Exceptions to Broad Tissue-Specific Transcriptomic Interdependence: Searching for Independence in Expression of Genes
Abstract
1. Introduction
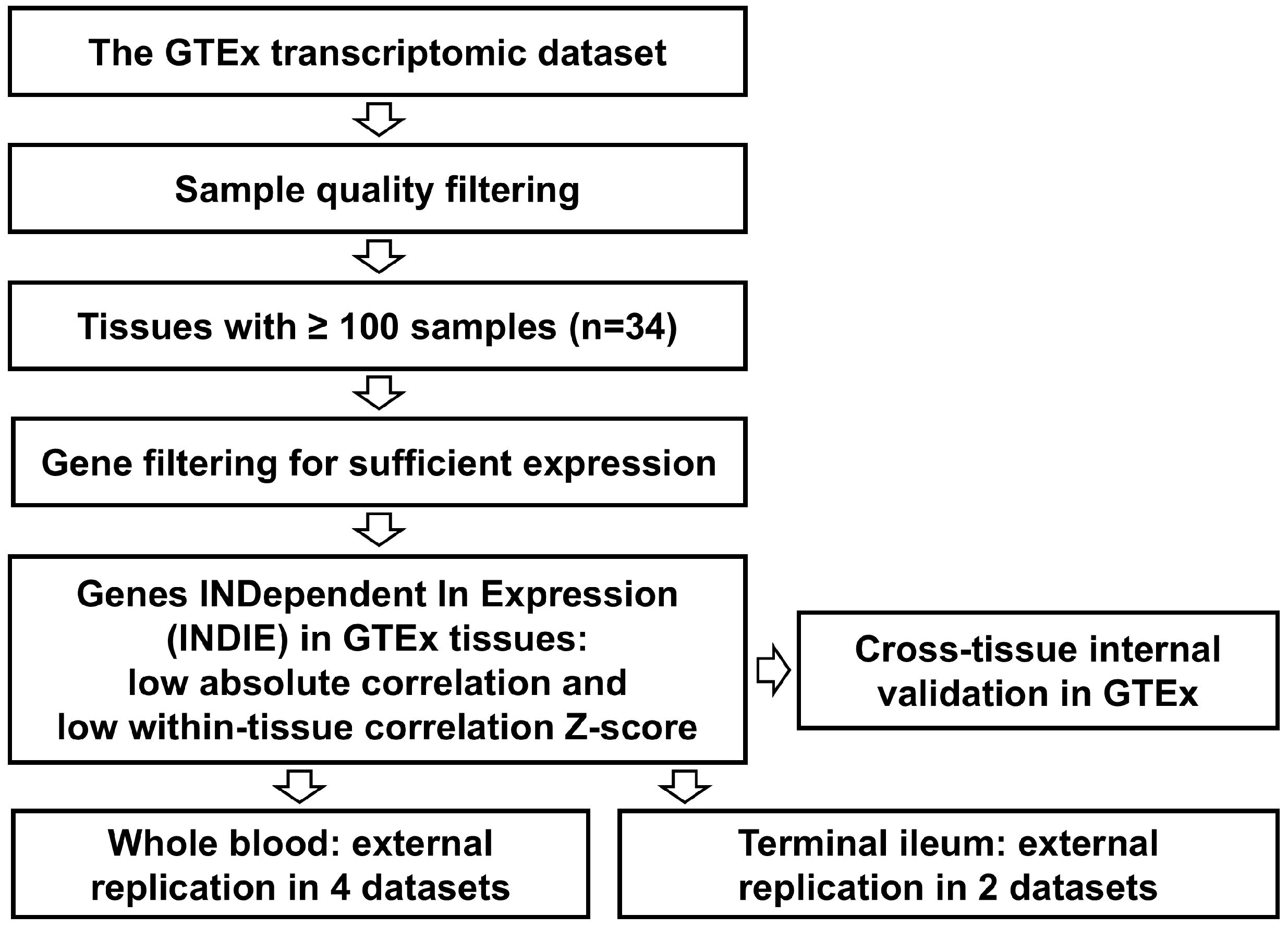
2. Methods
3. Results
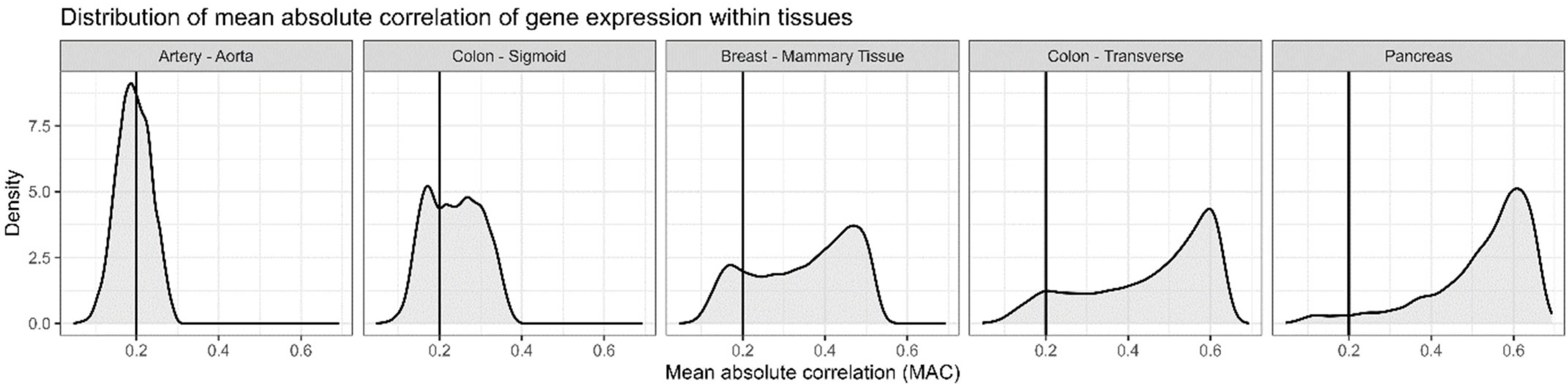
3.1. GTEx Correlations in Tissues—Descriptive
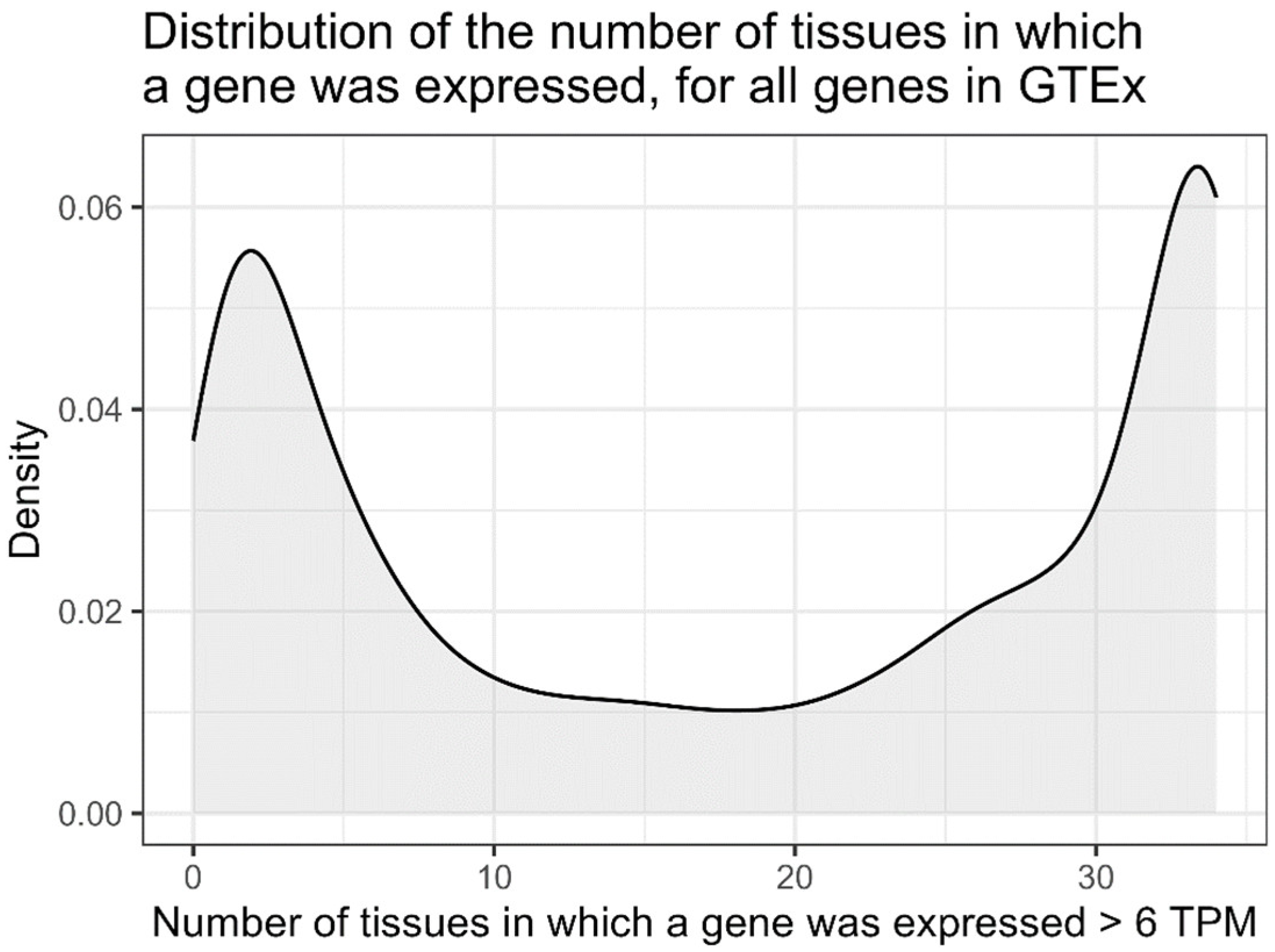
3.2. GTEx Correlations in Tissues—Genes That Were Present in More than n Tissues
3.3. Partial Replication of INDIE Genes in the Whole Blood Across Four External Datasets
3.4. Partial Replication of INDIE Genes in the Terminal Ileum in Two External Datasets
3.5. Other Remarks
4. Discussion
4.1. Distributions of Correlations Differ Between Tissues
4.2. Uncorrelated Genes Within GTEx
4.3. INDIE Genes Replicated in the Whole Blood
4.4. INDIE Genes Replicated in the Ileal Tissues
4.5. Main Correlation Coefficients in Transcriptomics
4.6. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Dam, S.; Võsa, U.; van der Graaf, A.; Franke, L.; de Magalhães, J.P. Gene Co-Expression Analysis for Functional Classification and Gene–Disease Predictions. Brief. Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef] [PubMed]
- Lelewer, D.A.; Hirschberg, D.S. Data Compression. ACM Comput. Surv. 1987, 19, 261–296. [Google Scholar] [CrossRef]
- Hyvärinen, A. Independent Component Analysis: Recent Advances. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2013, 371, 20110534. Available online: https://royalsocietypublishing.org/doi/full/10.1098/rsta.2011.0534 (accessed on 22 July 2025). [CrossRef]
