Molecular Structure, Comparative Analysis, and Phylogenetic Insights into the Complete Chloroplast Genomes of Fissidens crispulus
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.1.1. Sampling, DNA Extraction, and Sequencing
2.1.2. Chloroplast Genome Assembly and Annotation
2.2. Genome Feature Analyses
2.2.1. Analysis of Inverted Repeat (IR) Boundary
2.2.2. Repeat Analysis
2.2.3. Nucleotide Diversity (Pi) Analysis
2.3. Phylogenetic Inference
Phylogenetic Analysis
2.4. Codon Usage Bias Analyses
2.4.1. Calculation of Parameters Related to Codon Usage Bias
2.4.2. Neutrality Plot Analysis
2.4.3. ENC-Plot Analysis
2.4.4. Parity Rule 2 (PR2) Bias Plot Analysis
2.4.5. Relative Synonymous Codon Usage (RSCU) Analysis and Identification of Optimal Codons
3. Results
3.1. Chloroplast Genome Characteristics of Fissidens crispulus
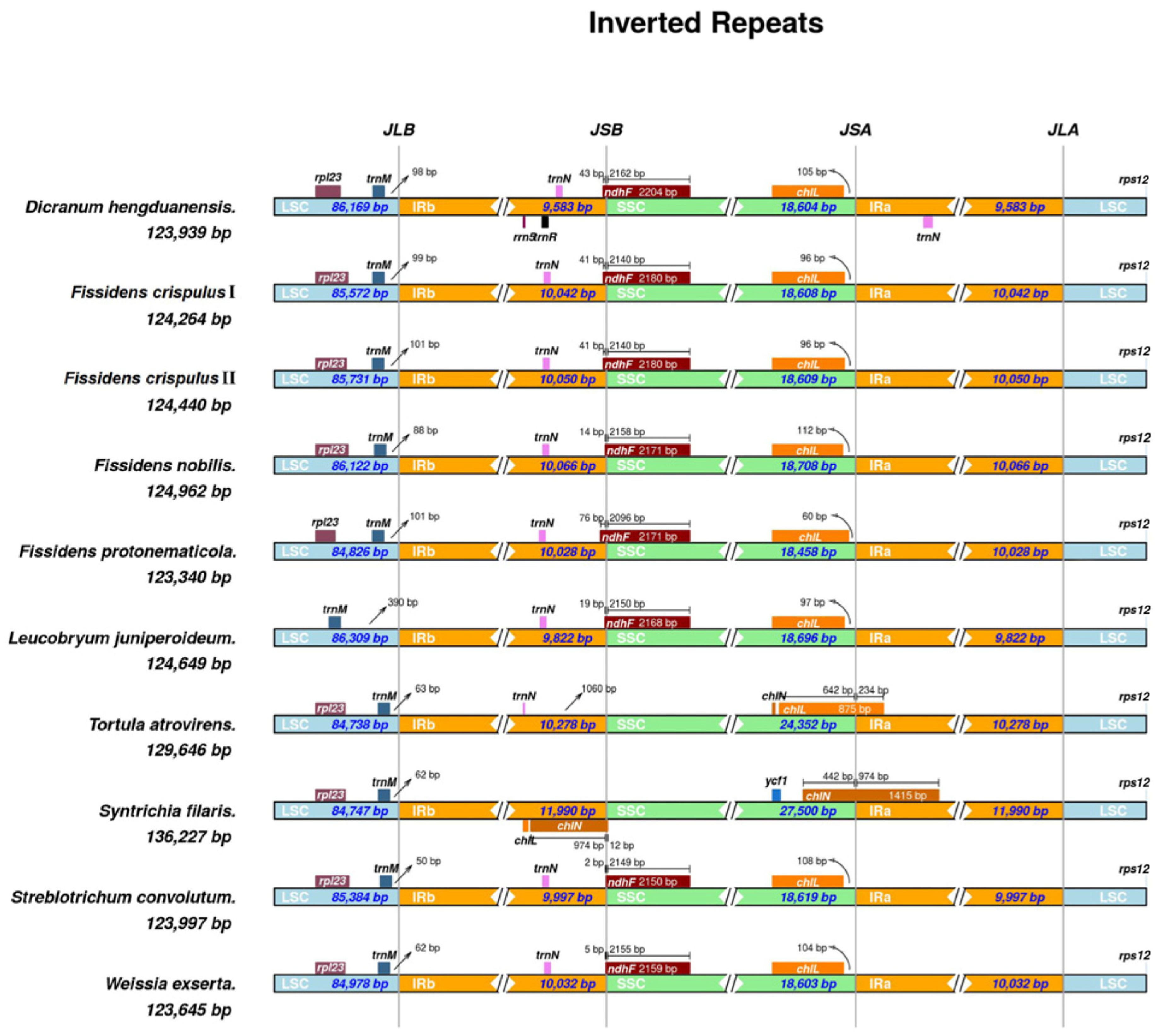
3.2. IR Expansion and Contraction in Chloroplast Genomes
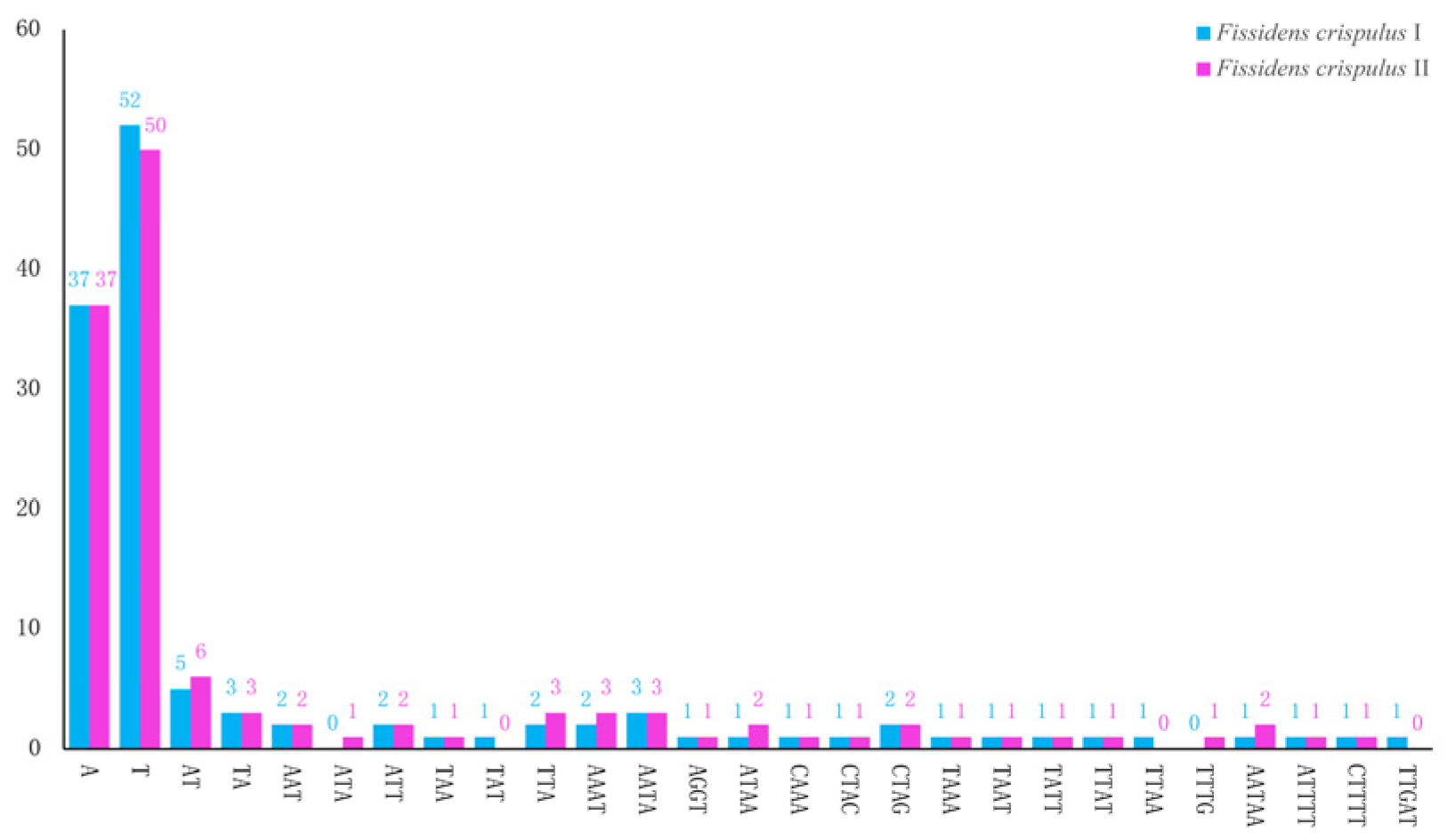
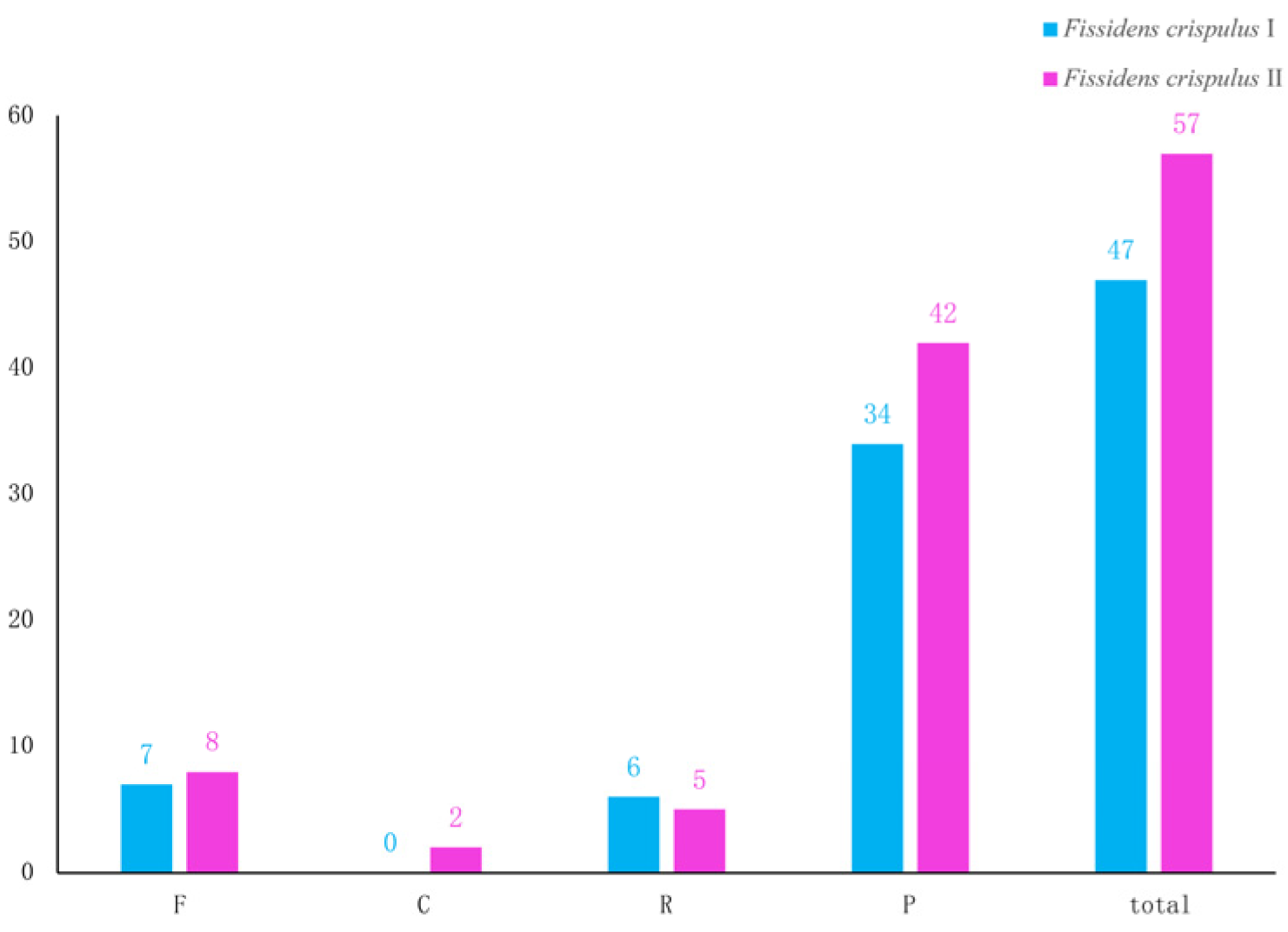
3.3. Repeat Sequence Analysis
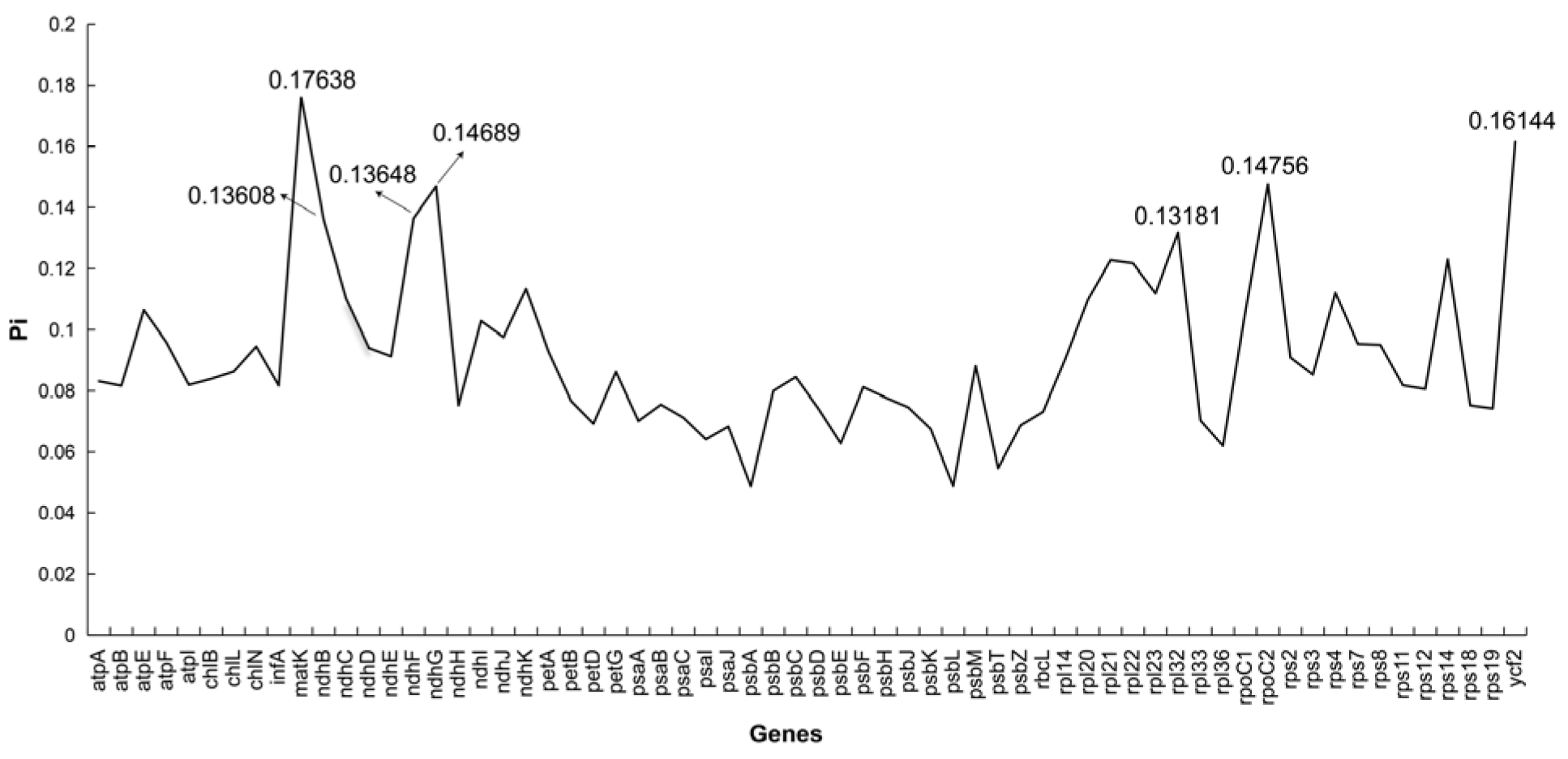
3.4. Nucleotide Diversity Analysis
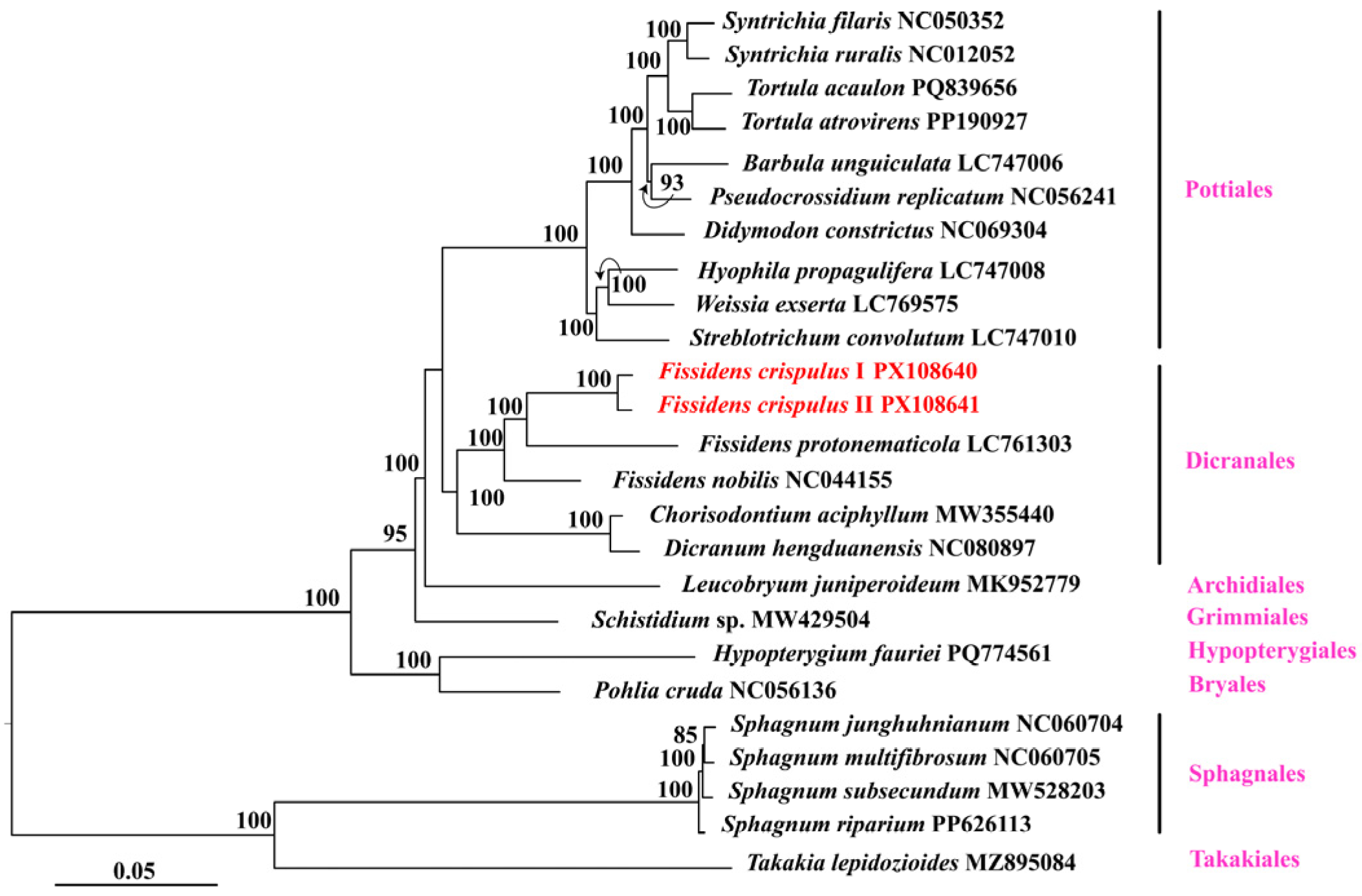
3.5. Phylogenetic Analysis
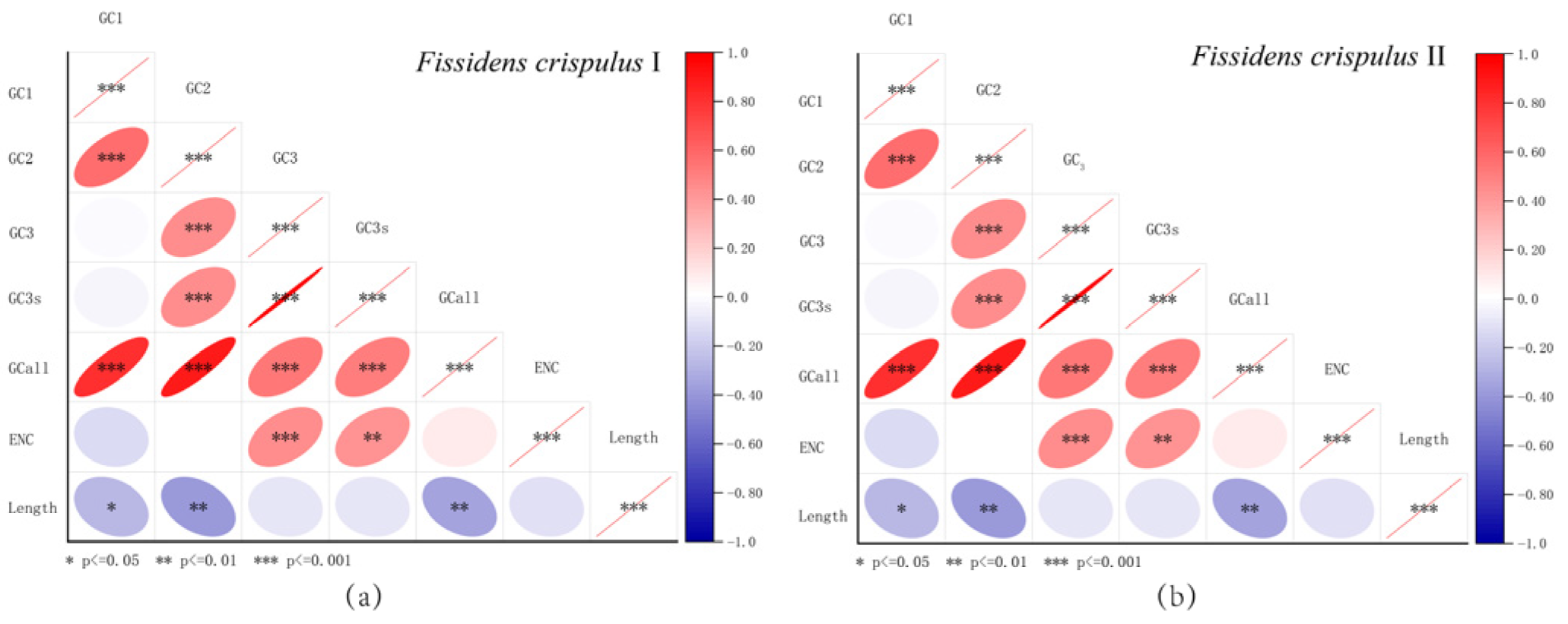
3.6. Correlation Analysis of Codon Usage Parameters
3.7. Codon Usage Bias Analyses
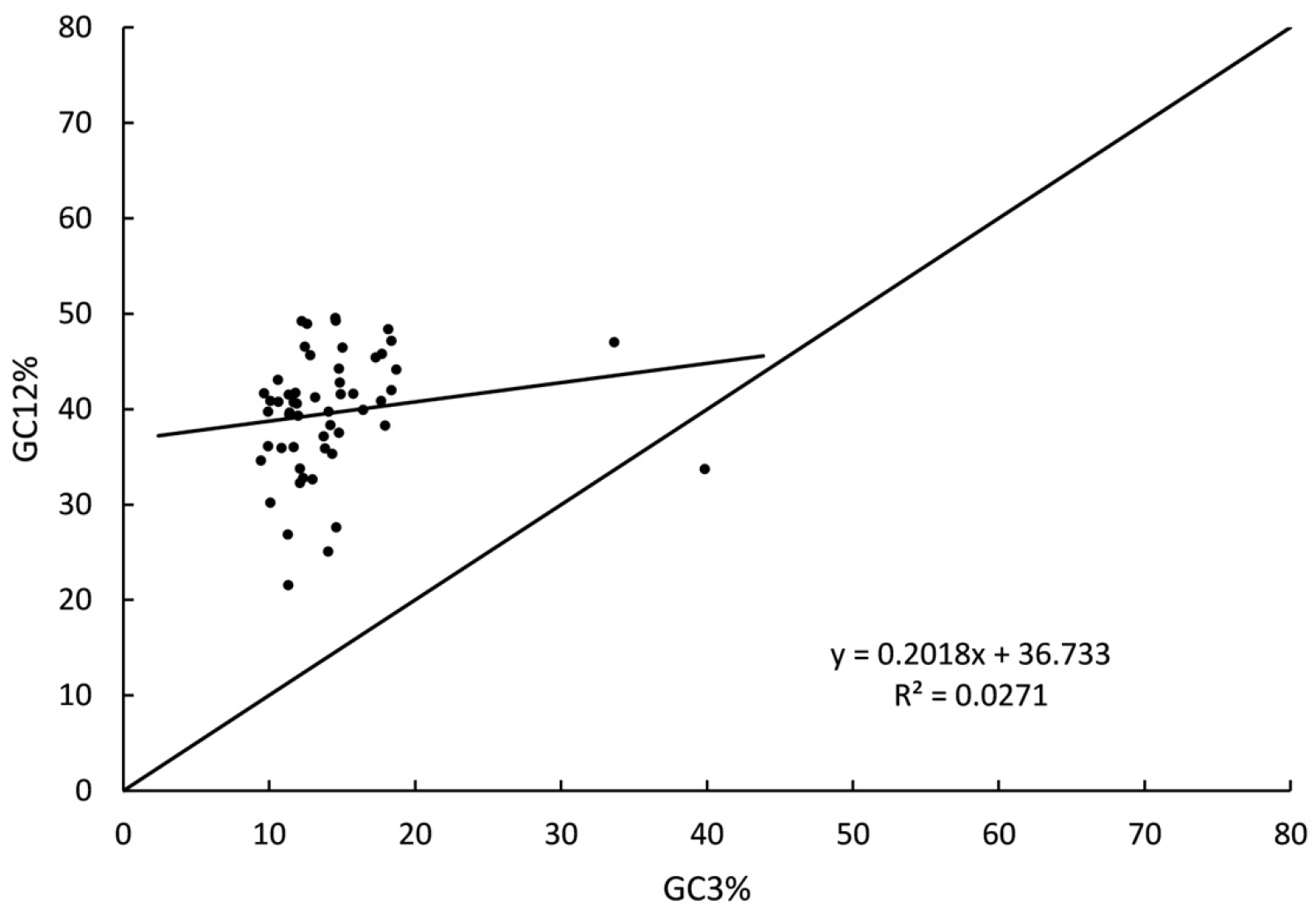
3.7.1. Neutrality Plot Analysis
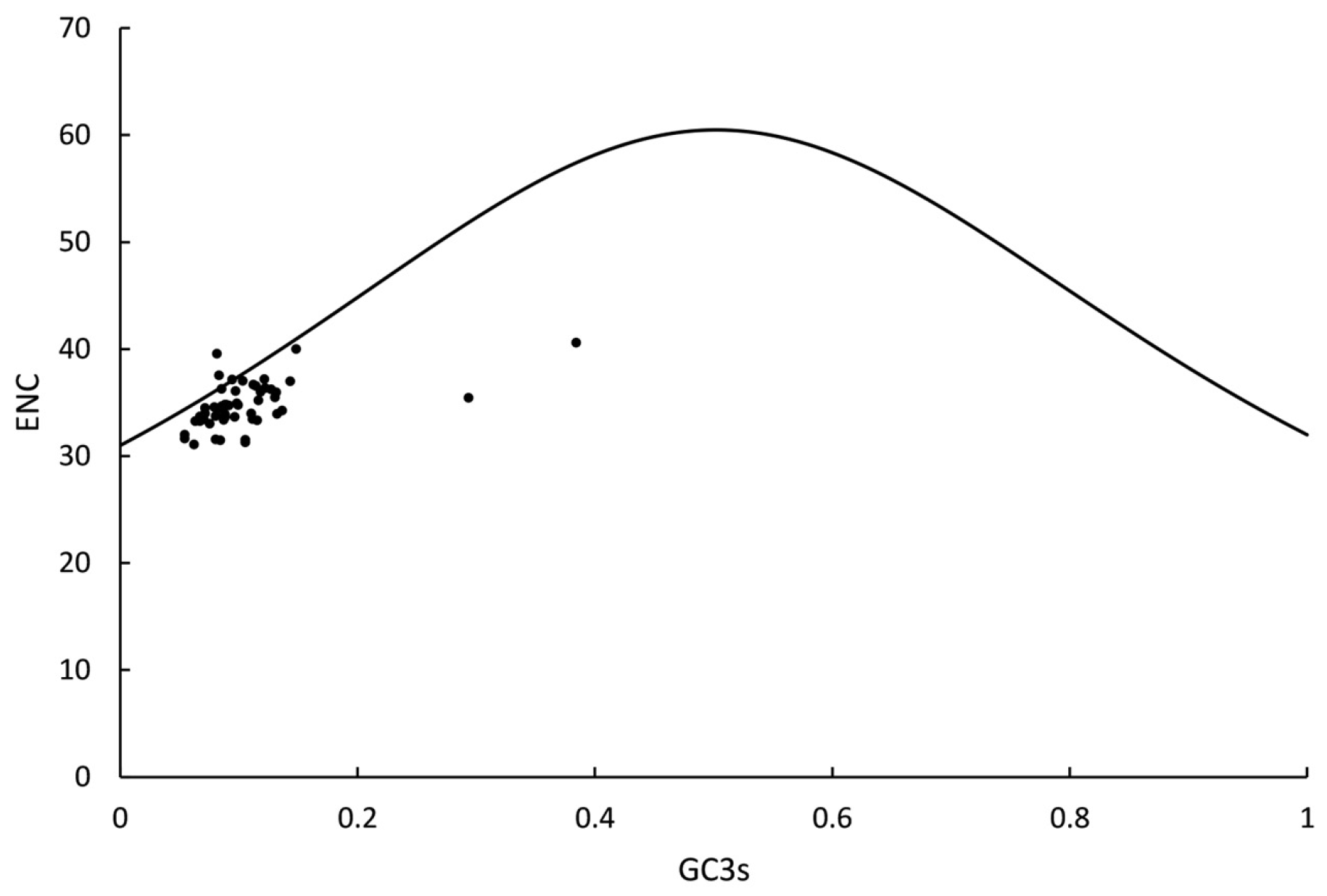
3.7.2. ENC-Plot Analysis
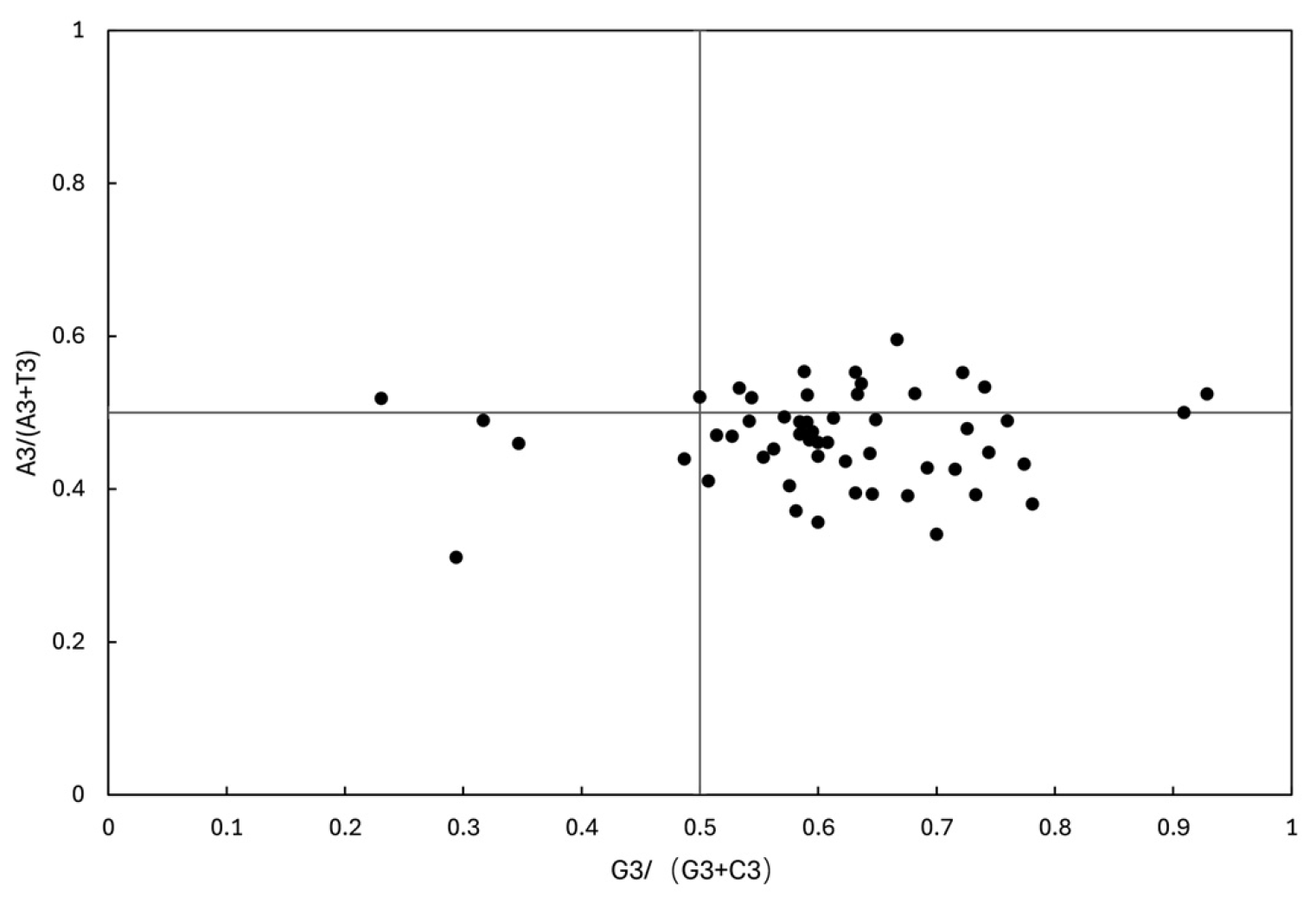
3.7.3. Parity-Rule 2 (PR2) Bias Plot Analysis
3.7.4. Analysis of RSCU and Optimal Codons
4. Discussion
4.1. Chloroplast Features of Fissidens crispulus
