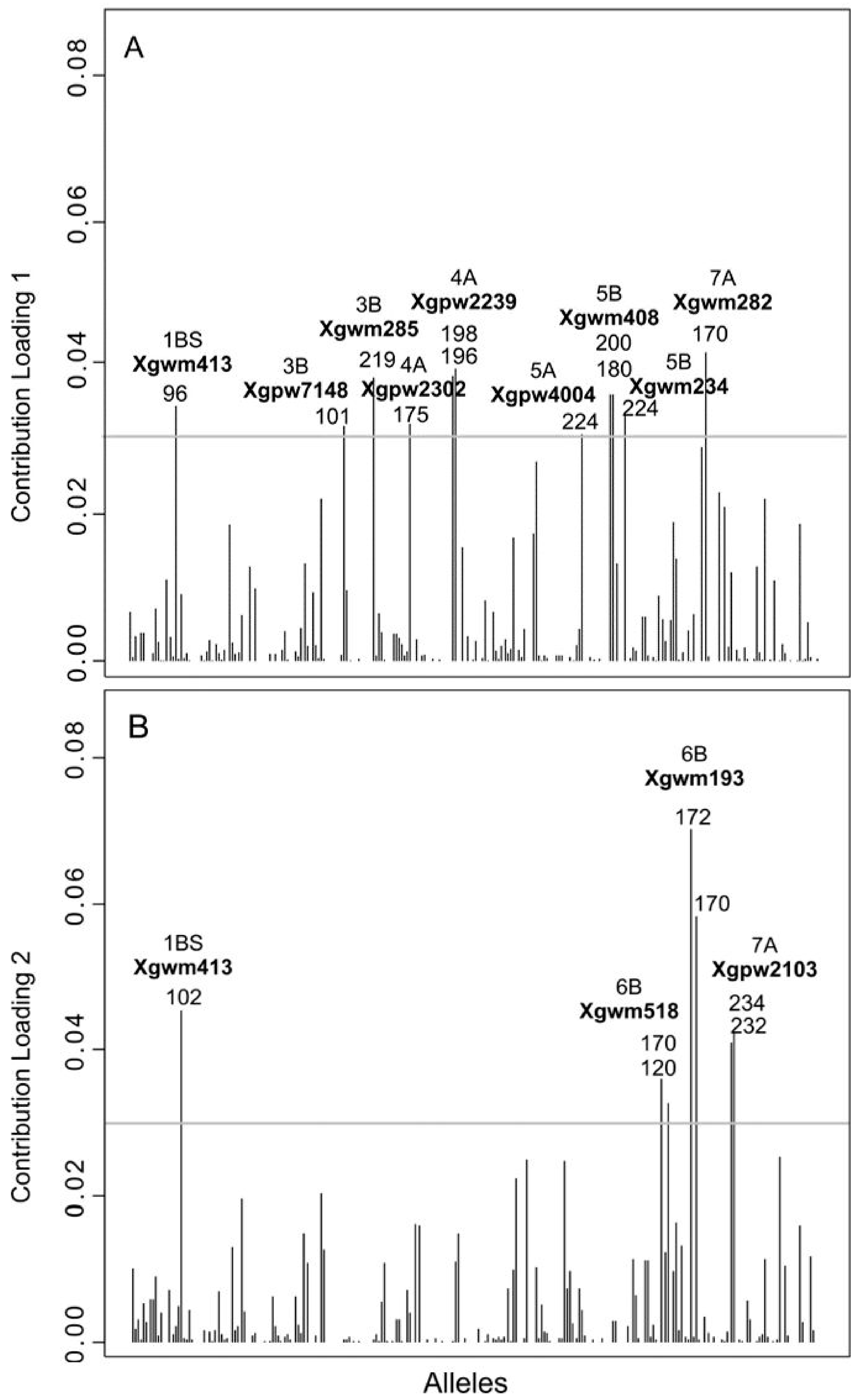
3.1. Overall Levels of Genetic Diversity
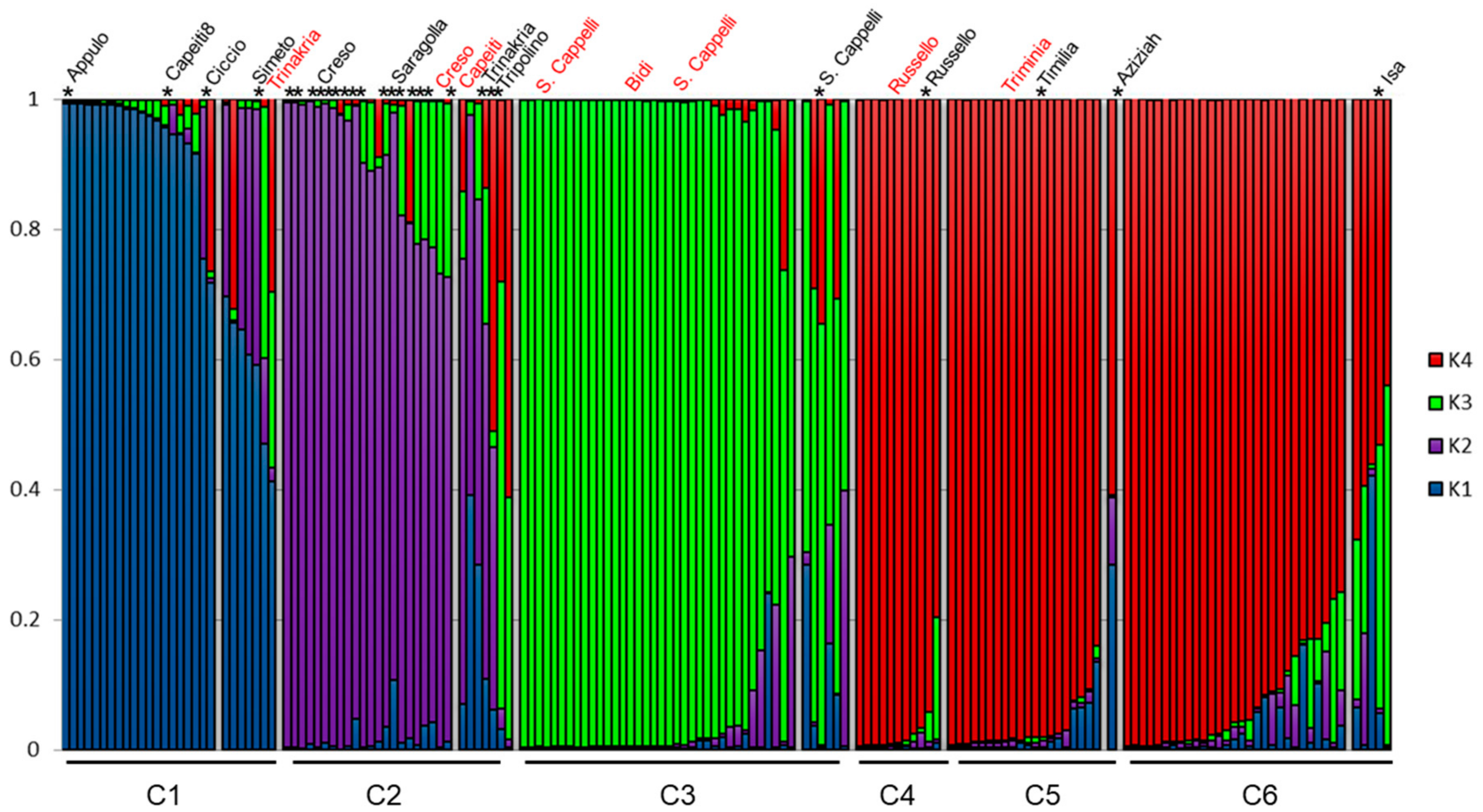
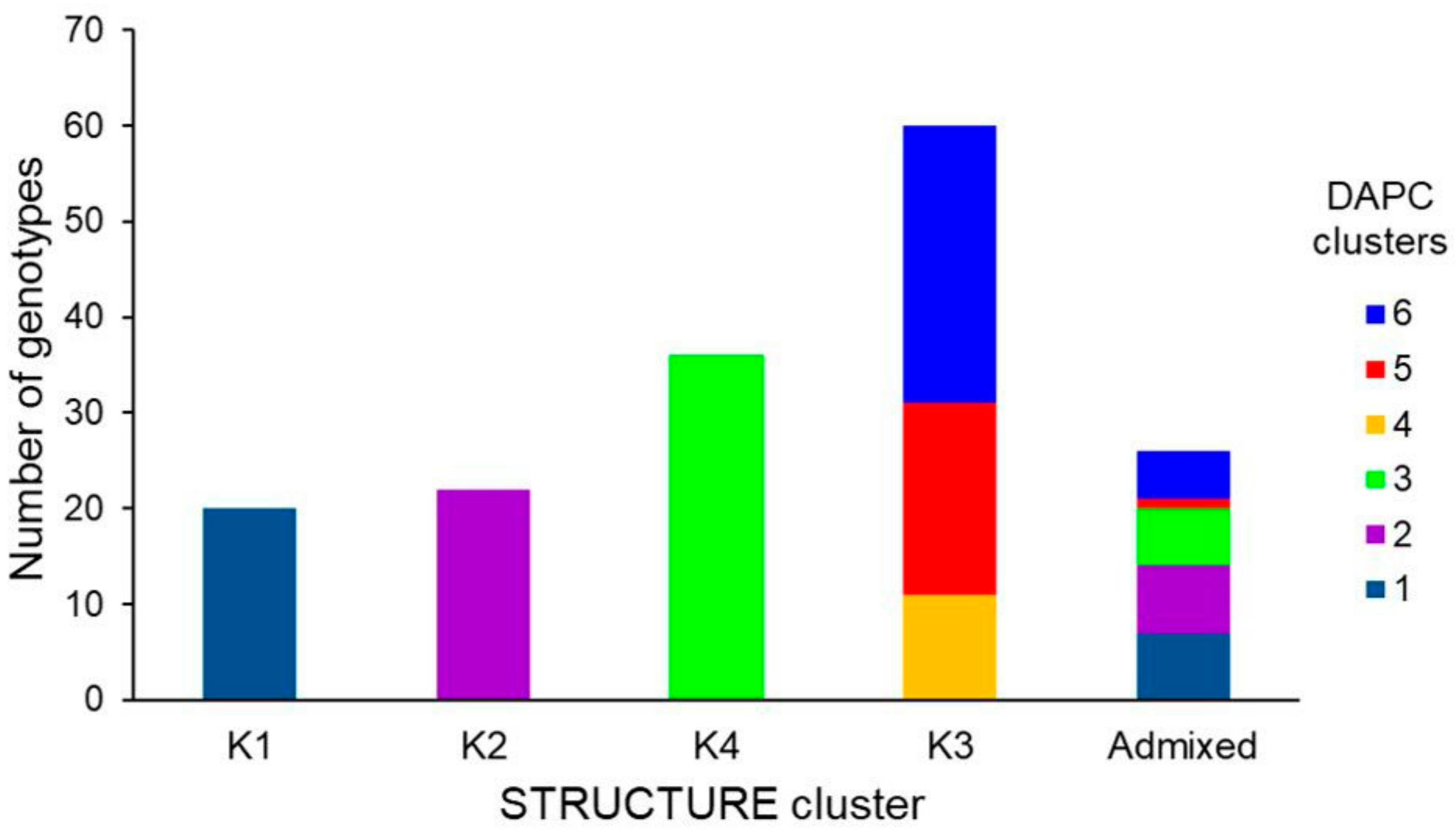
3.2. Genetic Structure of the Wheat Collection
3.3. Genetic Differentiation among DAPC Clusters
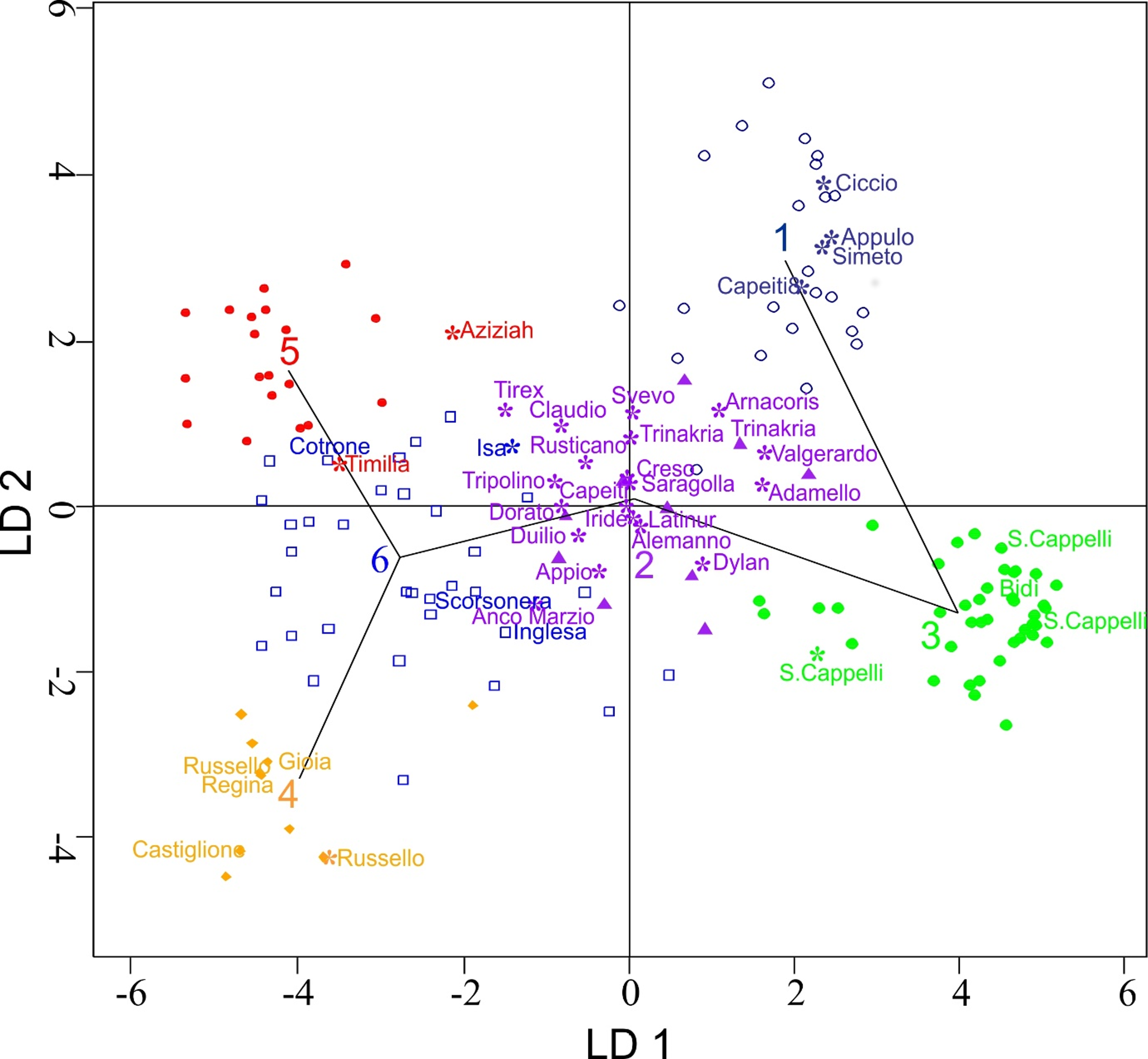
3.4. Morphological Characterization
3.5. Morphological Differentiation among Discriminant Analysis of Principal Components Clusters
4. Discussion
4.1. Diversity of South Italian Durum Wheat Germplasm
4.2. Analysis of Population Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Odong, T.L.; Jansen, J.; van Eeuwijk, F.; van Hintum, T.J.L. Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor. Appl. Genet. 2013, 126, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Deo Upadhyaya, H.; Varshney, R.K.; Gowda, C.L.L. Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legumes. Front. Plant Sci. 2013, 4, 309. [Google Scholar] [CrossRef] [PubMed]
- Paux, E.; Sourdille, P.; Mackay, I.; Feuillet, C. Sequence-Based marker development in wheat: Advances and applications to breeding. Biotechnol. Adv. 2012, 30, 1071–1088. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sun, D.; Chen, L.; You, F.M.; Wang, J.; Peng, Y.; Nevo, E.; Sun, D.; Luo, M.C.; Peng, J. Genetic diversity revealed by single nucleotide polymorphism markers in a worldwide germplasm collection of durum wheat. Int. J. Mol. Sci. 2013, 14, 7061–7088. [Google Scholar] [CrossRef] [PubMed]
- Thuillet, A.C.; Bataillon, T.; Poirier, S.; Santoni, S.; David, J.L. Estimation of long-term effective population sizes through the history of durum wheat using microsatellite data. Genetics 2005, 169, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Evenson, R.E.; Gollin, D. Assessing the impact of the Green revolution. Science 2003, 300, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Akar, T.; Baresel, J.P.; Bebeli, P.J.; Bettencourt, E.; Bladenopoulos, K.V.; Czembor, J.H.; Fasoula, D.A.; Katsiotis, A.; Koutis, K.; et al. Cereal landraces for sustainable agriculture. A review. Agron. Sustain. Dev. 2010, 30, 237–269. [Google Scholar] [CrossRef]
