The Role of Bromodomain Proteins in Regulating Gene Expression
Abstract
:1. Introduction
2. Histone Acetylation and the Regulation of Transcription
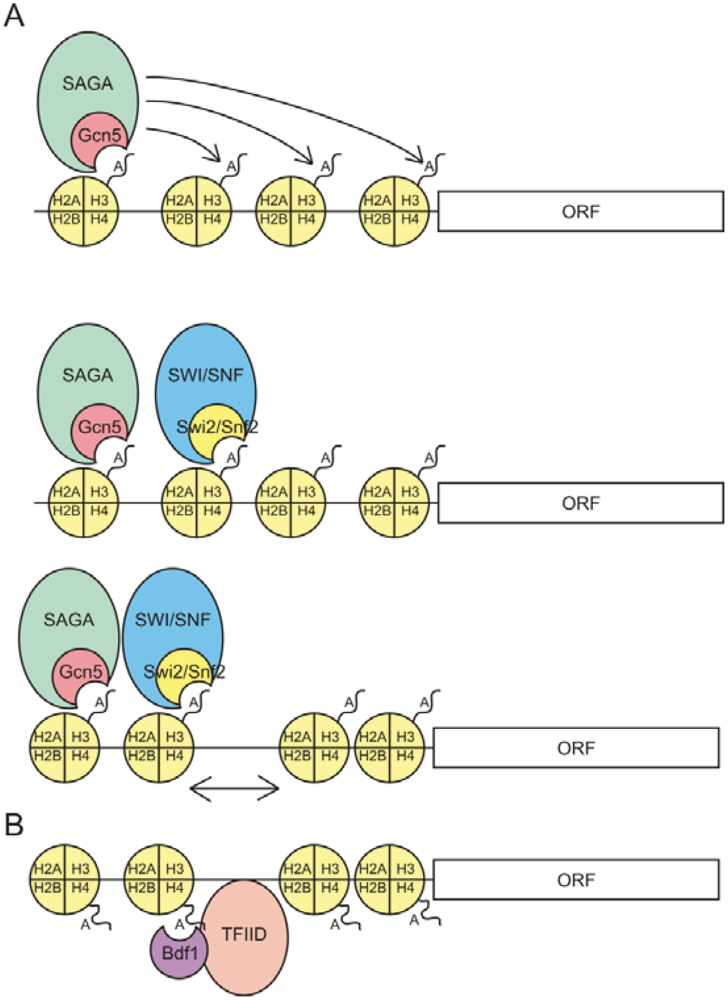
3. Recruitment of Trans Factors to Acetylated Histones by Bromodomains
| Protein | Organism | Complex | Histone-binding specificity | Other interactions | Function | |||
|---|---|---|---|---|---|---|---|---|
| Components of histone acetyltransferases: | ||||||||
| Gcn5 | Yeast | SAGA, SALSA/SLIK, ADA | H3ac, H4K16ac [45,53] | Required for acetylation of nucleosomal histones leading to gene activation [57,58,59,60,61,62,63] | ||||
| Spt7 | Yeast | SAGA | H3K9ac [53] | |||||
| p300/CBP | Mammals | p300: H3ac [64], H1K74ac, H2AK5ac, H2AK15ac, H2AK36ac, H2BK43ac, H2BK46ac, H3K56ac, H3K79ac, H3K115ac, H3K122ac, H4K5ac, H4K44ac [49] CBP: H2AK15ac, H3K56ac [49], H4K20ac [65] | Acetylated p53 [43], RNA polymerase II [66], TBP [67], TFIIB [68], RNA helicase A [69], c-Jun [70], c-Fos [71] | Acetylates histones [72,73], general and specific transcription factors [74,75]; recruitment of transcriptional machinery leading to transcription initiation [76,77] | ||||
| Components of chromatin remodeling complexes: | ||||||||
| Swi2/Snf2 | Yeast | SWI/SNF | H3ac, especially H3K14ac [52] | Catalytic component. Remodels chromatin in promoters and also evicts nucleosomes in elongation thus enhancing transcriptional elongation [78,79,80] | ||||
| Rsc1 | Yeast | RSC | H3ac (weakly) [52] | |||||
| Rsc2 | Yeast | RSC | H3ac (weakly) [52] | |||||
| Rsc4 | Yeast | RSC | H2Bac and H3ac, especially H3K14ac [52,81] | |||||
| Sth1 | Yeast | RSC | H3K14ac, H3K115ac, H2AK21ac [52] | Catalytic component. Remodels chromatin in promoters and plays a role in enhancing elongation [82,83,84] | ||||
| BET bromodomains: | ||||||||
| BDF1 | Yeast | SWR1 | H3ac, H4ac [52,53,85,86,87] | TFIID subunit Taf67 [86] | Recruitment of TFIID thus leading to transcription initiation [88], incorporation of H2A.Z into nucleosomes [89] | |||
| BDF2 | Yeast | H2Bac and H3ac [52] | TFIID subunit Taf67 [86] | Partially redundant with Bdf1 [88] | ||||
| Brd2 | Mammals | H4K12ac, H4K5/8ac [46,47,90,91,92], H1K74ac [49] | E2F transcription factors [93], TBP [94], unknown HAT [95], TAFII250, components of SWI/SNF complex [96] | Increases transcription of E2F- regulated genes [93] | ||||
| Brd4 | Mammals | H3ac, H4ac [47,51,97], H2AK5ac, H2AK36ac, H2AK75ac, H2BK43ac, H3K18ac, H3K36ac, H3K37ac, H3K56ac, H4K5ac, H4K20ac, H4K44ac [49] | Mediator complex [98,99], P-TEFb complex [100,101] | Increases transcription by RNA polymerase II [100,101] | ||||
3.1. Histone Acetyltransferase Complexes
3.2. Chromatin Remodeling Complexes
3.3. BET Bromodomain Proteins
4. Histone Acetylation Pathways as Drug Targets
5. Conclusions
References
- Ramakrishnan, V. Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 83–112. [Google Scholar] [CrossRef]
- Katan-Khaykovich, Y.; Struhl, K. Dynamics of global histone acetylation and deacetylation in vivo: Rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002, 16, 743–752. [Google Scholar] [CrossRef]
- Murr, R.; Loizou, J.I.; Yang, Y.G.; Cuenin, C.; Li, H.; Wang, Z.Q.; Herceg, Z. Histone acetylation by Trrap–Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2005, 8, 91–99. [Google Scholar]
- Unnikrishnan, A.; Gafken, P.R.; Tsukiyama, T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 2010, 17, 430–439. [Google Scholar]
- Verreault, A.; Kaufman, P.D.; Kobayashi, R.; Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 1996, 87, 95–104. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailey, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J., III.; Gingeras, T.R. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef]
- Schübeler, D.; MacAlpine, D.M.; Scalzo, D.; Wirbelauer, C.; Kooperberg, C.; van Leeuwen, F.; Gottschling, D.E.; O’Neill, L.P.; Turner, B.M.; Delrow, J.; Bell, S.P.; Groudine, M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004, 18, 1263–1271. [Google Scholar] [CrossRef]
