Characterization of the PHO1 Gene Family in Vigna radiata L. and Its Expression Analysis Under Phosphate-Deficient Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Procedures
2.2. Identification and Physicochemical Characterization of Members of the Mung Bean PHO1 Gene Family
2.3. Structural Analysis of the PHO1 Genes and Protein in Mung Beans
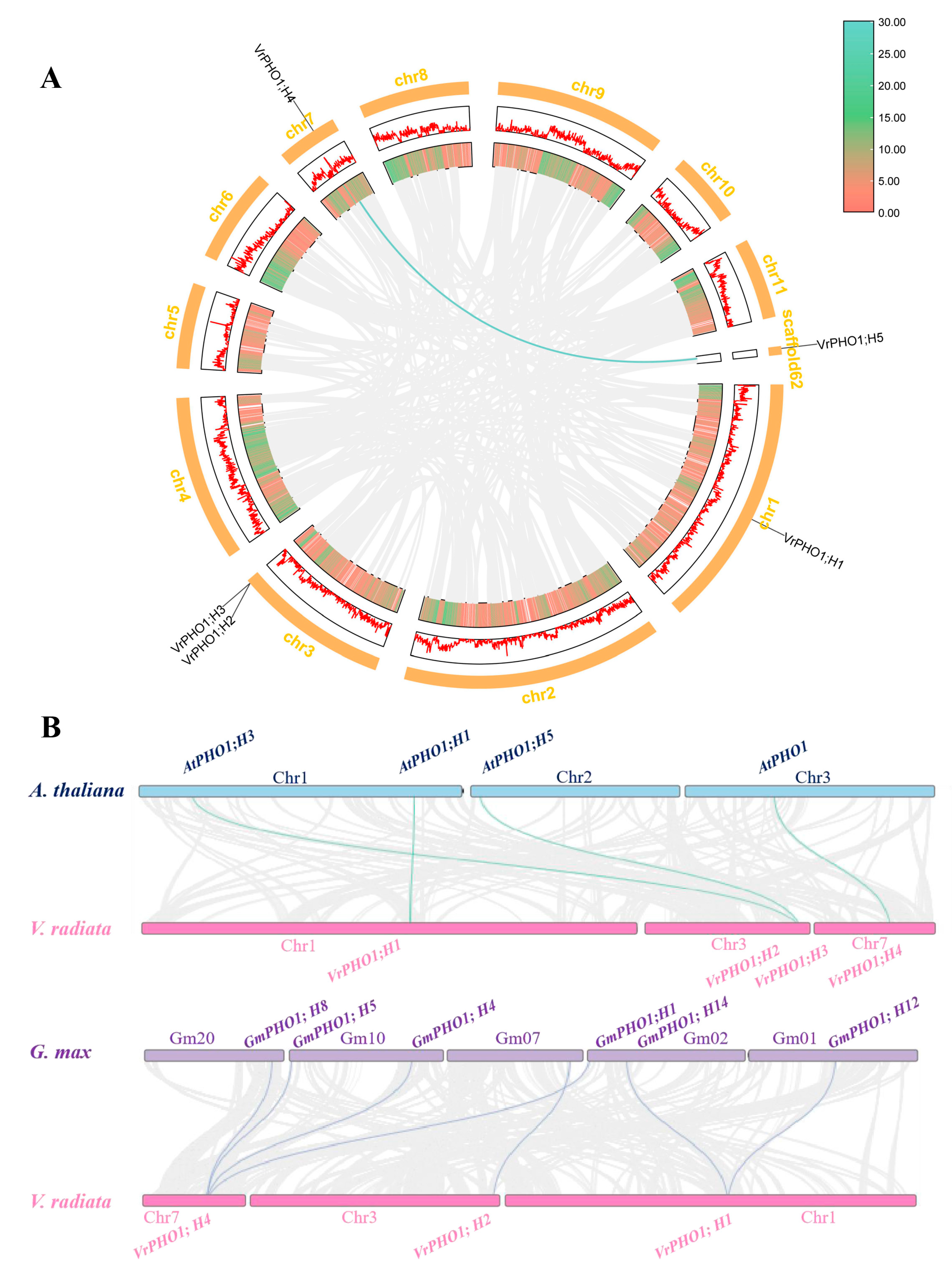
2.4. Systematic Evolution and Collinearity Analysis of the PHO1 Gene Family in Mung Bean
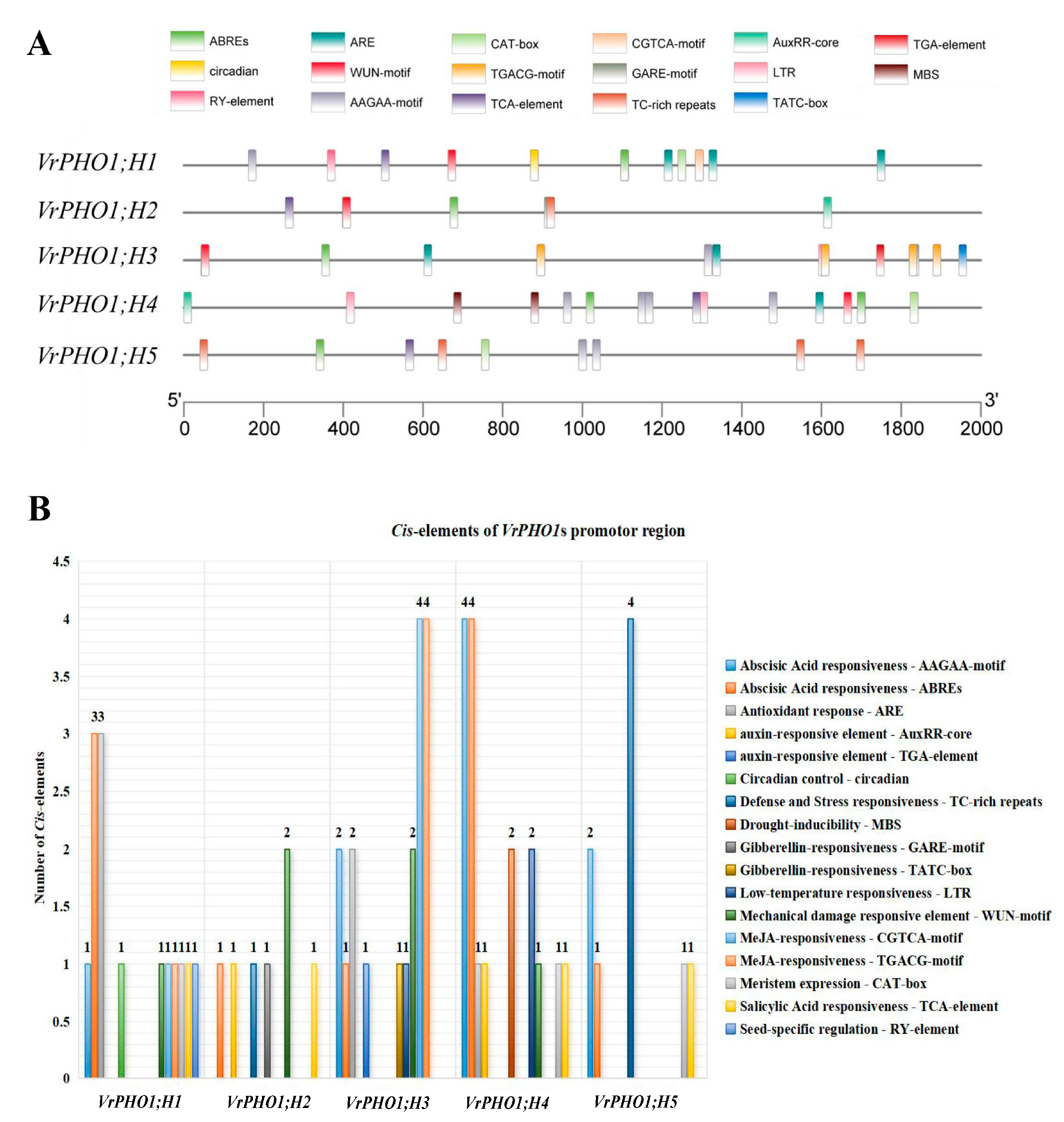
2.5. Analysis of Cis-Acting Elements in the PHO1 Genes Promoter of Mung Beans
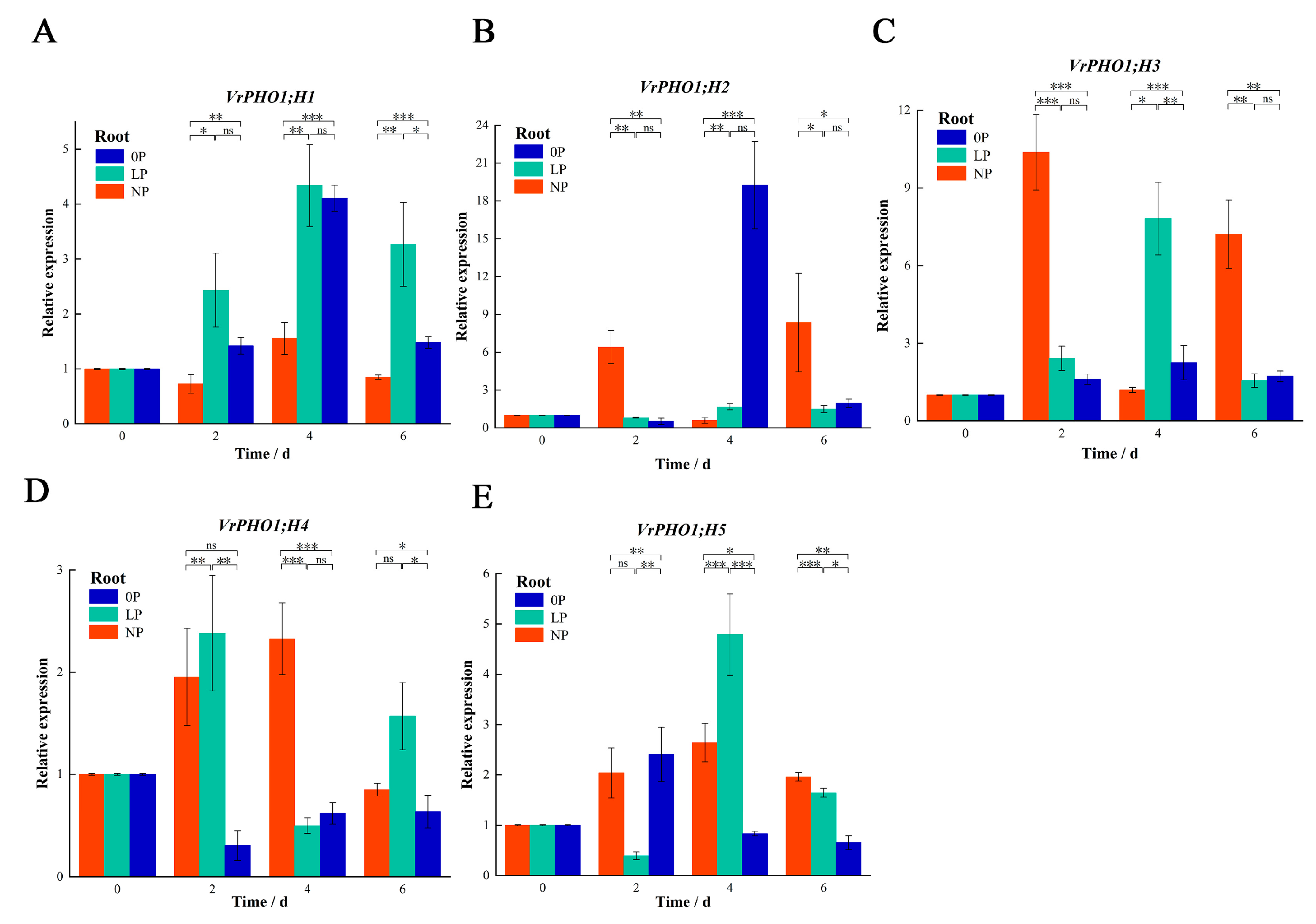
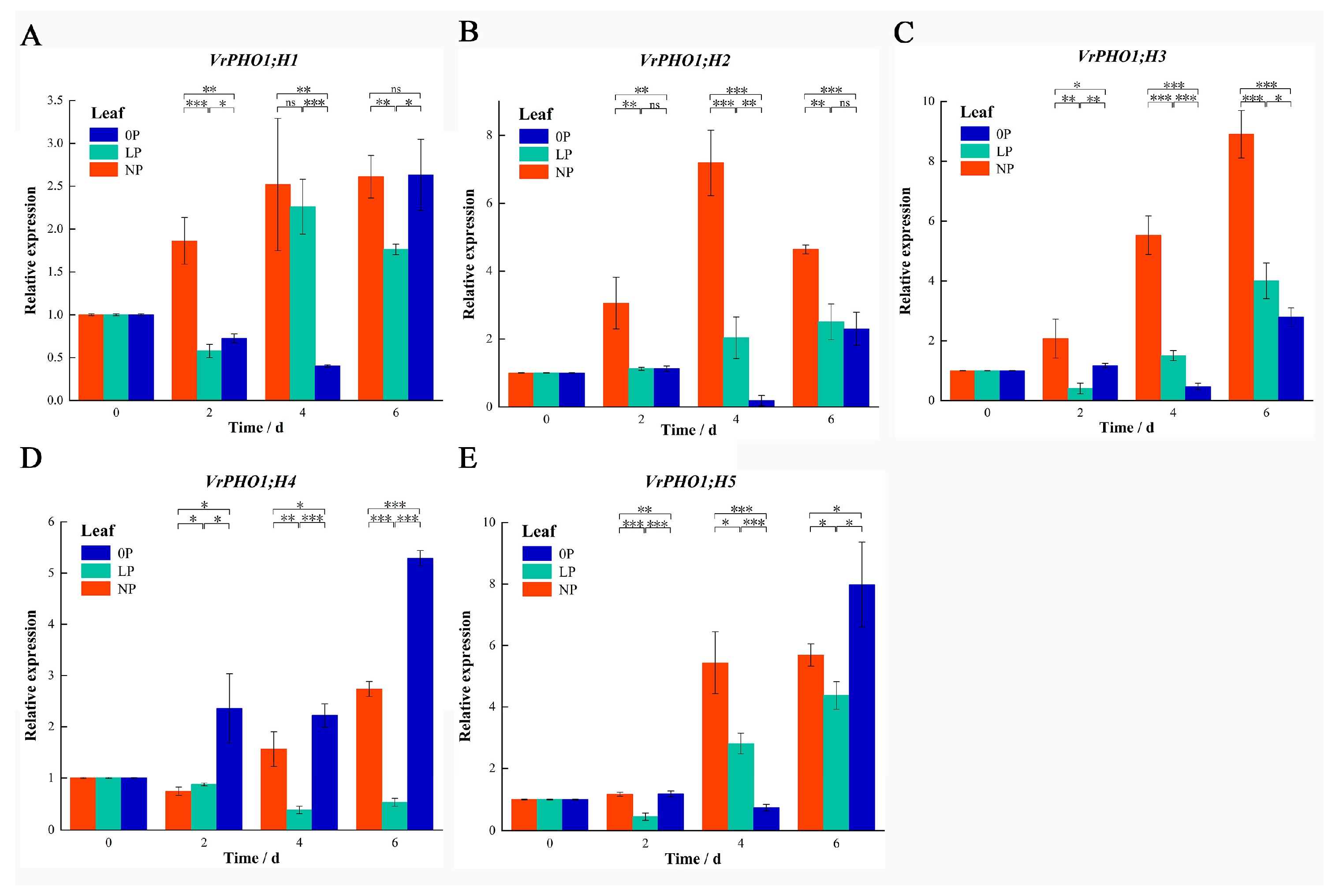
2.6. Gene Expression Analysis and Data Processing
3. Results and Analysis
3.1. Identification of Members of the Mung Bean PHO1 Gene Family and Analysis of Their Protein Physicochemical Properties
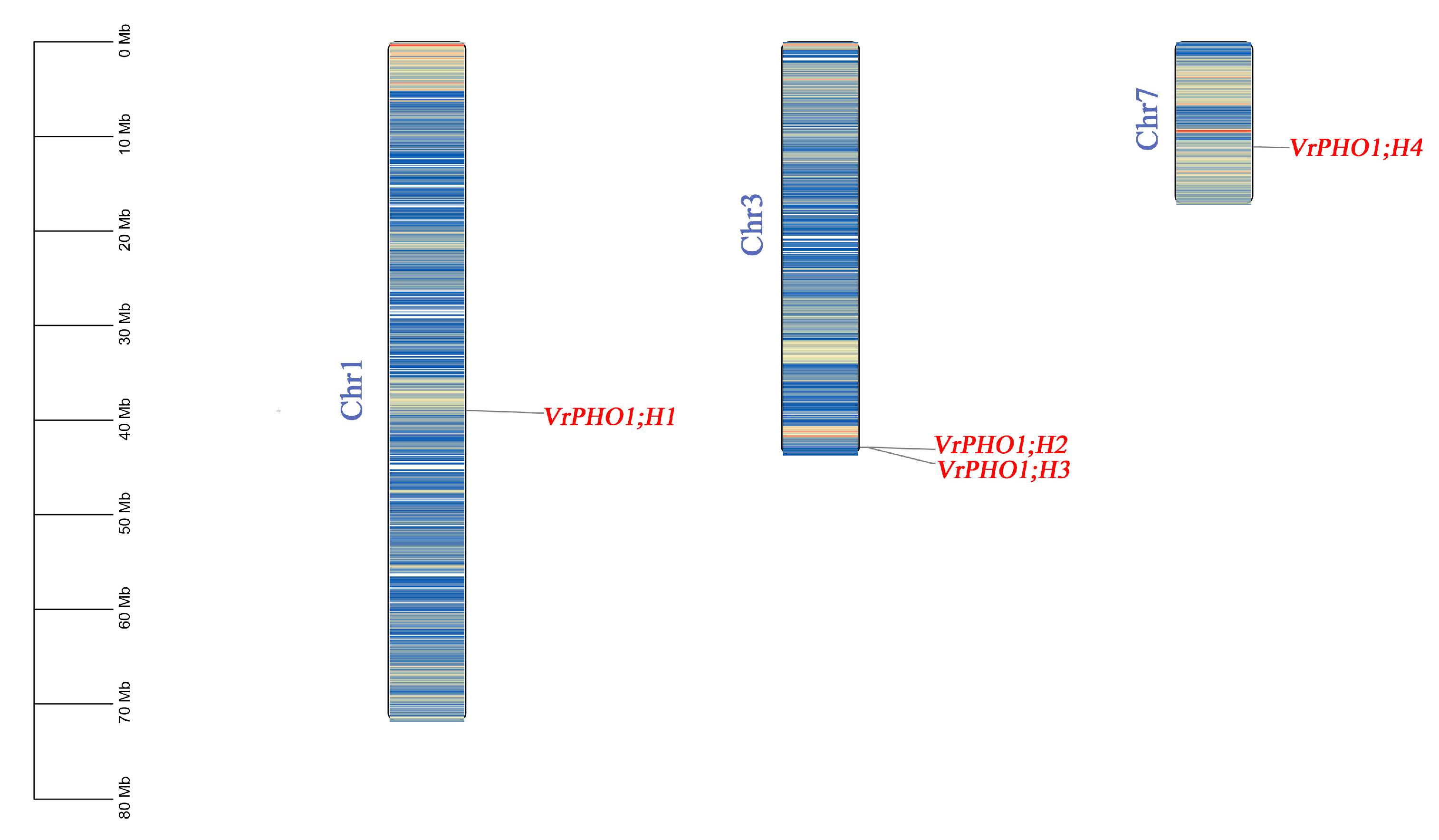
3.2. Chromosomal Localization Analysis of the PHO1 Gene Family in Mung Beans
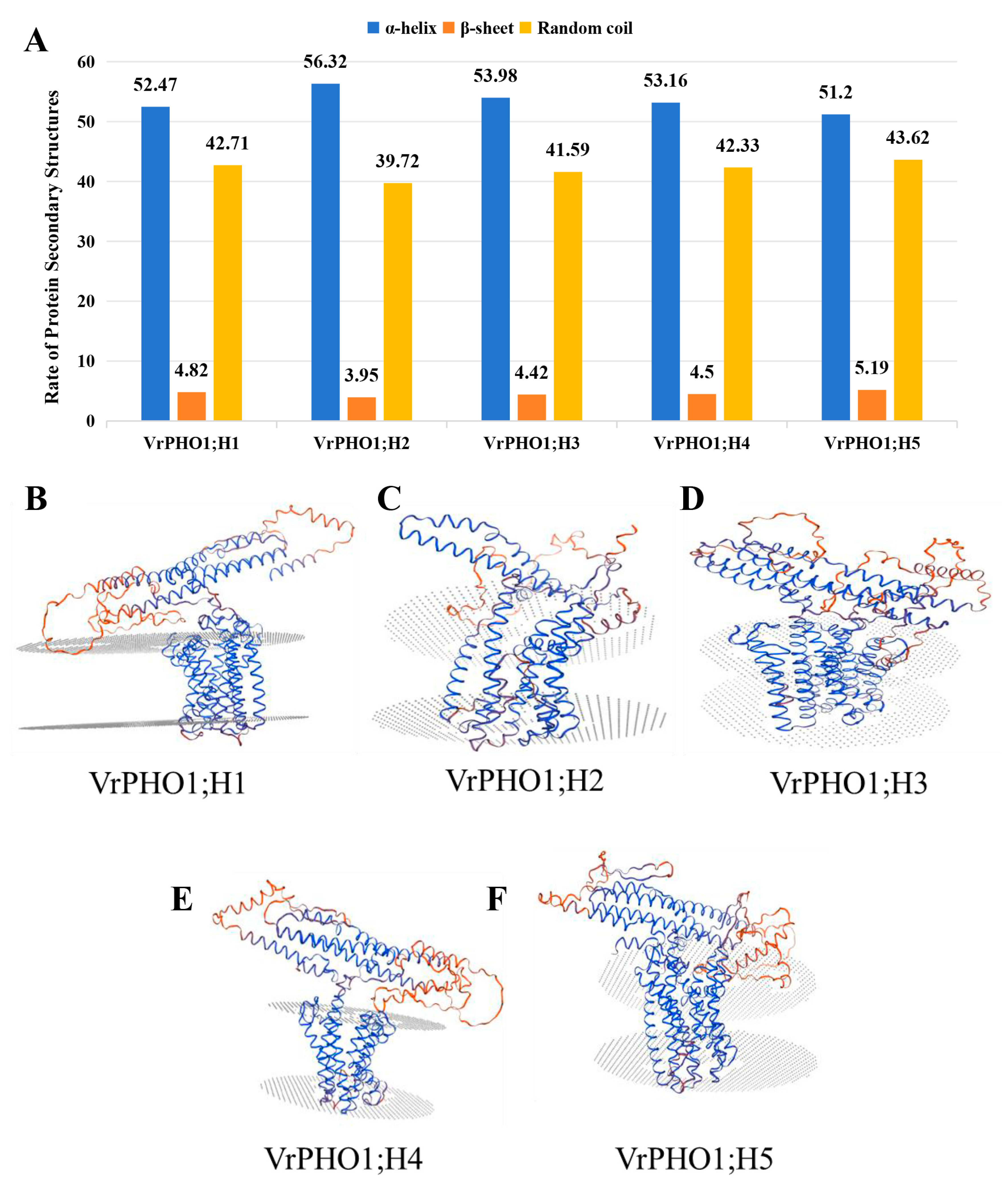
3.3. Structural Analysis Prediction of VrPHO1s Proteins
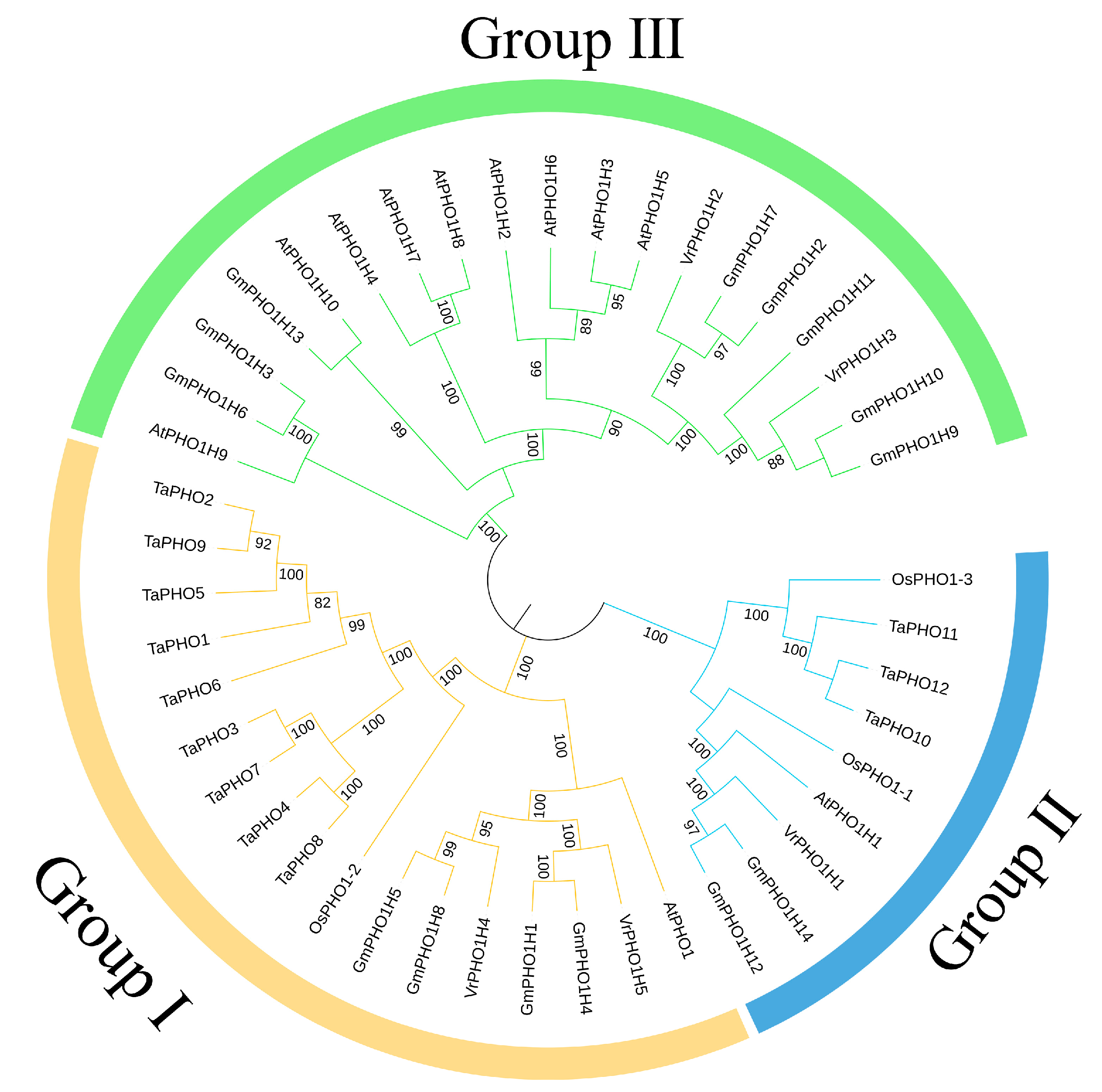
3.4. Phylogenetic Tree Analysis of the VrPHO1 Gene Family Members
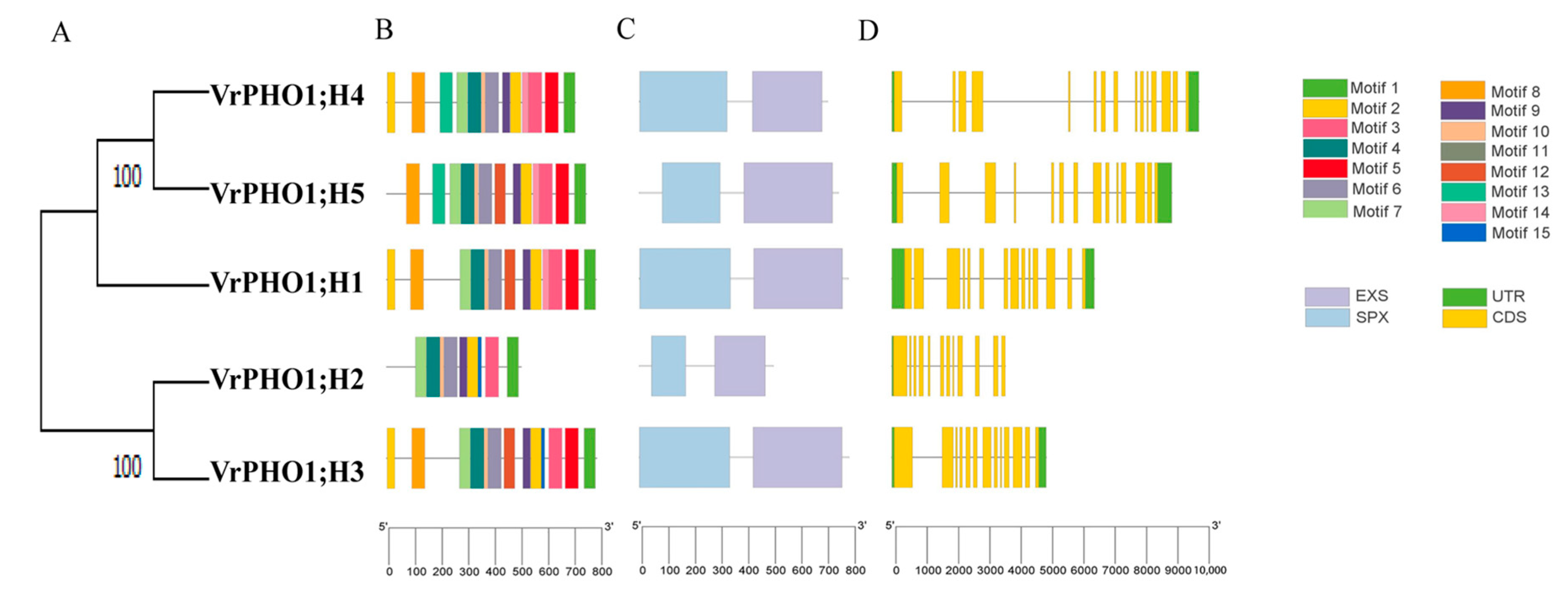
3.5. Genetic Structure and Analysis of Conserved Protein Motifs in Members of the VrPHO1 Gene Family
3.6. Analysis of Cis-Acting Elements in the VrPHO1 Genes Promoter
3.7. Inter-Species and Intra-Species Collinearity Analysis of the VrPHO1 Genes
3.8. Analysis of VrPHO1 Genes Expression Patterns Under Low-Phosphorus Stress
4. Discussion
4.1. Evolutionary Analysis of PHO1 Genes Exhibited Specificity Among Diverse Plants
4.2. Functional Conservation and Diversification of VrPHO1 Genes in Mung Beans