- Richiardi, J.; Altmann, A.; Milazzo, A.C.; Chang, C.; Chakravarty, M.M.; Banaschewski, T.; Barker, G.J.; Bokde, A.L.; Bromberg, U.; Büchel, C.; et al. Correlated Gene Expression Supports Synchronous Activity in Brain Networks. Science 2015, 348, 1241–1244. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Zhang, Y.; Fan, J.; Seelig, G.; Kannan, S. Scalable Preprocessing for Sparse scRNA-Seq Data Exploiting Prior Knowledge. Bioinformatics 2018, 34, i124–i132. [Google Scholar] [CrossRef]
- Mitić, N.S.; Malkov, S.N.; Maljković Ružičić, M.M.; Veljković, A.N.; Čukić, I.L.; Lin, X.; Lyu, M.; Brusić, V. Correlation-Based Feature Selection of Single Cell Transcriptomics Data from Multiple Sources. J. Big Data 2025, 12, 4. [Google Scholar] [CrossRef]
- Shainer, I.; Kappel, J.M.; Laurell, E.; Donovan, J.C.; Schneider, M.W.; Kuehn, E.; Arnold-Ammer, I.; Stemmer, M.; Larsch, J.; Baier, H. Transcriptomic Neuron Types Vary Topographically in Function and Morphology. Nature 2025, 638, 1023–1033. [Google Scholar] [CrossRef]
- Nowak, J.K.; Adams, A.T.; Kalla, R.; Lindstrøm, J.C.; Vatn, S.; Bergemalm, D.; Keita, Å.V.; Gomollón, F.; Jahnsen, J.; Vatn, M.H.; et al. Characterisation of the Circulating Transcriptomic Landscape in Inflammatory Bowel Disease Provides Evidence for Dysregulation of Multiple Transcription Factors Including NFE2, SPI1, CEBPB, and IRF2. J. Crohn’s Colitis 2022, 16, 1255–1268. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Gupta, R.K.; Rosenheim, J.; Bell, L.C.; Chandran, A.; Guerra-Assuncao, J.A.; Pollara, G.; Whelan, M.; Artico, J.; Joy, G.; Kurdi, H.; et al. Blood Transcriptional Biomarkers of Acute Viral Infection for Detection of Pre-Symptomatic SARS-CoV-2 Infection: A Nested, Case-Control Diagnostic Accuracy Study. Lancet Microbe 2021, 2, e508–e517. [Google Scholar] [CrossRef]