- Pokholok, D.K.; Harbison, C.T.; Levine, S.; Cole, M.; Hannett, N.M.; Lee, T.I.; Bell, G.W.; Walker, K.; Rolfe, P.A.; Herbolsheimer, E.; Zeitlinger, J.; Lewitter, F.; Gifford, D.K.; Young, R.A. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005, 122, 517–527. [Google Scholar] [CrossRef]
- Oppikofer, M.; Kueng, S.; Martino, F.; Soeroes, S.; Hancock, S.M.; Chin, J.W.; Fischle, W.; Gasser, S.M. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 2011, 30, 2610–2621. [Google Scholar] [CrossRef]
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; Zhao, K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903. [Google Scholar]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; Boyer, L.A.; Young, R.A.; Jaenisch, R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar]
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2010, 470, 279–283. [Google Scholar]
- Jin, C.; Felsenfeld, G. Nucleosome stability mediated by histone variants H3. 3 and H2A. Z. Genes Dev. 2007, 21, 1519–1529. [Google Scholar] [CrossRef]
- Bruce, K.; Myers, F.A.; Mantouvalou, E.; Lefevre, P.; Greaves, I.; Bonifer, C.; Tremethick, D.J.; Thorne, A.W.; Crane-Robinson, C. The replacement histone H2A. Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005, 33, 5633–5639. [Google Scholar]
- Hardy, S.; Jacques, P.; Gévry, N.; Forest, A.; Fortin, M.; Laflamme, L.; Gaudreau, L.; Robert, F. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A. Z deposition. PLoS Genet. 2009, 5, e1000687. [Google Scholar]
- Millar, C.B.; Xu, F.; Zhang, K.; Grunstein, M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006, 20, 711–722. [Google Scholar] [CrossRef]
- Ren, Q.; Gorovsky, M.A. Histone H2A. Z acetylation modulates an essential charge patch. Mol. Cell 2001, 7, 1329–1335. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Halley, J.E.; Rine, J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A. Z in Saccharomycescerevisiae. Genes Dev. 2006, 20, 700–710. [Google Scholar] [CrossRef]
- McKittrick, E.; Gafken, P.R.; Ahmad, K.; Henikoff, S. Histone H3. 3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 2004, 101, 1525–1530. [Google Scholar]
- Hake, S.B.; Garcia, B.A.; Duncan, E.M.; Kauer, M.; Dellaire, G.; Shabanowitz, J.; Bazett-Jones, D.P.; Allis, C.D.; Hunt, D.F. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006, 281, 559–568. [Google Scholar]
- Lee, D.Y.; Hayes, J.J.; Pruss, D.; Wolffe, A.P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 1993, 72, 73–84. [Google Scholar] [CrossRef]
- Tse, C.; Sera, T.; Wolffe, A.P.; Hansen, J.C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998, 18, 4629–4638. [Google Scholar]
- Clayton, A.L.; Hazzalin, C.A.; Mahadevan, L.C. Enhanced histone acetylation and transcription: A dynamic perspective. Mol. Cell 2006, 23, 289–296. [Google Scholar] [CrossRef]
- Allahverdi, A.; Yang, R.; Korolev, N.; Fan, Y.; Davey, C.A.; Liu, C.F.; Nordenskiöld, L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011, 39, 1680–1691. [Google Scholar]
- Vettese-Dadey, M.; Grant, P.A.; Hebbes, T.R.; Crane-Robinson, C.; Allis, C.D.; Workman, J.L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996, 15, 2508–2518. [Google Scholar]
- Liu, Y.; Lu, C.; Yang, Y.; Fan, Y.; Yang, R.; Liu, C.F.; Korolev, N.; Nordenskiold, L. Influence of Histone Tails and H4 Tail Acetylations on Nucleosome-Nucleosome Interactions. J. Mol. Biol. 2011, 414, 749–764. [Google Scholar]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311, 844–847. [Google Scholar]
- Agalioti, T.; Chen, G.; Thanos, D. Deciphering the transcriptional histone acetylation code for a human gene. Cell 2002, 111, 381–392. [Google Scholar] [CrossRef]
- Luebben, W.R.; Sharma, N.; Nyborg, J.K. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc. Natl. Acad. Sci. USA 2010, 107, 19254–19259. [Google Scholar]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.R.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2010, 30, 249–262. [Google Scholar]