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, L.; Bai, P.; Yuan, X.; Chen, H.; Wang, S.; Chen, X.; Cheng, X. Genetic diversity assessment of a set of introduced mung bean accessions (Vigna radiata L.). Crop J. 2018, 6, 207–213. [Google Scholar] [CrossRef]
- Zhou, R.; Yang, S.; Zhang, B.; Qi, Z.; Xin, D.; Su, A.; Li, S.; Cheng, P.; Bai, Y.; Yin, Z.; et al. Analysis of the genetic diversity of grain legume germplasm resources in China and the development of universal SSR primers. Biotechnol. Biotechnol. Equip. 2022, 35, 1706–1721. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus Uptake by Plants: From Soil to Cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Poirier, Y.; Thoma, S.; Somerville, C.; Schiefelbein, J. Mutant of Arabidopsis Deficient in Xylem Loading of Phosphate. Plant Physiol. 1991, 97, 1087–1093. [Google Scholar] [CrossRef]
- Han, C. Identification of the TaSPX Family in Wheat and Functional Analysis of the Low Phosphorus Tolerance-Associated Genes TaPHO1; H2; Henan Agricultural University: Zhengzhou, China, 2023. [Google Scholar]
- Li, Y.; Liu, J.; Li, B.; Li, J.; Yao, S.; Li, Z. Molecular Mechanisms of Self-Rescue in Higher Plants Under Phosphorus Deprivation. Biotechnol. Bull. 1999, 15, 8. [Google Scholar]
- Benedetti, E.L.; Wink, C.; Santin, D.; Sereda, F.; Roveda, L.F.; Serrat, B.M. Crescimento e sintomas em mudas de espinheira-santa com omissão de nitrogênio, fósforo e potássio. Floresta 2009, 39, 335–343. [Google Scholar] [CrossRef]
- Dumont, M.E.; Schlichter, J.B.; Cardillo, T.S.; Hayes, M.K.; Bethlendy, G.; Sherman, F. CYC2 encodes a factor involved in mitochondrial import of yeast cytochrome c. Mol. Cell. Biol. 1993, 13, 6442–6451. [Google Scholar]