4.2. Phylogenetic Relationships of Fissidens crispulus
4.3. Natural Selection Plays a Critical Role in Shaping Codon Usage Bias in Fissidens crispulus
4.4. Optimal Codons Provide Novel Insights for Chloroplast Genetic Engineering in Fissidens crispulus
4.5. Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, R.L.; Ma, X.Y.; Cao, C.; Cao, Z.Y. Advances in Research on Bryophyte Diversity in China. Biodivers. Sci. 2022, 30, 22378. [Google Scholar] [CrossRef]
- Slate, M.L.; Antoninka, A.; Bailey, L.; Berdugo, M.B.; Callaghan, D.A.; Cárdenas, M.; Chmielewski, M.W.; Fenton, N.J.; Holland-Moritz, H.; Hopkins, S.; et al. Impact of changing climate on bryophyte contributions to terrestrial water, carbon, and nitrogen cycles. New Phytol. 2024, 242, 2411–2429. [Google Scholar] [CrossRef]
- Dilrukshi, H.; Ruklani, N.; Rubasinghe, S. Cryptogams as bio-indicators for ecosystem monitoring in Sri Lanka: A comprehensive review and recommendations. Environ. Monit. Assess. 2024, 196, 1231. [Google Scholar] [CrossRef]
- Crosby, M.R.; Garden, M.B. A Checklist of the Mosses; Missouri Botanical Garden Press: St. Louis, MO, USA, 1999; pp. 27–28. [Google Scholar]
- Pursell, R.A.; Ronald, A. Fissidentaceae; Flora Neotropica; The New York Botanical Garden: New York, NY, USA, 2007; Volume 101, pp. 1–278. [Google Scholar]
- Suzuki, T.; Inoue, Y.; Tsubota, H. Molecular phylogeny of the genus (Fissidentaceae, Bryophyta) and a refinement of the infrageneric classification. Mol. Phylogenet. Evol. 2018, 127, 190–202. [Google Scholar] [CrossRef]
- Iwatsuki, Z.; Suzuki, T. A taxonomic revision of the Japanese species of Fissidens (Musci). J. Hattori Bot. Lab. 1982, 51, 329–508. [Google Scholar] [CrossRef]