- Rundlett, S.E.; Carmen, A.A.; Suka, N.; Turner, B.M.; Grunstein, M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 1998, 392, 831–835. [Google Scholar]
- Galarneau, L.; Nourani, A.; Boudreault, A.A.; Zhang, Y.; Héliot, L.; Allard, S.; Savard, J.; Lane, W.S.; Stillman, D.J.; Côté, J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 2000, 5, 927–937. [Google Scholar] [CrossRef]
- Holstege, F.C.P.; Jennings, E.G.; Wyrick, J.J.; Lee, T.I.; Hengartner, C.J.; Green, M.R.; Golub, T.R.; Lander, E.S.; Young, R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 1998, 95, 717–728. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Tong, J.K.; Schreiber, S.L. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 13708–13713. [Google Scholar]
- Dion, M.F.; Altschuler, S.J.; Wu, L.F.; Rando, O.J. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 2005, 102, 5501–5506. [Google Scholar]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Taverna, S.D.; Li, H.; Ruthenburg, A.J.; Allis, C.D.; Patel, D.J. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007, 14, 1025–1040. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Dhalluin, C.; Carlson, J.E.; Zeng, L.; He, C.; Aggarwal, A.K.; Zhou, M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999, 399, 491–496. [Google Scholar] [CrossRef]
- Mujtaba, S.; He, Y.; Zeng, L.; Farooq, A.; Carlson, J.E.; Ott, M.; Verdin, E.; Zhou, M.M. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell 2002, 9, 575–586. [Google Scholar] [CrossRef]
- Jacobson, R.H.; Ladurner, A.G.; King, D.S.; Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 2000, 288, 1422–1425. [Google Scholar] [CrossRef]
- Mujtaba, S.; He, Y.; Zeng, L.; Yan, S.; Plotnikova, O.; Sanchez, R.; Zeleznik-Le, N.J.; Ronai, Z.; Zhou, M.M. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell 2004, 13, 251–263. [Google Scholar] [CrossRef]
- Hudson, B.P.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain1. J. Mol. Biol. 2000, 304, 355–370. [Google Scholar] [CrossRef]
- Owen, D.J.; Ornaghi, P.; Yang, J.C.; Lowe, N.; Evans, P.R.; Ballario, P.; Neuhaus, D.; Filetici, P.; Travers, A.A. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000, 19, 6141–6149. [Google Scholar] [CrossRef]
- Nakamura, Y.; Umehara, T.; Nakano, K.; Jang, M.K.; Shirouzu, M.; Morita, S.; Uda-Tochio, H.; Hamana, H.; Terada, T.; Adachi, N.; Matsumoto, T.; Tanaka, A.; Horikoshi, M.; Ozato, K.; Padmanabhan, B.; Yokoyama, S. Crystal structure of the human BRD2 bromodomain. J. Biol. Chem. 2007, 282, 4193–4201. [Google Scholar]
- Vollmuth, F.; Blankenfeldt, W.; Geyer, M. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J. Biol. Chem. 2009, 284, 36547–36556. [Google Scholar] [CrossRef]
- Shen, W.; Xu, C.; Huang, W.; Zhang, J.; Carlson, J.E.; Tu, X.; Wu, J.; Shi, Y. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry 2007, 46, 2100–2110. [Google Scholar]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Muller, S.; Pawson, T.; Gingras, A.C.; Arrowsmith, C.H.; Knapp, S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar]
- Jeanmougin, F.; Wurtz, J.M.; Le Douarin, B.; Chambon, P.; Losson, R. The bromodomain revisited. Trends Biochem. Sci. 1997, 22, 151–153. [Google Scholar] [CrossRef]
- Philpott, M.; Yang, J.; Tumber, T.; Fedorov, O.; Uttarkar, S.; Filippakopoulos, P.; Picaud, S.; Keates, T.; Felletar, I.; Ciulli, A.; Knapp, S.; Heightman, T.D. Bromodomain-peptide displacement assays for interactome mapping and inhibitor discovery. Mol. BioSyst. 2011, 7, 2899–2908. [Google Scholar] [CrossRef]
- Zhang, Q.; Chakravarty, S.; Ghersi, D.; Zeng, L.; Plotnikov, A.N.; Sanchez, R.; Zhou, M.M. Biochemical profiling of histone binding selectivity of the yeast bromodomain family. PLoS One 2010, 5, e8903. [Google Scholar]
- Hassan, A.H.; Awad, S.; Al-Natour, Z.; Othman, S.; Mustafa, F.; Rizvi, T.A. Selective recognition of acetylated histones by bromodomains in transcriptional co-activators. Biochem. J. 2007, 402, 125. [Google Scholar] [CrossRef]