- Rosenheim, J.; Gupta, R.K.; Thakker, C.; Mann, T.; Bell, L.C.; Broderick, C.M.; Madon, K.; Papargyris, L.; Dayananda, P.; Kwok, A.J.; et al. SARS-CoV-2 Human Challenge Reveals Biomarkers That Discriminate Early and Late Phases of Respiratory Viral Infections. Nat. Commun. 2024, 15, 10434. [Google Scholar] [CrossRef]
- Momozawa, Y.; Dmitrieva, J.; Théâtre, E.; Deffontaine, V.; Rahmouni, S.; Charloteaux, B.; Crins, F.; Docampo, E.; Elansary, M.; Gori, A.S.; et al. IBD Risk Loci Are Enriched in Multigenic Regulatory Modules Encompassing Putative Causative Genes. Nat. Commun. 2018, 9, 2427. [Google Scholar] [CrossRef]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.O.; Tang, D.; Karns, R.; Baldassano, R.N.; Noe, J.D.; Rosh, J.; Markowitz, J.; et al. Pediatric Crohn Disease Patients Exhibit Specific Ileal Transcriptome and Microbiome Signature. J. Clin. Investig. 2014, 124, 3617–3633. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making Genome-Scale Phylogenetics Accessible to All. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Speir, M.L.; Bhaduri, A.; Markov, N.S.; Moreno, P.; Nowakowski, T.J.; Papatheodorou, I.; Pollen, A.A.; Raney, B.J.; Seninge, L.; Kent, W.J.; et al. UCSC Cell Browser: Visualize Your Single-Cell Data. Bioinformatics 2021, 37, 4578–4580. [Google Scholar] [CrossRef] [PubMed]
- Pratt, D.; Chen, J.; Welker, D.; Rivas, R.; Pillich, R.; Rynkov, V.; Ono, K.; Miello, C.; Hicks, L.; Szalma, S.; et al. NDEx, the Network Data Exchange. Cell Syst. 2015, 1, 302–305. [Google Scholar] [CrossRef]
- Pillich, R.T.; Chen, J.; Churas, C.; Fong, D.; Gyori, B.M.; Ideker, T.; Karis, K.; Liu, S.N.; Ono, K.; Pico, A.; et al. NDEx IQuery: A Multi-Method Network Gene Set Analysis Leveraging the Network Data Exchange. Bioinformatics 2023, 39, btad118. [Google Scholar] [CrossRef] [PubMed]