- Lv, R.Q.; Li, Z.H.; Li, M.P.; Dogra, V.; Lv, S.S.; Liu, R.Y.; Lee, K.P.; Kim, C.H. Uncoupled Expression of Nuclear and Plastid Photosynthesis-Associated Genes Contributes to Cell Death in a Lesion Mimic Mutant. Plant Cell 2019, 31, 210–230. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Dai, Z.W.; Dai, X.Y.; Lan, S.R.; Wu, S.S. The complete chloroplast genome of Pleione pleionoides (Orchidaceae). Mitochondrial DNA B Resour. 2019, 4, 2167–2168. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Kwak, S.Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef]
- Ruf, S.; Forner, J.; Hasse, C.; Kroop, X.; Seeger, S.; Schollbach, L.; Schadach, A.; Bock, R. High-efficiency generation of fertile transplastomic Arabidopsis plants. Nat. Plants 2019, 5, 282–289. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, 1003537. [Google Scholar] [CrossRef] [PubMed]
- Pahlich, E.; Chr, G. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; DePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, K.J. Review of originpro 8.5. J. Am. Chem. Soc. 2011, 133, 562. [Google Scholar] [CrossRef]
- Wei, L.; He, J.; Jia, X.; Qi, Q.; Liang, Z.; Zheng, H.; Ping, Y.; Liu, S.; Sun, J. Analysis of codon usage bias of mitochondrial genome in Bombyx mori and its relation to evolution. BMC Evol. Biol. 2014, 14, 262. [Google Scholar] [CrossRef]
- Song, H.; Liu, J.; Song, Q.; Zhang, Q.; Tian, P.; Nan, Z. Comprehensive analysis of codon usage bias in seven Epichloë species and their peramine-coding genes. Front. Microbiol. 2017, 8, 1419. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Sueoka, N. Translation-coupled violation of Parity Rule 2 in human genes is not the cause of heterogeneity of the DNA G+ C content of third codon position. Gene 1999, 238, 53–58. [Google Scholar] [CrossRef]