- Hoisington, D.; Khairallah, M.; Reeves, T.; Ribault, J.M.; Skovmand, B.; Taba, S.; Warburton, M. Plant genetic resources: What can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. USA 1999, 96, 5937–5943. [Google Scholar] [CrossRef] [PubMed]
- Moragues, M.; Moralejo, M.; Sorrells, M.E.; Royo, C. Dispersal of durum wheat [Triticum turgidum L. ssp. turgidum convar. durum (Desf.) MacKey] landraces across the Mediterranean basin assessed by AFLPs and microsatellites. Genet. Resour. Crop Evol. 2007, 54, 1133–1144. [Google Scholar] [CrossRef]
- Maccaferri, M.; Sanguineti, M.C.; Donini, P.; Tuberosa, R. Microsatellite analysis reveals a progressive widening of the genetic basis in the elite durum wheat germplasm. Theor. Appl. Genet. 2003, 107, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Figliuolo, G.; Mazzeo, M.; Greco, I. Temporal variation of diversity in Italian durum wheat germplasm. Genet. Resour. Crop Evol. 2007, 54, 615–626. [Google Scholar] [CrossRef]
- Ruiz, M.; Giraldo, P.; Royo, C.; Villegas, D.; Aranzana, M.J.; Carrillo, J.M. Diversity and genetic structure of a collection of Spanish durum wheat landraces. Crop Sci. 2012, 52, 2262–2275. [Google Scholar] [CrossRef]
- Laidò, G.; Mangini, G.; Taranto, F.; Gadaleta, A.; Blanco, A.; Cattivelli, L.; Marone, D.; Mastrangelo, A.M.; Papa, R.; De Vita, P. Genetic diversity and population structure of tetraploid wheats (Triticum turgidum L.) estimated by SSR, DArT and pedigree data. PLoS ONE 2013, 8, e67280. [Google Scholar] [CrossRef] [PubMed]
- Sahri, A.; Chentoufi, L.; Arbaoui, M.; Ardisson, M.; Belqadi, L.; Birouk, A.; Roumet, P.; Muller, M.H. Towards a comprehensive characterization of durum wheat landraces in Moroccan traditional agrosystems: Analysing genetic diversity in the light of geography, farmer’s taxonomy and tetraploid wheat domestication history. BMC Evol. Biol. 2014, 14, 264. [Google Scholar] [CrossRef] [PubMed]