- Pillich, R.T.; Chen, J.; Churas, C.; Liu, S.; Ono, K.; Otasek, D.; Pratt, D. NDEx: Accessing Network Models and Streamlining Network Biology Workflows. Curr. Protoc. 2021, 1, e258. [Google Scholar] [CrossRef] [PubMed]
- Pillich, R.T.; Chen, J.; Rynkov, V.; Welker, D.; Pratt, D. NDEx: A Community Resource for Sharing and Publishing of Biological Networks. In Protein Bioinformatics: From Protein Modifications and Networks to Proteomics; Wu, C.H., Arighi, C.N., Ross, K.E., Eds.; Springer: New York, NY, USA, 2017; pp. 271–301. ISBN 978-1-4939-6783-4. [Google Scholar]
- Pratt, D.; Chen, J.; Pillich, R.; Rynkov, V.; Gary, A.; Demchak, B.; Ideker, T. NDEx 2.0: A Clearinghouse for Research on Cancer Pathways. Cancer Res. 2017, 77, e58–e61. [Google Scholar] [CrossRef]
- Hsiao, L.L.; Dangond, F.; Yoshida, T.; Hong, R.; Jensen, R.V.; Misra, J.; Dillon, W.; Lee, K.F.; Clark, K.E.; Haverty, P.; et al. A Compendium of Gene Expression in Normal Human Tissues. Physiol. Genom. 2001, 7, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Sadeesh, E.M.; Lahamge, M.S. Unveiling the Tissue-Specific Landscape of Nuclear-Encoded Mitochondrial Genes Involved in Amino Acid Metabolism in Buffalo. Amino Acids 2025, 57, 17. [Google Scholar] [CrossRef]
- Wang, G.; Yang, E.; Mandhan, I.; Brinkmeyer-Langford, C.L.; Cai, J.J. Population-Level Expression Variability of Mitochondrial DNA-Encoded Genes in Humans. Eur. J. Hum. Genet. 2014, 22, 1093–1099. [Google Scholar] [CrossRef]