- McLean, M.J.; Wolfe, K.H.; Devine, K.M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 1998, 47, 691–696. [Google Scholar] [CrossRef]
- Wu, Y.-Q.; Li, Z.-Y.; Zhao, D.-Q.; Jun, T. Comparative analysis of flower-meristem-identity gene APETALA2 (AP2) codon in different plant species. J. Integr. Agric. 2018, 17, 867–877. [Google Scholar] [CrossRef]
- Mensah, R.A.; Sun, X.; Cheng, C.; Lai, Z. Analysis of codon usage pattern of banana Basic Secretory Protease gene. Plant Dis. Pests 2019, 10, 1–4. [Google Scholar] [CrossRef]
- Inoue, Y.; Nakahara-Tsubota, M.; Ogiso-Tanaka, E.; Tsubota, H. Complete chloroplast and mitochondrial genomes of Ditrichum rhynchostegium Kindb. (Ditrichaceae, Bryophyta). Mitochondrial DNA B Resour. 2023, 8, 383–388. [Google Scholar] [CrossRef]
- Yoon, Y.-J.; Kang, Y.; Kim, M.-K.; Lee, J.; Park, H.; Kim, J.H.; Lee, H. The complete mitochondrial genome of an Antarctic moss Syntrichia filaris (Müll. Hal.) RH Zander. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 2779–2780. [Google Scholar] [CrossRef]
- Min, J.; Kwon, W.; Xi, H.; Park, J. The complete chloroplast genome of Leucobryum juniperoideum (brid.) C. Müll. (Leucobryaceae, Bryophyta). Mitochondrial DNA B Resour. 2019, 4, 2962–2963. [Google Scholar] [CrossRef]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.-Y.; Gao, L.-Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-L.; Xie, F.-M.; Wang, C.-J.; Ding, Y.-H.; Yan, W.-C.; Fang, K.; Chen, H. Comparative analysis of plastome structure in Sphagnum species from China. BMC Genom. 2025, 26, 585. [Google Scholar] [CrossRef]