- Ko, S.S.; Lu, W.C.; Hung, J.C.; Chang, H.F.; Li, M.J.; Yeh, K.C.; Chiou, T.J. Maternal effect contributes to grain-filling defects of Ospho1;2 rice mutants. New Phytol. 2024, 244, 351–357. [Google Scholar] [CrossRef]
- Guo, B.; Jin, Y.; Wussler, C.; Blancaflor, E.B.; Motes, C.M.; Versaw, W.K. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2007, 177, 889–898. [Google Scholar] [CrossRef]
- Hassler, S.; Jung, B.; Lemke, L.; Novák, O.; Strnad, M.; Martinoia, E.; Neuhaus, H.E. Function of the Golgi-located phosphate transporter PHT4;6 is critical for senescence-associated processes in Arabidopsis. J. Exp. Bot. 2016, 67, 4671–4684. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Xu, L.; Sun, M.; Yi, K.; Zhao, H. Functional analysis of phosphate transporter OsPHT4 family members in rice. Rice Sci. 2020, 27, 493–503. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Luan, M.; Wang, Y.; Zhang, C.; Zhang, B.; Shi, J.; Zhao, F.; Lan, W.; Luan, S. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E6571–E6578. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Cai, Q.; Zhang, X.; Li, T.; Yu, H.; Guo, J.; Chen, G. Response characteristics of root morphology and root exudates in phosphorus-efficient wild barley to low-level phytate-bound organic phosphorus. J. Plant Nutr. Fertil. 2016, 22, 10. [Google Scholar]
- Wang, Y.; Ribot, C.c.; Rezzonico, E.; Poirier, Y. Structure and Expression Profile of the Arabidopsis PHO1 Gene Family Indicates a Broad Role in Inorganic Phosphate Homeostasis. Plant Physiol. 2004, 135, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Yu, R. Expression and Functional Analysis of Phosphorus Transporter Genes in Soybean Tissues in Response to Low-Phosphorus Drought Stress; Jilin Agricultural University: Changchun, China, 2019. [Google Scholar]