- Yekula, A.; Hsia, T.; Kitchen, R.R.; Chakrabortty, S.K.; Yu, W.; Batool, S.M.; Lewis, B.; Szeglowski, A.J.; Weissleder, R.; Lee, H.; et al. Longitudinal Analysis of Serum-Derived Extracellular Vesicle RNA to Monitor Dacomitinib Treatment Response in EGFR-Amplified Recurrent Glioblastoma Patients. Neuro-Oncol. Adv. 2023, 5, vdad104. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J.; Zhang, W.; Wang, Y.; Zhai, Y.; Li, Y.; Li, W.; Chang, J.; Zhao, X.; Huang, M.; et al. A Liquid Biopsy Signature of Circulating Extracellular Vesicles-Derived RNAs Predicts Response to First Line Chemotherapy in Patients with Metastatic Colorectal Cancer. Mol. Cancer 2023, 22, 199. [Google Scholar] [CrossRef]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R.F. Pseudogenes: Pseudo-Functional or Key Regulators in Health and Disease? RNA 2011, 17, 792–798. [Google Scholar] [CrossRef]
- Sanij, E.; Diesch, J.; Lesmana, A.; Poortinga, G.; Hein, N.; Lidgerwood, G.; Cameron, D.P.; Ellul, J.; Goodall, G.J.; Wong, L.H.; et al. A Novel Role for the Pol I Transcription Factor UBTF in Maintaining Genome Stability through the Regulation of Highly Transcribed Pol II Genes. Genome Res. 2015, 25, 201–212. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Dhama, K.; Lee, S.S. Evaluation of Differentially Expressed Genes during Replication Using Gene Expression Landscape of Monkeypox-Infected MK2 Cells: A Bioinformatics and Systems Biology Approach to Understanding the Genomic Pattern of Viral Replication. J. Infect. Public Health 2023, 16, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, A. Next Generation Sequencing to Identify New Genetic Causes of Familial Craniosynostosis. Doctoral Dissertation, University of Oxford, Oxford, UK, 2017. Available online: https://ora.ox.ac.uk/objects/uuid:e72022d1-d77d-4d0f-8419-950599bd1b2c (accessed on 24 August 2025).