- Kuang, D.-Y.; Wu, H.; Wang, Y.-L.; Gao, L.-M.; Zhang, S.-Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef]
- McCoy, S.R.; Kuehl, J.V.; Boore, J.L.; Raubeson, L.A. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 2008, 8, 130. [Google Scholar] [CrossRef]
- Niu, Z.; Xue, Q.; Wang, H.; Xie, X.; Zhu, S.; Liu, W.; Ding, X. Mutational biases and GC-biased gene conversion affect GC content in the plastomes of Dendrobium genus. Int. J. Mol. Sci. 2017, 18, 2307. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.; Lin, Q.; Gerelle, W.K.; Joya, S.; Chang, Y.; Taylor, Z.N.; Rothfels, C.J.; Larsson, A.; Villarreal, J.C.; Li, F.W.; et al. Organellomic data sets confirm a cryptic consensus on (unrooted) land-plant relationships and provide new insights into bryophyte molecular evolution. Am. J. Bot. 2020, 107, 91–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-Z.; Xu, H.; Ma, X.-Y.; Zhu, R.-L. Dicranum hengduanensis (Dicranaceae, Bryophyta), a new species with fragile leaves from the Hengduan Mountains in China. Bryologist 2023, 126, 226–235. [Google Scholar] [CrossRef]
- Ren, Z.; Chen, H.; Zhang, S. The complete plastid genome of an Antarctic moss Chorisodontium aciphyllum (Hook. f. & Wilson) Broth (Dicranaceae, Dicranales). Mitochondrial DNA B Resour. 2022, 7, 683–685. [Google Scholar] [CrossRef]
- Bechteler, J.; Peñaloza-Bojacá, G.; Bell, D.; Gordon Burleigh, J.; McDaniel, S.F.; Christine Davis, E.; Sessa, E.B.; Bippus, A.; Christine Cargill, D.; Chantanoarrapint, S. Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. Am. J. Bot. 2023, 110, e16249. [Google Scholar] [CrossRef]