- Ruthenburg, A.J.; Li, H.; Patel, D.J.; Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007, 8, 983–994. [Google Scholar] [CrossRef]
- Ruthenburg, A.J.; Li, H.; Milne, T.A.; Dewell, S.; McGinty, R.K.; Yuen, M.; Ueberheide, B.; Dou, Y.; Muir, T.W.; Patel, D.J.; Allis, C.D. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 2011, 145, 692–706. [Google Scholar] [CrossRef]
- Morinière, J.; Rousseaux, S.; Steuerwald, U.; Soler-López, M.; Curtet, S.; Vitte, A.L.; Govin, J.; Gaucher, J.; Sadoul, K.; Hart, D.J.; Krijgsveld, D.J.; Khochbin, S.; Muller, C.W.; Petosa, C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 2009, 461, 664–668. [Google Scholar]
- Robert, F.; Pokholok, D.K.; Hannett, N.M.; Rinaldi, N.J.; Chandy, M.; Rolfe, A.; Workman, J.L.; Gifford, D.K.; Young, R.A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 2004, 16, 199–209. [Google Scholar] [CrossRef]
- Grant, P.A.; Duggan, L.; Côté, J.; Roberts, S.M.; Brownell, J.E.; Candau, R.; Ohba, R.; Owen-Hughes, T.; Allis, C.D.; Winston, F.; Berger, S.L.; Workman, J.L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997, 11, 1640–1650. [Google Scholar] [CrossRef]
- Candau, R.; Zhou, J.X.; Allis, C.D.; Berger, S.L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997, 16, 555–565. [Google Scholar] [CrossRef]
- Marcus, G.A.; Silverman, N.; Berger, S.L.; Horiuchi, J.; Guarente, L. Functional similarity and physical association between GCN5 and ADA2: Putative transcriptional adaptors. EMBO J. 1994, 13, 4807–4815. [Google Scholar]
- Sterner, D.E.; Grant, P.A.; Roberts, S.M.; Duggan, L.J.; Belotserkovskaya, R.; Pacella, L.A.; Winston, F.; Workman, J.L.; Berger, S.L. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999, 19, 86–98. [Google Scholar]
- Hassan, A.H.; Prochasson, P.; Neely, K.E.; Galasinski, S.C.; Chandy, M.; Carrozza, M.J.; Workman, J.L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 2002, 111, 369–379. [Google Scholar] [CrossRef]
- Pray-Grant, M.G.; Schieltz, D.; McMahon, S.J.; Wood, J.M.; Kennedy, E.L.; Cook, R.G.; Workman, J.L.; Yates, J.R., III.; Grant, P.A. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 2002, 22, 8774–8786. [Google Scholar]
- Manning, E.T.; Ikehara, T.; Ito, T.; Kadonaga, J.T.; Kraus, W.L. p300 forms a stable, template-committed complex with chromatin: Role for the bromodomain. Mol. Cell. Biol. 2001, 21, 3876–3887. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, Q.; Gerona-Navarro, G.; Moshkina, N.; Zhou, M.M. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 2008, 16, 643–652. [Google Scholar] [CrossRef]
- Kee, B.L.; Arias, J.; Montminy, M.R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J. Biol. Chem. 1996, 271, 2373–2375. [Google Scholar]
- Swope, D.L.; Mueller, C.L.; Chrivia, J.C. CREB-binding protein activates transcription through multiple domains. J. Biol. Chem. 1996, 271, 28138–28145. [Google Scholar]
- Kwok, R.P.S.; Lundblad, J.R.; Chrivia, J.C.; Richards, J.P.; Bächinger, H.P.; Brennan, R.G.; Roberts, S.G.E.; Green, M.R.; Goodman, R.H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 1994, 370, 223–226. [Google Scholar]
- Nakajima, T.; Uchida, C.; Anderson, S.F.; Lee, C.G.; Hurwitz, J.; Parvin, J.D.; Montminy, M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 1997, 90, 1107–1112. [Google Scholar]
- Bannister, A.J.; Oehler, T.; Wilhelm, D.; Angel, P.; Kouzarides, T. Stimulation of c-Jun activity by CBP: C-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene 1995, 11, 2509–2514. [Google Scholar]
- Bannister, A.J.; Kouzarides, T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995, 14, 4758–4762. [Google Scholar]
- Ogryzko, V.V.; Schiltz, R.L.; Russanova, V.; Howard, B.H.; Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996, 87, 953–959. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. The CBP co-activator is a histone acetyltransferase. Nature 1996, 384, 641–643. [Google Scholar] [CrossRef]