- Motzo, R.; Giunta, F. The effect of breeding on the phenology of Italian durum wheats: From landraces to modern cultivars. Eur. J. Agron. 2007, 26, 462–470. [Google Scholar] [CrossRef]
- De Vita, P.; Li Destri Nicosia, O.; Nigro, F.; Platani, C.; Riefolo, C.; Di Fonzo, N.; Cattivelli, L. Breeding progress in morphophysiological, agronomical and qualitative traits of durum wheat cultivars released in Italy during the 20th century. Eur. J. Agron. 2007, 26, 39–53. [Google Scholar] [CrossRef]
- Giunta, F.; Motzo, R.; Pruneddu, G. Trends since 1900 in the yield potential of Italian-bred durum wheat cultivars. Eur. J. Agron. 2007, 27, 12–24. [Google Scholar] [CrossRef]
- Kabbaj, H.; Sall, A.T.; Al-Abdallat, A.; Geleta, M.; Amri, A.; Filali-Maltouf, A.; Belkadi, B.; Ortiz, R.; Bassi, F.M. Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front. Plant Sci. 2017, 8, 1277. [Google Scholar] [CrossRef] [PubMed]
- Martos, V.; Royo, C.; Rharrabti, Y.; Garcia del Moral, L.F. Using AFLPs to determine phylogenetic relationships and genetic erosion in durum wheat cultivars released in Italy and Spain throughout the 20th century. Field Crops Res. 2005, 91, 107–116. [Google Scholar] [CrossRef]
- De Cillis, E. I Grani d’Italia; Tipografia della Camera dei Deputati: Roma, Italy, 1927; p. 173. [Google Scholar]
- Deidda, M.; Motzo, R.; Giunta, F.; Fois, S. Evoluzione Varietale e Qualità in Frumento Duro (Triticum turgidum subsp. Durum): Dalle Vecchie Popolazioni Alle Attuali Cultivar; Dipartimento di Scienze agronomiche e Genetica vegetale agraria, Facoltà di Agraria, Università degli Studi di Sassari: Sassari, Italy, 2001; p. 3. [Google Scholar]
- Grignac, P. Contribution à L’étude de Triticum durum Desf. Thése Doctorat, Université de Toulouse, Toulouse, France, 1965; p. 151. [Google Scholar]