- Dong, S.; Zhang, S.; Zhang, L.; Wu, H.; Goffinet, B.; Liu, Y. Plastid genomes and phylogenomics of liverworts (Marchantiophyta): Conserved genome structure but highest relative plastid substitution rate in land plants. Mol. Phylogenet. Evol. 2021, 161, 107171. [Google Scholar] [CrossRef]
- Frangedakis, E.; Shimamura, M.; Villarreal, J.C.; Li, F.W.; Tomaselli, M.; Waller, M.; Sakakibara, K.; Renzaglia, K.S.; Szövényi, P. The hornworts: Morphology, evolution and development. New Phytol. 2021, 229, 735–754. [Google Scholar] [CrossRef]
- Chen, J.; Wang, F.; Zhao, Z.; Li, M.; Liu, Z.; Peng, D. Complete chloroplast genomes and comparative analyses of three Paraphalaenopsis (Aeridinae, Orchidaceae) species. Int. J. Mol. Sci. 2023, 24, 11167. [Google Scholar] [CrossRef]
- Kong, W.Q.; Yang, J.H. The complete chloroplast genome sequence of Morus cathayana and Morus multicaulis, and comparative analysis within genus Morus L. PeerJ 2017, 5, e3037. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, B.; Li, B.; Zhou, Q.; Wang, G.; Jiang, X.; Wang, C.; Xu, Z. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 2020, 8, e8251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, T.; Zhou, Q.; Sheng, Q.; Zhu, Z. Complete chloroplast genomes and phylogenetic relationships of Bougainvillea spectabilis and Bougainvillea glabra (Nyctaginaceae). Int. J. Mol. Sci. 2023, 24, 13044. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liu, J.; Li, H.; Yue, C.; Gao, M. Complete chloroplast genome structural characterization and comparative analysis of Viburnum japonicum (Adoxaceae). Forests 2023, 14, 1819. [Google Scholar] [CrossRef]