- Ribot, C.; Wang, Y.; Poirier, Y. Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 2007, 227, 1025–1036. [Google Scholar] [CrossRef]
- Hao, X.; Wang, W.; Wang, D.; Fang, Z.; Liu, S.; Zhu, W.; Pang, Z.; Zhang, S.; Zhao, C.; Zhang, F. Genome-Wide Identification and Expression Analysis of PHO Gene Family in Wheat. J. Triticeae Crops 2023, 10, 26–35. [Google Scholar]
- Ma, B.; Zhang, L.; Gao, Q.; Wang, J.; Li, X.; Wang, H.; Liu, Y.; Lin, H.; Liu, J.; Wang, X.; et al. A plasma membrane transporter coordinates phosphate reallocation and grain filling in cereals. Nat. Genet. 2021, 53, 906–915. [Google Scholar] [CrossRef]
- Ren, Q.; An, L.; Yao, Y.; Yao, X.; Liu, Y.; Wu, K. Cloning, Subcellular Localization and Expression Analysis of Phosphate Transporter Gene HvPHO1;2 in Hulless Barely. Acta Agric. Boreali-Occident. Sin. 2021, 12, 1461–1472. [Google Scholar]
- Liu, L.; Fu, Y.; Yu, R.; He, X.; Yang, X.; Yang, M. Cloning, Expression Analysis and Function of the Soybean Phosphate Transporter Gene GmPHO1;7. Jilin. Agric. Univ. 2023, 45, 14–21. [Google Scholar]
- He, L. Molecular Evolution and Functional Differentiation of the Soybean PHO1 Gene Family; University of Chinese Academy of Sciences: Beijing, China, 2013. [Google Scholar]
- Cui, W. Cloning and Functional Analysis of the FvPHO1; H9 Gene in Wild Strawberry; Shenyang Agricultural University: Shenyang, China, 2020. [Google Scholar]