- Bozzini, A. Genetics and breeding of durum wheats. Genet. Agraria 1970, 24, 145–193. [Google Scholar]
- Porceddu, E.; De Pace, C.; Spagnoletti Zeuli, P.L.; Scarascia Mugnozza, G.T. Genetic variability in segregating populations of durum wheat (Triticum durum). Genet. Agraria XXXIV 1980, 34, 182–183. [Google Scholar]
- Blanco, A.; Scarascia-Mugnozza, G.T. Messapia. A new early and productive variety of durum wheat. L’Inf. Agr. XLV 1989, 39, 61–63. [Google Scholar]
- Blanco, A.; Simeone, R.; Resta, P. The addition of Dasypirum villosum (L.) Candargy chromosome in durum wheat (Triticum durum Desf.). Theor. Appl. Genet. 1987, 74, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Giorgi, B.; Perrino, P.; Simeone, R. Genetic resources and breeding of durum wheat quality. Agricoltura e Ricerca 1991, 144, 41–58. [Google Scholar]
- Hammer, K.; Laghetti, G.; Cifarelli, S.; Perrino, P. Collection of plant genetic resources in Italy 1989. Die Kulturpflanze 1990, 38, 311–323. [Google Scholar] [CrossRef]
- Somers, D.J.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Korzun, V.; Börner, A.; Worland, A.J.; Law, C.N.; Röder, M.S. Application of microsatellite markers to distinguish inter-varietal chromosome substitution lines of wheat (Triticum aestivum L.). Euphytica 1997, 95, 149–155. [Google Scholar] [CrossRef]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixer, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [PubMed]
- Sourdille, P.; Gandon, B.; Chiquet, V.; Nicot, N.; Somers, D.; Murigneux, A.; Bernard, M. Wheat Genoplante SSR Mapping Data Release: A New Set of Markers and Comprehensive Genetic and Physical Mapping Data. Graingenes. 2004. Available online: http://wheat.pw.usda.gov/GG2/index.shtml (accessed on 15 March 2018).