- Sablok, G.; Nayak, K.C.; Vazquez, F.; Tatarinova, T.V. Synonymous codon usage, GC 3, and evolutionary patterns across plastomes of three pooid model species: Emerging grass genome models for monocots. Mol. Biotechnol. 2011, 49, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.-K.; Song, X.; Chen, C.-G.; Li, G.; Xie, S.-Q. Codon usage profiling of chloroplast genome in Magnoliaceae. J. Agric. Sci. Technol. 2020, 22, 52–62. [Google Scholar] [CrossRef]
- Sun, C.Q.; Chen, F.; Teng, N.J.; Xu, Y.C.; Dai, Z.L. Comparative analysis of the complete chloroplast genome of seven Nymphaea species. Aquat. Bot. 2021, 170, 103353. [Google Scholar] [CrossRef]

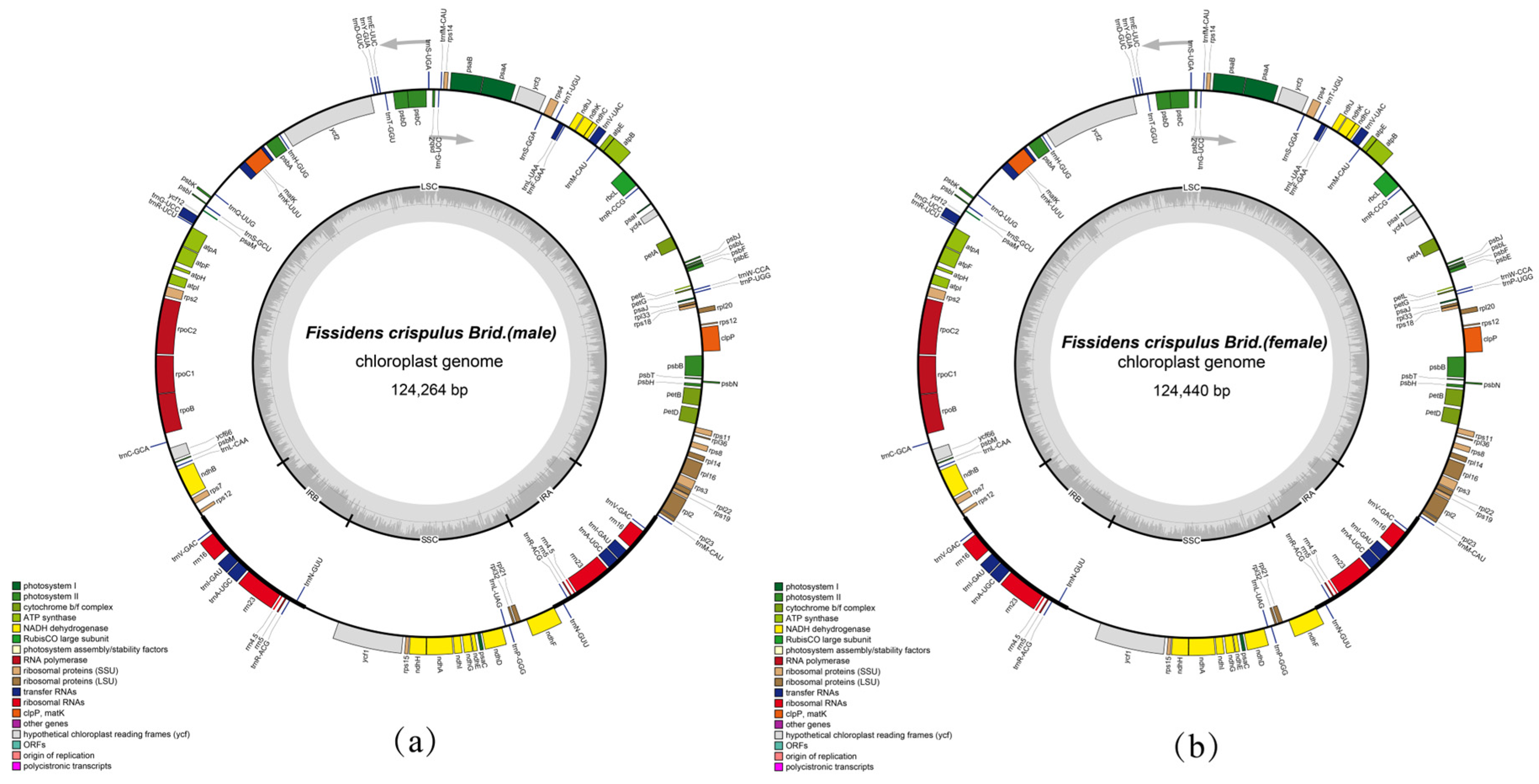

| Species | Size (bp) | LSC (bp) | SSC (bp) | IR (bp) | Total GC (%) | LSC GC (%) | SSC GC (%) | IR GC (%) |
|---|---|---|---|---|---|---|---|---|
| Fissidens crispulus I | 124,264 | 85,572 | 18,608 | 10,042 | 28.7 | 26.0 | 25.1 | 43.3 |
| Fissidens crispulus II | 124,440 | 85,731 | 18,609 | 10,050 | 28.7 | 26.0 | 25.2 | 43.4 |
| Species | Variable | Correlation Coefficient | |||||
|---|---|---|---|---|---|---|---|
| GC1 | GC2 | GC3 | GC3s | GCall | ENC | ||
| F. crispulus I | GC2 | 0.560 | |||||
| GC3 | −0.061 | 0.436 | |||||
| GC3s | −0.121 | 0.369 | 0.963 | ||||
| GCall | 0.786 | 0.890 | 0.502 | 0.426 | |||
| ENC | −0.124 | 0.08 | 0.428 | 0.511 | 0.114 | ||
| Length | −0.308 | −0.425 | −0.108 | −0.067 | −0.391 | −0.052 | |
| F. crispulus II | GC2 | 0.579 | |||||
| GC3 | −0.001 | 0.441 | |||||
| GC3s | −0.023 | 0.444 | 0.996 | ||||