- Reddy, V.R.P.; Das, S.; Dikshit, H.K.; Mishra, G.P.; Aski, M.; Meena, S.K.; Singh, A.; Pandey, R.; Singh, M.P.; Tripathi, K.; et al. Genome-Wide Association Analysis for Phosphorus Use Efficiency Traits in Mungbean (Vigna radiata L. Wilczek) Using Genotyping by Sequencing Approach. Front. Plant Sci. 2020, 11, 537766. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Yang, N.; Zhu, C.; Xia, Y. Screening and Validation of Internal Reference Genes for Real-Time Fluorescent Quantitative PCR in Mung Beans. Plant Physiol. J. 2021, 57, 10. [Google Scholar]
- Poirier, Y.; Jaskolowski, A.; Clúa, J. Phosphate acquisition and metabolism in plants. Curr. Biol. 2022, 32, R623–R629. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, X.; Jiao, W.; Mao, J.; Ye, W.; Cao, Y.; Chen, Q.; Song, Q. Pangenome analysis provides insights into legume evolution and breeding. Nat. Genet. 2025, 57, 2052–2061. [Google Scholar] [CrossRef]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A.; et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Lavin, M.; Sanderson, M.J. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 2004, 91, 1846–1862. [Google Scholar] [CrossRef]
- Cannon, S.B.; Sterck, L.; Rombauts, S.; Sato, S.; Cheung, F.; Gouzy, J.; Wang, X.; Mudge, J.; Vasdewani, J.; Schiex, T.; et al. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 14959–14964. [Google Scholar] [CrossRef]
- Zhuang, Y.; Wang, X.; Li, X.; Hu, J.; Fan, L.; Landis, J.B.; Cannon, S.B.; Grimwood, J.; Schmutz, J.; Jackson, S.A. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat. Plants 2022, 8, 233–244. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, R.; Jiang, K.-W.; Qi, J.; Hu, Y.; Guo, J.; Zhu, R.; Zhang, T.; Egan, A.N.; Yi, T.-S. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol. Plant 2021, 14, 748–773. [Google Scholar] [CrossRef]