- Imhof, A.; Yang, X.J.; Ogryzko, V.V.; Nakatani, Y.; Wolffe, A.P.; Ge, H. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 1997, 7, 689–692. [Google Scholar] [CrossRef]
- Boyes, J.; Byfield, P.; Nakatani, Y.; Ogryzko, V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 1998, 396, 594–598. [Google Scholar]
- Merika, M.; Williams, A.J.; Chen, G.; Collins, T.; Thanos, D. Recruitment of CBP/p300 by the IFN [beta] enhanceosome is required for synergistic activation of transcription. Mol. Cell 1998, 1, 277–287. [Google Scholar] [CrossRef]
- Kim, T.K.; Kim, T.H.; Maniatis, T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-β enhanceosome in vitro. Proc. Natl. Acad. Sci. USA 1998, 95, 12191–12196. [Google Scholar]
- Peterson, C.L.; Workman, J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000, 10, 187–192. [Google Scholar] [CrossRef]
- Hassan, A.H.; Neely, K.E.; Workman, J.L. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 2001, 104, 817–827. [Google Scholar] [CrossRef]
- Schwabish, M.A.; Struhl, K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 2007, 27, 6987–6995. [Google Scholar] [CrossRef]
- Kasten, M.; Szerlong, H.; Erdjument-Bromage, H.; Tempst, P.; Werner, M.; Cairns, B.R. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004, 23, 1348–1359. [Google Scholar] [CrossRef]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002, 16, 806–819. [Google Scholar] [CrossRef]
- Parnell, T.J.; Huff, J.T.; Cairns, B.R. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2007, 27, 100–110. [Google Scholar]
- Carey, M.; Li, B.; Workman, J.L. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 2006, 24, 481–487. [Google Scholar] [CrossRef]
- Ladurner, A.G.; Inouye, C.; Jain, R.; Tjian, R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 2003, 11, 365–376. [Google Scholar] [CrossRef]
- Matangkasombut, O.; Buratowski, R.M.; Swilling, N.W.; Buratowski, S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000, 14, 951–962. [Google Scholar]
- Matangkasombut, O.; Buratowski, S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 2003, 11, 353–363. [Google Scholar] [CrossRef]
- Durant, M.; Pugh, B.F. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2007, 27, 5327–5335. [Google Scholar] [CrossRef]
- Krogan, N.J.; Keogh, M.C.; Datta, N.; Sawa, C.; Ryan, O.W.; Ding, H.; Haw, R.A.; Pootoolal, J.; Tong, A.; Canadien, V.; Richards, D.P.; Wu, X.; Emili, A.; Hughes, T.R.; Buratowski, S.; Greenblatt, J.F. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 2003, 12, 1565–1576. [Google Scholar] [CrossRef]
- Kanno, T.; Kanno, Y.; Siegel, R.M.; Jang, M.K.; Lenardo, M.J.; Ozato, K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 2004, 13, 33–43. [Google Scholar] [CrossRef]
- Umehara, T.; Nakamura, Y.; Jang, M.K.; Nakano, K.; Tanaka, A.; Ozato, K.; Padmanabhan, B.; Yokoyama, S. Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J. Biol. Chem. 2010, 285, 7610. [Google Scholar]
- Umehara, T.; Nakamura, Y.; Wakamori, M.; Ozato, K.; Yokoyama, S.; Padmanabhan, B. Structural implications for K5/K12-di-acetylated histone H4 recognition by the second bromodomain of BRD2. FEBS Lett. 2010, 584, 3901–3908. [Google Scholar] [CrossRef]
- Denis, G.V.; Vaziri, C.; Guo, N.; Faller, D.V. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000, 11, 417. [Google Scholar]
- Peng, J.; Dong, W.; Chen, L.; Zou, T.; Qi, Y.; Liu, Y. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol. Cell. Biochem. 2007, 294, 45–54. [Google Scholar] [CrossRef]
- Sinha, A.; Faller, D.V.; Denis, G.V. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem. J. 2005, 387, 257–269. [Google Scholar] [CrossRef]
- Denis, G.V.; McComb, M.E.; Faller, D.V.; Sinha, A.; Romesser, P.B.; Costello, C.E. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 2006, 5, 502–511. [Google Scholar] [CrossRef]
- Dey, A.; Chitsaz, F.; Abbasi, A.; Misteli, T.; Ozato, K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 2003, 100, 8758. [Google Scholar]