| GCall | 0.803 | 0.889 | 0.526 | 0.513 | |||
| ENC | −0.122 | 0.005 | 0.447 | 0.432 | 0.088 | ||
| Length | −0.279 | −0.387 | −0.099 | −0.098 | −0.351 | −0.102 | |
| Category | Codons | Count | Number of U-Ending Codons (Percentage) |
|---|---|---|---|
| Shared Codons | GCA, UGU, UUU, GGU, UUA, AAU, CCU, CGA, CGU, AGU, UCA, ACU | 12 | 8 (66.67%) |
| Fissidens crispulus I-Specific | AUG, ACA, UGG | 3 | 0 (0%) |
| Fissidens crispulus II-Specific | GCU, AUU, UCU, GUU | 4 | 4 (100%) |
| Total | 19 | 12 (63.16%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.-Q.; Kang, K.-L.; Chen, J.; Wei, Y.-M.; Xiang, Y.-L.; Peng, T. Molecular Structure, Comparative Analysis, and Phylogenetic Insights into the Complete Chloroplast Genomes of Fissidens crispulus. Genes 2025, 16, 1103. https://doi.org/10.3390/genes16091103
Song Y-Q, Kang K-L, Chen J, Wei Y-M, Xiang Y-L, Peng T. Molecular Structure, Comparative Analysis, and Phylogenetic Insights into the Complete Chloroplast Genomes of Fissidens crispulus. Genes. 2025; 16(9):1103. https://doi.org/10.3390/genes16091103
Chicago/Turabian StyleSong, Yun-Qi, Kai-Li Kang, Jin Chen, Yu-Mei Wei, You-Liang Xiang, and Tao Peng. 2025. "Molecular Structure, Comparative Analysis, and Phylogenetic Insights into the Complete Chloroplast Genomes of Fissidens crispulus" Genes 16, no. 9: 1103. https://doi.org/10.3390/genes16091103
APA StyleSong, Y.-Q., Kang, K.-L., Chen, J., Wei, Y.-M., Xiang, Y.-L., & Peng, T. (2025). Molecular Structure, Comparative Analysis, and Phylogenetic Insights into the Complete Chloroplast Genomes of Fissidens crispulus. Genes, 16(9), 1103. https://doi.org/10.3390/genes16091103