- Victoria, S.; Castro, A.; Pittini, A.; Olivera, D.; Russo, S.; Cebrian, I.; Mombru, A.W.; Osinaga, E.; Pardo, H.; Segovia, M.; et al. Formulating a TMEM176B Blocker in Chitosan Nanoparticles Uncouples Its Paradoxical Roles in Innate and Adaptive Antitumoral Immunity. Int. J. Biol. Macromol. 2024, 279, 135327. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Luo, B.; Fei, L.; Zhang, Q.; Liang, X.; Chen, Y.; Zhou, X. MS4A Superfamily Molecules in Tumors, Alzheimer’s and Autoimmune Diseases. Front. Immunol. 2024, 15, 1481494. [Google Scholar] [CrossRef]
- Jiang, L.; Yi, R.; Chen, H.; Wu, S. Quercetin Alleviates Metabolic-Associated Fatty Liver Disease by Tuning Hepatic Lipid Metabolism, Oxidative Stress and Inflammation. Anim. Biotechnol. 2025, 36, 2442351. [Google Scholar] [CrossRef]
- Zheng, Z.; Chen, R.; Liu, M.; Ding, Y.; Xu, S.; Hou, C.; Li, S. Identification of Novel Therapeutic Targets for Hypertension. Hypertension 2025, 82, 1056–1070. [Google Scholar] [CrossRef]
- Harada, K.; Sakamoto, N.; Kitaoka, T.; Nakamura, Y.; Kondo, R.; Morisue, R.; Hashimoto, H.; Yamamoto, Y.; Ukai, S.; Maruyama, R.; et al. PI3 Expression Predicts Recurrence after Chemotherapy with DNA-damaging Drugs in Gastric Cancer. J. Pathol. 2025, 265, 472–485. [Google Scholar] [CrossRef]
- Zhang, M.; Cai, F.; Guo, J.; Liu, S.; Ma, G.; Cai, M.; Zhang, R.; Deng, J. ACAT2 Suppresses the Ubiquitination of YAP1 to Enhance the Proliferation and Metastasis Ability of Gastric Cancer via the Upregulation of SETD7. Cell Death Dis. 2024, 15, 297. [Google Scholar] [CrossRef]
- Latiano, A.; Palmieri, O.; Pastorelli, L.; Vecchi, M.; Pizarro, T.T.; Bossa, F.; Merla, G.; Augello, B.; Latiano, T.; Corritore, G.; et al. Associations between Genetic Polymorphisms in IL-33, IL1R1 and Risk for Inflammatory Bowel Disease. PLoS ONE 2013, 8, e62144. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Jiménez, D.; Núñez, L.; De la Fuente, M.; Dubois-Camacho, K.; Sepúlveda, H.; Montecino, M.; Torres-Riquelme, A.; García-González, P.; Chnaiderman, J.; Vossenkamper, A.; et al. A Functional IL1RL1 Variant Regulates Corticosteroid-Induced sST2 Expression in Ulcerative Colitis. Sci. Rep. 2017, 7, 10180. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.J.; Plichta, D.; Hogstrom, L.; Borren, N.Z.; Lau, H.; Gregory, S.M.; Tan, W.; Khalili, H.; Clish, C.; Vlamakis, H.; et al. Multi-Omics Reveal Microbial Determinants Impacting Responses to Biologic Therapies in Inflammatory Bowel Disease. Cell Host Microbe 2021, 29, 1294–1304.e4. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.T.; Kennedy, N.A.; Hansen, R.; Ventham, N.T.; O’Leary, K.R.; Drummond, H.E.; Noble, C.L.; El-Omar, E.; Russell, R.K.; Wilson, D.C.; et al. Two-Stage Genome-Wide Methylation Profiling in Childhood-Onset Crohn’s Disease Implicates Epigenetic Alterations at the VMP1/MIR21 and HLA Loci. Inflamm. Bowel Dis. 2014, 20, 1784–1793. [Google Scholar] [CrossRef]
- Joustra, V.; Hageman, I.L.; Satsangi, J.; Adams, A.; Ventham, N.T.; de Jonge, W.J.; Henneman, P.; D’Haens, G.R.; Li Yim, A.Y. Systematic Review and Meta-Analysis of Peripheral Blood DNA Methylation Studies in Inflammatory Bowel Disease. J. Crohn’s Colitis 2023, 17, 185–198. [Google Scholar] [CrossRef]
- Codrich, M.; Bertuzzi, M.; Russo, R.; Francescatto, M.; Espinoza, S.; Zentilin, L.; Giacca, M.; Cesselli, D.; Beltrami, A.P.; Ascenzi, P.; et al. Neuronal Hemoglobin Affects Dopaminergic Cells’ Response to Stress. Cell Death Dis. 2018, 8, e2538. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Yan, Y.; Pu, J.; Zhang, B. Physiological and Pathological Functions of Neuronal Hemoglobin: A Key Underappreciated Protein in Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 9088. [Google Scholar] [CrossRef]
- Richter, F.; Meurers, B.H.; Zhu, C.; Medvedeva, V.P.; Chesselet, M.F. Neurons Express Hemoglobin α- and β-Chains in Rat and Human Brains. J. Comp. Neurol. 2009, 515, 538–547. [Google Scholar] [CrossRef]
- Jafari, S.; Ravan, M.; Karimi-Sani, I.; Aria, H.; Hasan-Abad, A.M.; Banasaz, B.; Atapour, A.; Sarab, G.A. Screening and Identification of Potential Biomarkers for Pancreatic Cancer: An Integrated Bioinformatics Analysis. Pathol. Res. Pract. 2023, 249, 154726. [Google Scholar] [CrossRef] [PubMed]
- Pelia, R.; Venkateswaran, S.; Matthews, J.D.; Haberman, Y.; Cutler, D.J.; Hyams, J.S.; Denson, L.A.; Kugathasan, S. Profiling Non-Coding RNA Levels with Clinical Classifiers in Pediatric Crohn’s Disease. BMC Med. Genom. 2021, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Onwuegbuzie, A.J.; Daniel, L.G. Uses and Misuses of the Correlation Coefficient; U.S. Department of Education: Washington, DC, USA, 1999.