- He, L.; Zhao, M.; Wang, Y.; Gai, J.; He, C. Phylogeny, structural evolution and functional diversification of the plant PHOSPHATE1 gene family: A focus on Glycine max. BMC Evol. Biol. 2013, 13, 103. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, H.; He, L.; Zhu, W.; Yan, L.; Chen, Q.; He, C. The PHOSPHATE1 genes participate in salt and Pi signaling pathways and play adaptive roles during soybean evolution. BMC Plant Biol. 2019, 19, 353. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Baumann, A.; Poirier, Y. Characterization of the Rice PHO1 Gene Family Reveals a Key Role for OsPHO1;2 in Phosphate Homeostasis and the Evolution of a Distinct Clade in Dicotyledons. Plant Physiol. 2010, 152, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Yang, Y.; Zhang, X.; Yang, Z.; Zhang, M.; Zhao, Y.; Zhang, C.; Yu, F.; Wang, Y.-F.; Zhang, P. Structural mechanism underlying PHO1; H1-mediated phosphate transport in Arabidopsis. Nat. Plants 2025, 11, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Wild, R.; Gerasimaite, R.; Jung, J.-Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef]
- Vetal, P.V.; Poirier, Y. The Arabidopsis PHOSPHATE 1 exporter undergoes constitutive internalization via clathrin-mediated endocytosis. Plant J. 2023, 116, 1477–1491. [Google Scholar] [CrossRef]
- Mani, B.; Maurya, K.; Kohli, P.S.; Giri, J. Chickpea (Cicer arietinum) PHO1 family members function redundantly in Pi transport and root nodulation. Plant Physiol. Bioch 2024, 211, 13. [Google Scholar] [CrossRef]