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- International Board for Plant Genetic Resources (IPGRI). Descriptors for Wheat (Revised); International Plant Genetic Resources Institute: Rome, Italy, 1985. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [PubMed]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.02, Logiciel Sous WindowsTM pour la Génétique des Populations; Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II: Montpellier, France, 2001. [Google Scholar]
- El Mousadik, A.; Petit, R.J. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3). 2002. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 15 March 2018).
- Yang, M.H.; Fu, Y.B. An R version of FPTest for testing differences in allelic count. Conserv. Genet. Resour. 2017, 9, 313–318. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Weir, B.; Cockerham, C.C. Estimating F statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Institute Inc. SAS. SAS/STAT® 9.2. User’s Guide, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2009. [Google Scholar]
- Al-Khanjari, S.; Hammer, K.; Buerkert, A.; Röder, M.S. Molecular diversity of Omani wheat revealed by microsatellites: I. Tetraploid landraces. Genet. Resour. Crop Evol. 2007, 54, 1291–1300. [Google Scholar] [CrossRef]
- Reif, J.C.; Zhang, P.; Dreisigacker, S.; Warburton, M.L.; van Ginkel, M.; Hoisington, D.; Bohn, M.; Melchinger, A.E. Wheat genetic diversity trends during domestication and breeding. Theor. Appl. Genet. 2005, 110, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Vellvè, R. The decline of diversity in European agriculture. Ecologist 1993, 23, 64–69. [Google Scholar]
- Medini, M.; Hamza, S.; Rebai, A.; Baum, M. Analysis of genetic diversity in Tunisian durum wheat cultivars and related wild species by SSR and AFLP markers. Genet. Resour. Crop Evol. 2005, 52, 21–31. [Google Scholar] [CrossRef]
- Dalrymple, D.G. Development and Spread of High-Yielding Wheat Varieties in Developing Countries, 7th ed.; US Agency for International Development: Washington, DC, USA, 1986.