- Jiang, Y.W.; Veschambre, P.; Erdjument-Bromage, H.; Tempst, P.; Conaway, J.W.; Conaway, R.C.; Kornberg, R.D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 1998, 95, 8538–8543. [Google Scholar]
- Houzelstein, D.; Bullock, S.L.; Lynch, D.E.; Grigorieva, E.F.; Wilson, V.A.; Beddington, R.S.P. Growth and Early Postimplantation Defects in Mice Deficient for the Bromodomain-Containing Protein Brd4. Mol. Cell. Biol. 2002, 22, 3794–3802. [Google Scholar] [CrossRef]
- Jang, M.K.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar] [CrossRef]
- Yang, Z.; Yik, J.H.N.; Chen, R.; He, N.; Jang, M.K.; Ozato, K.; Zhou, Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 2005, 19, 535–545. [Google Scholar] [CrossRef]
- Grant, P.A.; Eberharter, A.; John, S.; Cook, R.G.; Turner, B.M.; Workman, J.L. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 1999, 274, 5895–5900. [Google Scholar]
- Eberharter, A.; Sterner, D.E.; Schieltz, D.; Hassan, A.; Yates III, J.R.; Berger, S.L.; Workman, J.L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 6621–6631. [Google Scholar]
- Wu, P.Y.J.; Winston, F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 2002, 22, 5367–5379. [Google Scholar] [CrossRef]
- Sterner, D.E.; Belotserkovskaya, R.; Berger, S.L. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11622. [Google Scholar]
- Gansheroff, L.J.; Dollard, C.; Tan, P.; Winston, F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 1995, 139, 523–536. [Google Scholar]
- Arany, Z.; Sellers, W.R.; Livingston, D.M.; Eckner, R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 1994, 77, 799–800. [Google Scholar] [CrossRef]
- Wang, Z.; Zang, C.; Cui, K.; Schones, D.E.; Barski, A.; Peng, W.; Zhao, K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009, 138, 1019–1031. [Google Scholar] [CrossRef]
- Kawasaki, H.; Eckner, R.; Yao, T.P.; Taira, K.; Chiu, R.; Livingston, D.M.; Yokoyama, K.K. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 1998, 393, 284–289. [Google Scholar]
- Kasper, L.H.; Fukuyama, T.; Biesen, M.A.; Boussouar, F.; Tong, C.; De Pauw, A.; Murray, P.J.; van Deursen, J.M.A.; Brindle, P.K. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell. Biol. 2006, 26, 789–809. [Google Scholar]
- Kraus, W.L.; Manning, E.T.; Kadonaga, J.T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 1999, 19, 8123–8135. [Google Scholar]
- Neish, A.S.; Anderson, S.F.; Schlegel, B.P.; Wei, W.; Parvin, J.D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998, 26, 847–853. [Google Scholar]
- Tamkun, J.W.; Deuring, R.; Scott, M.P.; Kissinger, M.; Pattatucci, A.M.; Kaufman, T.C.; Kennison, J.A. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 1992, 68, 561–572. [Google Scholar] [CrossRef]
- Khavari, P.A.; Peterson, C.L.; Tamkun, J.W.; Mendel, D.B.; Crabtree, G.R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 1993, 366, 170–174. [Google Scholar]
- Wang, W.; Cote, J.; Xue, Y.; Zhou, S.; Khavari, P.; Biggar, S.; Muchardt, C.; Kalpana, G.; Goff, S.; Yaniv, M.; Workman, J.L.; Crabtree, G.R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996, 15, 5370–5382. [Google Scholar]
- Wang, W.; Xue, Y.; Zhou, S.; Kuo, A.; Cairns, B.R.; Crabtree, G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996, 10, 2117–2130. [Google Scholar] [CrossRef]
- Sudarsanam, P.; Iyer, V.R.; Brown, P.O.; Winston, F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 3364–3369. [Google Scholar]
- Chatterjee, N.; Sinha, D.; Lemma-Dechassa, M.; Tan, S.; Shogren-Knaak, M.A.; Bartholomew, B. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 2011, 39, 8378–8391. [Google Scholar]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Awad, S.; Hassan, A.H. The Swi2/Snf2 Bromodomain Is Important for the Full Binding and Remodeling Activity of the SWI/SNF Complex on H3- and H4-acetylated Nucleosomes. Ann. N. Y. Acad. Sci. 1138, 366–375. [Google Scholar]
- Roberts, S.M.; Winston, F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 1997, 147, 451–465. [Google Scholar]