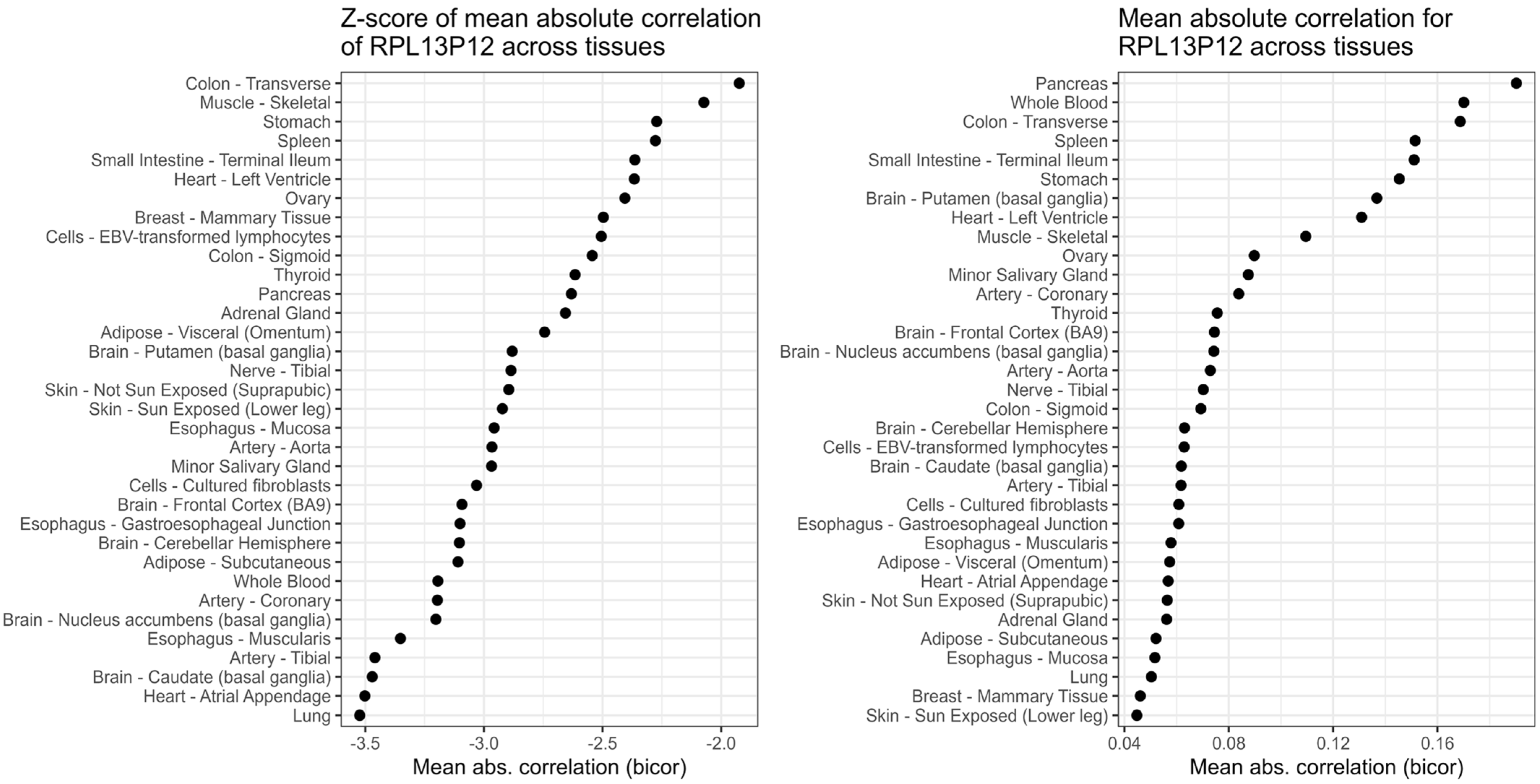
| Gene | Name | % of Validation Across Tissues | n of Tissues in Which Gene Was INDIE | n of Tissues in Which Gene Had TPM ≥ 6 |
|---|---|---|---|---|
| RPL13P12 | Ribosomal Protein L13 Pseudogene 12 | 100% | 34 | 34 |
| NPIPA5 | Nuclear Pore Complex Interacting Protein Family Member A5 | 100% | 23 | 23 |
| AC018738.2 | - | 86% | 18 | 21 |
| RN7SL2 | RNA Component of Signal Recognition Particle 7SL2 | 85% | 29 | 34 |
| RPL24P4 | Ribosomal Protien Large 24 Pseudogene 4 | 84% | 16 | 19 |
| RPSAP58 | Ribosomal Protein SA Pseudogene 58 | 82% | 27 | 33 |
| MTND4P12 | Mitochondrially Encoded NADH:Ubiquinone Oxidoreductase Core Subunit 4 Pseudogene 12 | 79% | 23 | 29 |
| ACTA1 | Actin Alpha 1 | 79% | 15 | 19 |
| HBB | Haemoglobin Subunit Beta | 78% | 25 | 32 |
| HBA2 | Haemoglobin Subunit Alpha 2 | 78% | 21 | 27 |
| n | n | Z-Score of Mean Absolute Value of r (Bicor) in Datasets | ||||
|---|---|---|---|---|---|---|
| Gene | datasets | validations | IBD Character | Migraine | COVID-19 d.1 | COVID-19 d.2 |
| HIST1H2AD—Histone Cluster 1 H2A Family Member D | 4 | 4 | −1.90 | −2.29 | −2.11 | −2.67 |
| TMEM176B—Transmembrane Protein 176B | 4 | 4 | −2.77 | −3.60 | −1.95 | −2.22 |
| GPX1—Glutathione Peroxidase 1 | 4 | 3 | −0.42 | −3.02 | −2.09 | −2.73 |