- Maccaferri, M.; Cane’, M.A.; Sanguineti, M.C.; Salvi, S.; Colalongo, M.C.; Massi, A.; Clarke, F.; Knox, R.; Pozniak, C.J.; Clarke, J.M.; et al. A consensus framework map of durum wheat (Triticum durum Desf.) suitable for linkage disequilibrium analysis and genome-wide association mapping. BMC Genom. 2014, 15, 873. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Porceddu, E. Varietal evolution and current problems of breeding winter cereals. Rivista di Agronomia 1987, 21, 33–54. [Google Scholar]
- Tóth, B.; Galiba, G.; Fehěr, E.; Sutka, J.; Snape, J.W. Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theor. Appl. Genet. 2003, 107, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Snape, J.W.; Butterworth, K.; Whitechurch, E.; Worland, A.J. Waiting for fine times: Genetics of flowering time in wheat. Euphytica 2001, 119, 185–190. [Google Scholar] [CrossRef]
- Leonova, I.; Pestsova, E.; Salina, E.; Efremova, T.; Röder, M.S.; Börner, A.; Fischbeck, G. Mapping of the Vrn-B1 gene in Triticum aestivum using microsatellite marker. Plant Breed. 2003, 122, 209–212. [Google Scholar] [CrossRef]
- Jia, G.; Chen, P.; Qin, G.; Bai, G.; Wang, X.; Wang, S.; Zhou, B.; Zhang, S.; Liu, D. QTLs for Fusarium head blight response in a wheat DH population of Wangshuibai/Alondra‘s’. Euphytica 2005, 146, 183–191. [Google Scholar] [CrossRef]
- Zanetti, S.; Winzeler, M.; Feuillet, G.; Keller, B.; Messmer, M. Genetic analysis of bread-making quality in wheat and spelt. Plant Breed. 2001, 120, 13–20. [Google Scholar] [CrossRef]
- Arbelbide, M.; Bernardo, R. Mixed-model QTL mapping for kernel hardness and dough strength in bread wheat. Theor. Appl. Genet. 2006, 112, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Gonzalez-Hernandez, J.L.; Mergoum, M.; Ali, S.; Adhikari, T.B.; Kianian, S.F.; Elias, E.M.; Hughes, G.R. Identification and molecular mapping of a gene conferring resistance to Pyrenophora tritici-repentis race 3 in tetraploid wheat. Phytopathology 2006, 96, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.A.; Procunier, J.D.; Humphreys, D.G.; Tranquilli, G.; Schlatter, A.R.; Marcucci-Poltri, S.; Frohberg, R.; Dubcovsky, J. Development of PCR based markers for a high grain protein content gene from Triticum turgidum ssp. dicoccoides transferred to bread wheat. Crop Sci. 2000, 40, 518–524. [Google Scholar] [CrossRef]
- Olmos, S.; Distelfeld, A.; Chicaiza, O.; Schlatter, A.R.; Fahima, T.; Echenique, V.; Dubcovsky, J. Precise mapping of a locus affecting grain protein content in durum wheat. Theor. Appl. Genet. 2003, 107, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, Z.; He, Z.; Wu, L.; Bai, B.; Lan, C.; Wang, C.; Zhou, G.; Zhu, H.; Xia, X. QTL mapping of adult-plant resistances to stripe rust and leaf rust in Chinese wheat cultivar Bainong 64. Theor. Appl. Genet. 2012, 125, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.R.; Kumar, N.; Jaiswal, V.; Girdharwal, N.; Prasad, M.; Balyan, H.S.; Gupta, P.K. Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Mol. Breed. 2012, 29, 963–972. [Google Scholar] [CrossRef]