- Neely, K.E.; Hassan, A.H.; Brown, C.E.; Howe, L.A.; Workman, J.L. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 2002, 22, 1615–1625. [Google Scholar] [CrossRef]
- Syntichaki, P.; Topalidou, I.; Thireos, G. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 2000, 404, 414–417. [Google Scholar]
- Cosma, M.P.; Tanaka, T.; Nasmyth, K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle–and developmentally regulated promoter. Cell 1999, 97, 299–311. [Google Scholar] [CrossRef]
- Kim, J.H.; Saraf, A.; Florens, L.; Washburn, M.; Workman, J.L. Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes Dev. 2010, 24, 2766–2771. [Google Scholar] [CrossRef]
- Cairns, B.R.; Lorch, Y.; Li, Y.; Zhang, M.; Lacomis, L.; Erdjument-Bromage, H.; Tempst, P.; Du, J.; Laurent, B.; Kornberg, R.D. RSC, an essential, abundant chromatin-remodeling complex. Cell 1996, 87, 1249–1260. [Google Scholar] [CrossRef]
- Xue, Y.; Canman, J.C.; Lee, C.S.; Nie, Z.; Yang, D.; Moreno, G.T.; Young, M.K.; Salmon, E.; Wang, W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13015–13020. [Google Scholar]
- Damelin, M.; Simon, I.; Moy, T.I.; Wilson, B.; Komili, S.; Tempst, P.; Roth, F.P.; Young, R.A.; Cairns, B.R.; Silver, P.A. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 2002, 9, 563–573. [Google Scholar] [CrossRef]
- Cao, Y.; Cairns, B.R.; Kornberg, R.D.; Laurent, B.C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 1997, 17, 3323–3334. [Google Scholar]
- Du, J.; Nasir, I.; Benton, B.K.; Kladde, M.P.; Laurent, B.C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics 1998, 150, 987–1005. [Google Scholar]
- Cairns, B.R.; Schlichter, A.; Erdjument-Bromage, H.; Tempst, P.; Kornberg, R.D.; Winston, F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 1999, 4, 715–723. [Google Scholar] [CrossRef]
- Pamblanco, M.; Poveda, A.; Sendra, R.; Rodri’guez-Navarro, S.; Perez-Orti’n, J.E.; Tordera, V. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 2001, 496, 31–35. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar]
- Zhang, H.; Roberts, D.N.; Cairns, B.R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 2005, 123, 219–231. [Google Scholar] [CrossRef]
- LeRoy, G.; Rickards, B.; Flint, S. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell 2008, 30, 51–60. [Google Scholar] [CrossRef]
- Crowley, T.E.; Kaine, E.M.; Yoshida, M.; Nandi, A.; Wolgemuth, D.J. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol. Endocrinol. 2002, 16, 1727–1737. [Google Scholar] [CrossRef]
- Rahman, S.; Sowa, M.E.; Ottinger, M.; Smith, J.A.; Shi, Y.; Harper, J.W.; Howley, P.M. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011, 31, 2641–2652. [Google Scholar] [CrossRef]
- Price, D.H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000, 20, 2629–2634. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Lewis, B.A.; Cherman, N.; Hewitt, M.C.; Albrecht, B.K.; Robey, P.G.; Ozato, K.; Sims, R.J., III.; Singer, D.S. BRD4 is an atypical kinase that phosphorylates Serine2 of the RNA Polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA 2012, 20, 2629–2634. [Google Scholar]
- Wang, R.; Li, Q.; Helfer, C.M.; Jiao, J.; You, J. The bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J. Biol. Chem. 2012, 287, 10738–10752. [Google Scholar]
- Lin, R.J.; Nagy, L.; Inoue, S.; Shao, W.; Miller, W.H.; Evans, R.M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 1998, 391, 811–814. [Google Scholar]
- Grignani, F.; De Matteis, S.; Nervi, C.; Tomassoni, L.; Gelmetti, V.; Cioce, M.; Fanelli, M.; Ruthardt, M.; Ferrara, F.F.; Zamir, I.; Seiser, C.; Grignani, F.; Lazar, M.A.; Minucci, S.; Pellici, P.G. Fusion proteins of the retinoic acid receptor-a recruit histone deacetylase in promyelocytic leukaemia. Nature 1998, 391, 815–817. [Google Scholar]
- Thurn, K.T.; Thomas, S.; Moore, A.; Munster, P.N. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Futur. Oncol. 2011, 7, 263–283. [Google Scholar] [CrossRef]