| PI3—Peptidase Inhibitor 3 | 4 | 3 | −2.53 | −2.80 | −1.02 | −2.70 |
| TMEM176A—Transmembrane Protein 176A | 3 | 2 | −3.49 | −1.44 | −2.30 | |
| HIST1H2BD—Histone Cluster 1 H2B Family Member D | 2 | 2 | −1.97 | −2.32 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danielewski, M.; Walkowiak, J.; Wielgus, K.; Nowak, J.K. Exceptions to Broad Tissue-Specific Transcriptomic Interdependence: Searching for Independence in Expression of Genes. Genes 2025, 16, 1067. https://doi.org/10.3390/genes16091067
Danielewski M, Walkowiak J, Wielgus K, Nowak JK. Exceptions to Broad Tissue-Specific Transcriptomic Interdependence: Searching for Independence in Expression of Genes. Genes. 2025; 16(9):1067. https://doi.org/10.3390/genes16091067
Chicago/Turabian StyleDanielewski, Mikołaj, Jarosław Walkowiak, Karolina Wielgus, and Jan Krzysztof Nowak. 2025. "Exceptions to Broad Tissue-Specific Transcriptomic Interdependence: Searching for Independence in Expression of Genes" Genes 16, no. 9: 1067. https://doi.org/10.3390/genes16091067
APA StyleDanielewski, M., Walkowiak, J., Wielgus, K., & Nowak, J. K. (2025). Exceptions to Broad Tissue-Specific Transcriptomic Interdependence: Searching for Independence in Expression of Genes. Genes, 16(9), 1067. https://doi.org/10.3390/genes16091067