- Salazar-Vidal, M.N.; Acosta-Segovia, E.; Sánchez-León, N.; Ahern, K.R.; Brutnell, T.P.; Sawers, R.J.H. Characterization and Transposon Mutagenesis of the Maize (Zea mays) Pho1 Gene Family. PLoS ONE 2016, 11, e0161882. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, Y.; Fan, Y.; Zhang, L.; Li, X.; Zhang, Q.; Shu, Q.; Huang, J.; Chen, G.; Li, Q.; et al. Genetic improvement of phosphate- limited photosynthesis for high yield in rice. Proc. Natl. Acad. Sci. USA 2024, 121, e2404199121. [Google Scholar] [CrossRef]
- Alonso-Nieves, A.L.; Salazar-Vidal, M.N.; Torres-Rodríguez, J.V.; Pérez-Vázquez, L.M.; Massange-Sánchez, J.A.; Gillmor, C.S.; Sawers, R.J.H. The pho1;2a′-m1.1 allele of Phosphate1 conditions misregulation of the phosphorus starvation response in maize (Zea mays ssp. mays L.). Plant Direct 2022, 6, e416. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, J.; Satheesh, V.; Meng, F.; Gao, W.; Dong, J.; Zheng, Z.; An, G.; Nussaume, L.; Liu, D.; et al. SHORT-ROOT stabilizes PHOSPHATE1 to regulate phosphate allocation in Arabidopsis Nat. Plants 2021, 8, 1074–1081. [Google Scholar]
| Gene | Gene ID | Chromosome Localization | Strand | Number of Amino Acids/aa | Molecular Weight/kD | PI | Aliphatic INDEX | Lipolysis Index | Hydrophobicity | Prediction of Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| VrPHO1; H1 | Vradi01g00002120 | 38992032.. 38998514 | + | 789 | 91.66 | 9.09 | 43.69 | 88.95 | −0.193 | Cytoplasmic membrane |
| VrPHO1; H2 | Vradi03g00002410 | 42859336.. 42862967 | + | 506 | 58.68 | 9.38 | 36.56 | 92.65 | −0.115 | Cytoplasmic membrane |
| VrPHO1; H3 | Vradi03g00002411 | 42866638.. 42871577 | + | 791 | 92.02 | 9.55 | 42.8 | 91.47 | −0.188 | Cytoplasmic membrane |
| VrPHO1; H4 | Vradi07g00000873 | 11103676.. 11113505 | + | 711 | 83.13 | 9.05 | 45.21 | 94.29 | −0.173 | Cytoplasmic membrane |