- Vojinovic, J.; Damjanov, N. HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol. Med. 2011, 17, 397–403. [Google Scholar]
- Guidotti, A.; Auta, J.; Chen, Y.; Davis, J.; Dong, E.; Gavin, D.; Grayson, D.; Matrisciano, F.; Pinna, G.; Satta, R.; Sharma, R.P.; Tremolizzo, L.; Tueting, P. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology 2011, 60, 1007–1016. [Google Scholar] [CrossRef]
- Zeng, L.; Li, J.; Muller, M.; Yan, S.; Mujtaba, S.; Pan, C.; Wang, Z.; Zhou, M.M. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J. Am. Chem. Soc. 2005, 127, 2376–2377. [Google Scholar]
- French, C.A.; Ramirez, C.L.; Kolmakova, J.; Hickman, T.T.; Cameron, M.J.; Thyne, M.E.; Kutok, J.L.; Toretsky, J.A.; Tadavarthy, A.K.; Kees, U.R.; Fletcher, J.A.; Aster, J.C. BRD–NUT oncoproteins: A family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 2007, 27, 2237–2242. [Google Scholar]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; Philpott, M.; Munro, S.; McKeown, M.K.; Wang, Y.; Christie, A.L.; West, N.; Cameron, M.J.; Schwartz, B.; Heightman, T.D.; La Thangue, N.B.; French, C.A.; Wiest, O.; Kung, A.L.; Knapp, S.; Bradner, J.E. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; Chesi, M.; Schinze, A.C.; McKeown, M.R.; Heffernan, T.P.; Vakoc, C.R.; Bergsagel, P.L.; Ghobrial, I.M.; Richardson, P.G.; Young, R.A.; Hahn, W.C.; Anderson, K.C.; Kung, A.L.; Bradner, J.E.; Mitsiades, C.S. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell 2011, 146, 904–917. [Google Scholar]
- Zuber, J.; Shi, J.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; Taylor, M.J.; Johns, C.; Chicas, A.; Mulloy, M.C.; Kogan, S.C.; Brown, P.; Valent, P.; Bradner, J.E.; Lowe, S.W.; Vakoc, C.R. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar]
- Mertz, J.A.; Conery, A.R.; Bryant, B.M.; Sandy, P.; Balasubramanian, S.; Mele, D.A.; Bergeron, L.; Sims, R.J., III. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 2011, 108, 16669–16674. [Google Scholar]
- Dawson, M.A.; Prinjha, R.K.; Dittman, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; Huthmacher, C.; Gudgin, E.; Lugo, D.; Beinke, S.; Chapman, T.D.; Roberts, E.J.; Soden, P.E.; Auger, K.R.; Mirguet, O.; Doehner, K.; Delwel, R.; Burnett, A.K.; Jeffrey, P.; Drewes, G.; Lee, K.; Huntly, B.J.; Kouzarides, T. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; White, J.; Kirilovsky, J.; Rice, C.M.; Lora, J.M.; Prinjha, R.K.; Lee, K.; Tarakhovsky, A. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar]
- Seal, J.; Lamotte, Y.; Donche, F.; Bouillot, A.; Mirguet, O.; Gellibert, F.; Nicodeme, E.; Krysa, G.; Kirilovsky, J.; Beinke, S.; McCleary, S.; Rioja, I.; Bamborough, P.; Chung, C.W.; Gordon, L.; Lewis, T.; Walker, A.L.; Cutler, L.; Lugo, D.; Wilson, D.M.; Witherington, J.; Lee, K.; Prinjha, R.K. Identification of a novel series of BET family bromodomain inhibitors: Binding mode and profile of I-BET151 (GSK1210151A). Bioorg. Med. Chem. Lett. 2012, 22, 2968–2972. [Google Scholar]
- Templeton, T.J.; Iyer, L.M.; Anantharaman, V.; Enomoto, S.; Abrahante, J.E.; Subramanian, G.M.; Hoffman, S.L.; Abrahamsen, M.S.; Aravind, L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004, 14, 1686–1695. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Josling, G.A.; Selvarajah, S.A.; Petter, M.; Duffy, M.F. The Role of Bromodomain Proteins in Regulating Gene Expression. Genes 2012, 3, 320-343. https://doi.org/10.3390/genes3020320
Josling GA, Selvarajah SA, Petter M, Duffy MF. The Role of Bromodomain Proteins in Regulating Gene Expression. Genes. 2012; 3(2):320-343. https://doi.org/10.3390/genes3020320
Chicago/Turabian StyleJosling, Gabrielle A., Shamista A. Selvarajah, Michaela Petter, and Michael F. Duffy. 2012. "The Role of Bromodomain Proteins in Regulating Gene Expression" Genes 3, no. 2: 320-343. https://doi.org/10.3390/genes3020320
APA StyleJosling, G. A., Selvarajah, S. A., Petter, M., & Duffy, M. F. (2012). The Role of Bromodomain Proteins in Regulating Gene Expression. Genes, 3(2), 320-343. https://doi.org/10.3390/genes3020320