- Van Bueren, E.L.; Jones, S.S.; Tamm, L.; Murphy, K.M.; Myers, J.R.; Leifert, C.; Messmer, M.M. The need to breed crop varieties suitable for organic farming, using wheat, tomato and broccoli as examples: A review. NJAS Wagening. J. Life Sci. 2011, 58, 193–205. [Google Scholar] [CrossRef]
- Van Bueren, E.L.; Struik, P.C.; Jacobsen, E. Ecological concepts in organic farming and their consequences for an organic crop ideotype. NJAS Wagening J. Life Sci. 2002, 50, 1–26. [Google Scholar] [CrossRef]
- Anderson, W.K.; Seymour, M.; D’Antuono, M.F. Evidence for differences between cultivars in responsiveness of wheat to applied nitrogen. Aust. J. Agric. Res. 1991, 42, 363–377. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Sylvester-Bradley, R.; Scott, R.K. Evidence for differences between winter wheat cultivars in acquisition of soil mineral nitrogen and uptake and utilization of applied fertilizer nitrogen. J. Agric. Sci. 1998, 130, 29–44. [Google Scholar] [CrossRef]
- Dawson, J.C.; Murphy, K.M.; Jones, S.S. Decentralized selection and participatory approaches in plant breeding for low-input systems. Euphytica 2008, 160, 143–154. [Google Scholar] [CrossRef]
- Gioia, T.; Nagel, K.A.; Beleggia, R.; Fragasso, M.; Ficco, D.B.M.; Pieruschka, R.; De Vita, P.; Fiorani, F.; Papa, R. Impact of domestication on the phenotypic architecture of durum wheat under contrasting nitrogen fertilization. J. Exp. Bot. 2015, 66, 5519–5530. [Google Scholar] [CrossRef] [PubMed]
| Germplasm Group | n | Na | He | Rs |
|---|---|---|---|---|
| Accessions | 136 | 5.39 | 0.601 | 4.562 |
| Group1 (1947–1950) | 37 | 4.34 | 0.579 | 3.715 |
| Group2 (1973–1982) | 77 | 5.00 | 0.603 | 3.882 |
| Group3 (1983–2003) | 22 | 3.57 | 0.531c | 3.303 |
| Varieties | 28 | 3.86 | 0.576 | 3.830 |
| Old and intermediate varieties (1915–1973) | 9 | 3.45 | 0.604 | 3.434 |
| Modern varieties (1974–2007) | 19 | 3.09 | 0.534 | 3.011 |
| Germplasm | Year | C1 | C2 | C3 | C4 | C5 | C6 | Total |
|---|---|---|---|---|---|---|---|---|
| Accession | Collection | |||||||
| Group1 (1947–1950) | 1947–1950 | 16 | 7 | 2 | 12 | 37 | ||
| Group2 (1973–1982) | 1973–1982 | 17 | 5 | 19 | 1 | 17 | 18 | 77 |
| Group3 (1983–2003) | 1983–2007 | 6 | 5 | 6 | 2 | - | 3 | 22 |
| Total | 23 | 10 | 41 | 10 | 19 | 33 | 136 | |
| Varieties | Release | |||||||
| Old and intermediate | 1915–1973 | 2 | 3 | 1 | 1 | 2 | - | 9 |
| Modern | 1974–2007 | 2 | 16 | - | - | - | 1 | 19 |
| Total | 4 | 19 | 1 | 1 | 2 | 1 | 28 |
| DAPC Cluster | C1 | C2 | C3 | C4 | C5 | C6 |
|---|---|---|---|---|---|---|
| C1 | 0.225 ** | 0.284 ** | 0.415 ** | 0.317 ** | 0.220 ** | |
| C2 | 0.256 ** | 0.276 ** | 0.218 ** | 0.145 ** | ||
| C3 | 0.502 ** | 0.458 ** | 0.327 ** | |||
| C4 | 0.251 ** | 0.158 ** | ||||
| C5 | 0.122 ** | |||||
| C6 |
| Source of Variations | ||||
|---|---|---|---|---|
| Traits | Between Clusters | Between Germplasm Types (Clusters) | Between Accessions (Germplasm Type × Clusters) | Error |
| Heading date (from 10 April) | 268.1 *** | 186.2 *** | 126.4 *** | 8.7 |