| VrPHO1; H5 | VradiU00000886 | 564256.. 573220 | + | 752 | 86.54 | 9.09 | 46.66 | 88.64 | −0.103 | Cytoplasmic membrane |
| Gene1 | Gene2 | Ka | Ks | Ka/Ks | T(MYA) |
|---|---|---|---|---|---|
| VrPHO1; H4 | VrPHO1; H5 | 0.16 | 0.75 | 0.21 | 61.48 |
| GmPHO1; H1 | VrPHO1; H4 | 0.13 | 0.65 | 0.21 | 106.61 |
| GmPHO1; H2 | VrPHO1; H2 | 0.1 | 0.28 | 0.36 | 45.65 |
| GmPHO1; H4 | VrPHO1; H4 | 0.13 | 0.65 | 0.19 | 107.23 |
| GmPHO1; H5 | VrPHO1; H4 | 0.06 | 0.37 | 0.16 | 60.68 |
| GmPHO1; H12 | VrPHO1; H1 | 0.04 | 0.22 | 0.17 | 36.22 |
| GmPHO1; H14 | VrPHO1; H1 | 0.03 | 0.2 | 0.14 | 32.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Jiang, L.; Sun, P.; Zhou, T.; Liu, Y.; Kong, Z.; Zhang, N.; He, H.; Zhang, X. Characterization of the PHO1 Gene Family in Vigna radiata L. and Its Expression Analysis Under Phosphate-Deficient Stress. Genes 2026, 17, 25. https://doi.org/10.3390/genes17010025
Jiang L, Sun P, Zhou T, Liu Y, Kong Z, Zhang N, He H, Zhang X. Characterization of the PHO1 Gene Family in Vigna radiata L. and Its Expression Analysis Under Phosphate-Deficient Stress. Genes. 2026; 17(1):25. https://doi.org/10.3390/genes17010025
Chicago/Turabian StyleJiang, Lina, Ping Sun, Tingting Zhou, Yang Liu, Zihan Kong, Nan Zhang, Hongli He, and Xingzheng Zhang. 2026. "Characterization of the PHO1 Gene Family in Vigna radiata L. and Its Expression Analysis Under Phosphate-Deficient Stress" Genes 17, no. 1: 25. https://doi.org/10.3390/genes17010025
APA StyleJiang, L., Sun, P., Zhou, T., Liu, Y., Kong, Z., Zhang, N., He, H., & Zhang, X. (2026). Characterization of the PHO1 Gene Family in Vigna radiata L. and Its Expression Analysis Under Phosphate-Deficient Stress. Genes, 17(1), 25. https://doi.org/10.3390/genes17010025