| Plant height (cm) | 13,786.8 *** | 394.1 *** | 519.5 *** | 95.1 |
| Spike length (cm) | 98.7 *** | 28.9 *** | 9.4 *** | 2.2 |
| Spikelets number/spike (n) | 244.2 *** | 47.6 *** | 16.9 *** | 1.8 |
| Fertile spikelets number/spike (n) | 81.9 *** | 25.2 *** | 20.3 *** | 6.3 |
| Kernel length (mm) | 1.7 *** | 0.6 *** | 0.7 *** | 0.1 |
| Kernel width (mm) | 0.06 ns | 0.3 *** | 0.4 *** | 0.05 |
| Kernels number/spike (n) | 263.0ns | 39.1 ** | 182.4 *** | 65.4 |
| Kernels weight/spike (g) | 1.7 *** | 1.1 *** | 1.0 *** | 0.2 |
| 1000 kernels weight (g) | 370.7 *** | 569.9 *** | 323.9 *** | 46.5 |
| Kernels number/spikelet (n) | 2.6 *** | 0.7 *** | 0.3 *** | 0.1 |
| Kernels number/fertile spikelet (n) | 1.2 *** | 0.5 *** | 0.2 *** | 0.1 |
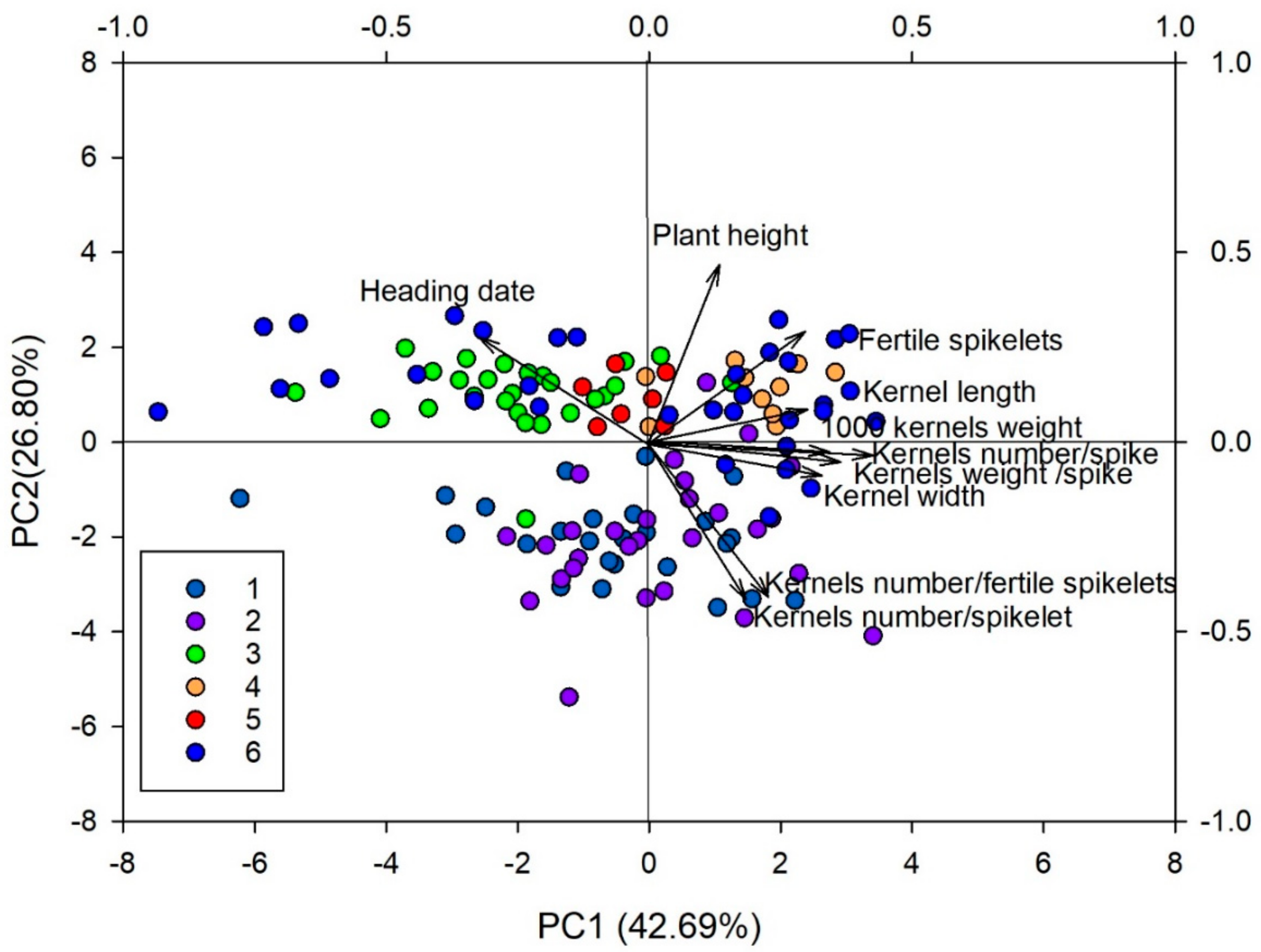
| Principal Components | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| Kernels weight/spike | 0.42 | −0.04 | −0.01 | 0.11 |
| Heading date | −0.31 | 0.27 | 0.21 | 0.37 |
| Plant height | 0.14 | 0.46 | 0.08 | −0.14 |
| Spikelets number/spike | 0.23 | 0.43 | −0.03 | −0.02 |
| Kernels number/fertile spikelets | 0.22 | −0.42 | 0.25 | 0.04 |
| Kernels number/spikelet | 0.19 | −0.41 | 0.38 | 0.09 |
| Kernels number/spike | 0.37 | −0.05 | 0.39 | 0.07 |
| Spike length | 0.17 | 0.29 | 0.34 | −0.25 |
| Fertile spikelets number/spike | 0.29 | 0.29 | 0.29 | 0.03 |
| 1000 kernels weight | 0.33 | −0.03 | −0.44 | 0.06 |
| Kernel length | 0.30 | 0.09 | −0.28 | 0.72 |
| Kernel width | 0.34 | −0.08 | −0.35 | −0.48 |
| Eigenvalues | ||||
| Variation explained (%) | 42.7 | 26.8 | 14.9 | 4.5 |
| Cumulative proportion of total variance | 42.7 | 69.5 | 84.4 | 88.9 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzario, S.; Logozzo, G.; David, J.L.; Zeuli, P.S.; Gioia, T. Molecular Genotyping (SSR) and Agronomic Phenotyping for Utilization of Durum Wheat (Triticum durum Desf.) Ex Situ Collection from Southern Italy: A Combined Approach Including Pedigreed Varieties. Genes 2018, 9, 465. https://doi.org/10.3390/genes9100465
Marzario S, Logozzo G, David JL, Zeuli PS, Gioia T. Molecular Genotyping (SSR) and Agronomic Phenotyping for Utilization of Durum Wheat (Triticum durum Desf.) Ex Situ Collection from Southern Italy: A Combined Approach Including Pedigreed Varieties. Genes. 2018; 9(10):465. https://doi.org/10.3390/genes9100465
Chicago/Turabian StyleMarzario, Stefania, Giuseppina Logozzo, Jacques L. David, Pierluigi Spagnoletti Zeuli, and Tania Gioia. 2018. "Molecular Genotyping (SSR) and Agronomic Phenotyping for Utilization of Durum Wheat (Triticum durum Desf.) Ex Situ Collection from Southern Italy: A Combined Approach Including Pedigreed Varieties" Genes 9, no. 10: 465. https://doi.org/10.3390/genes9100465
APA StyleMarzario, S., Logozzo, G., David, J. L., Zeuli, P. S., & Gioia, T. (2018). Molecular Genotyping (SSR) and Agronomic Phenotyping for Utilization of Durum Wheat (Triticum durum Desf.) Ex Situ Collection from Southern Italy: A Combined Approach Including Pedigreed Varieties. Genes, 9(10), 465. https://doi.org/10.3390/genes9100465





