Mitochondrial Genome and RNA Editing Tissue Specificity of Centella asiatica
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Acquisition and Sequencing
2.2. Mitochondrial Genome Assembly and Annotation
2.3. Codon Usage Analysis and Repetitive Sequence Identification
2.4. Mitochondrial-to-Plastid Sequence (MTPTs) Transfer Analysis
2.5. Phylogenetic and Synteny Analysis
2.6. Prediction and Identification of RNA Editing Sites
3. Results
3.1. General Features of the C. asiatica Mitochondrial Genome
3.2. Codon Usage Bias in PCGs
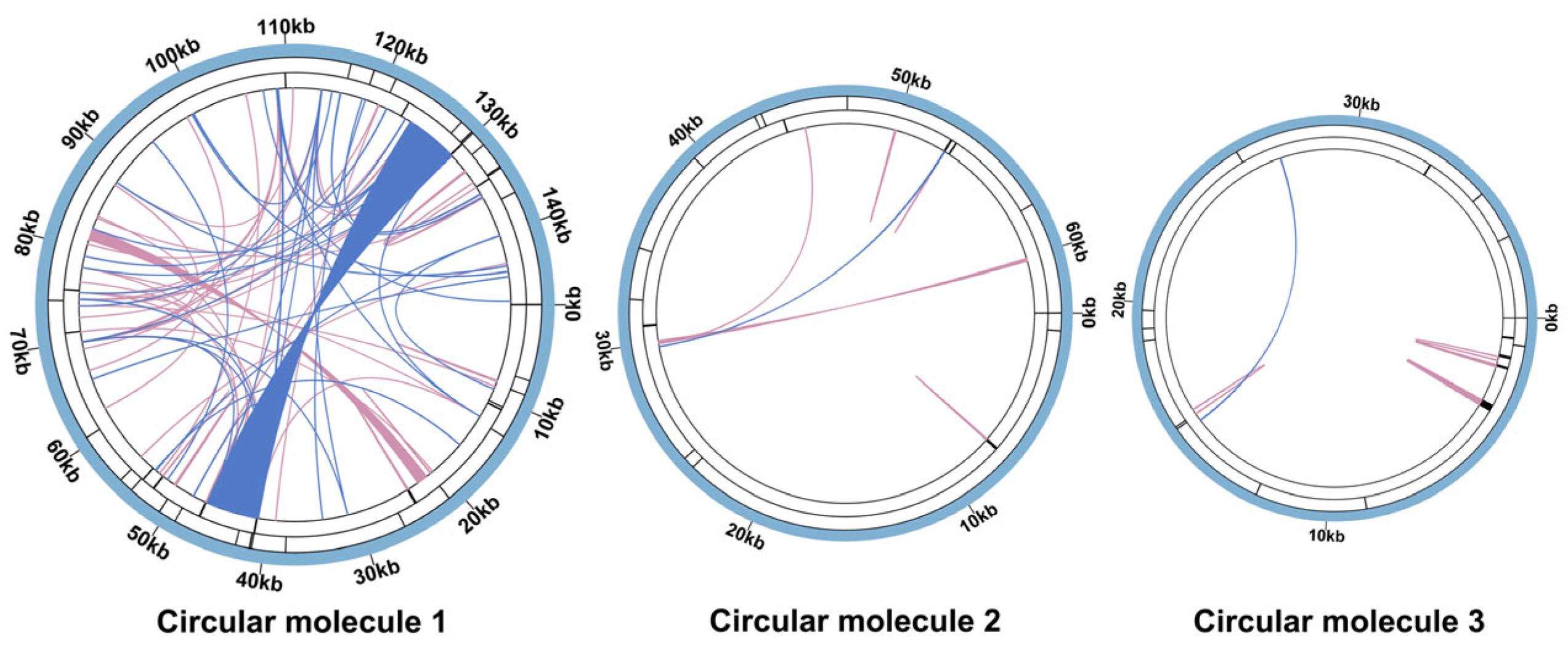
3.3. Repetitive Sequences
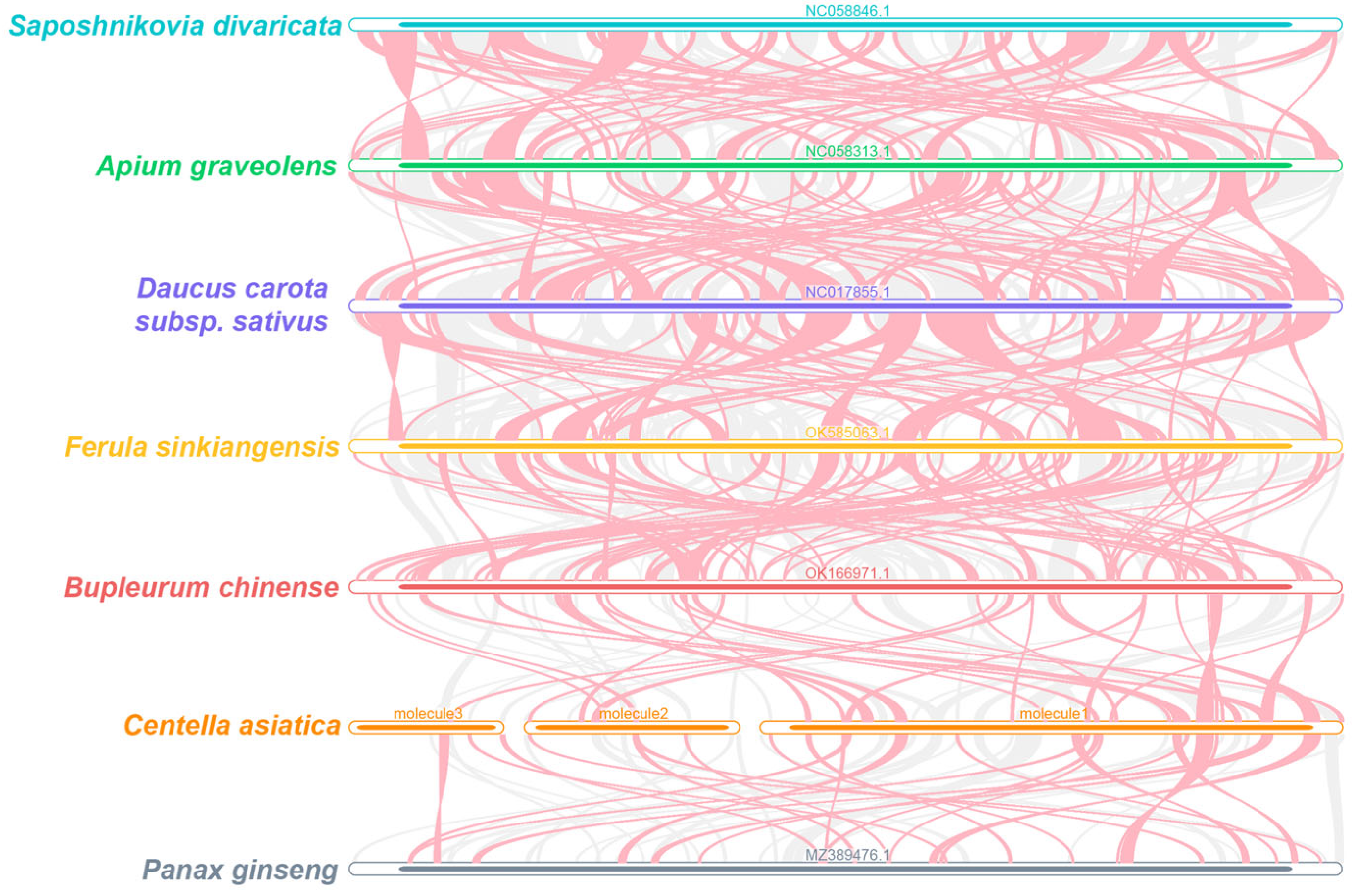
3.4. Sequence Transfer and Synteny Analysis
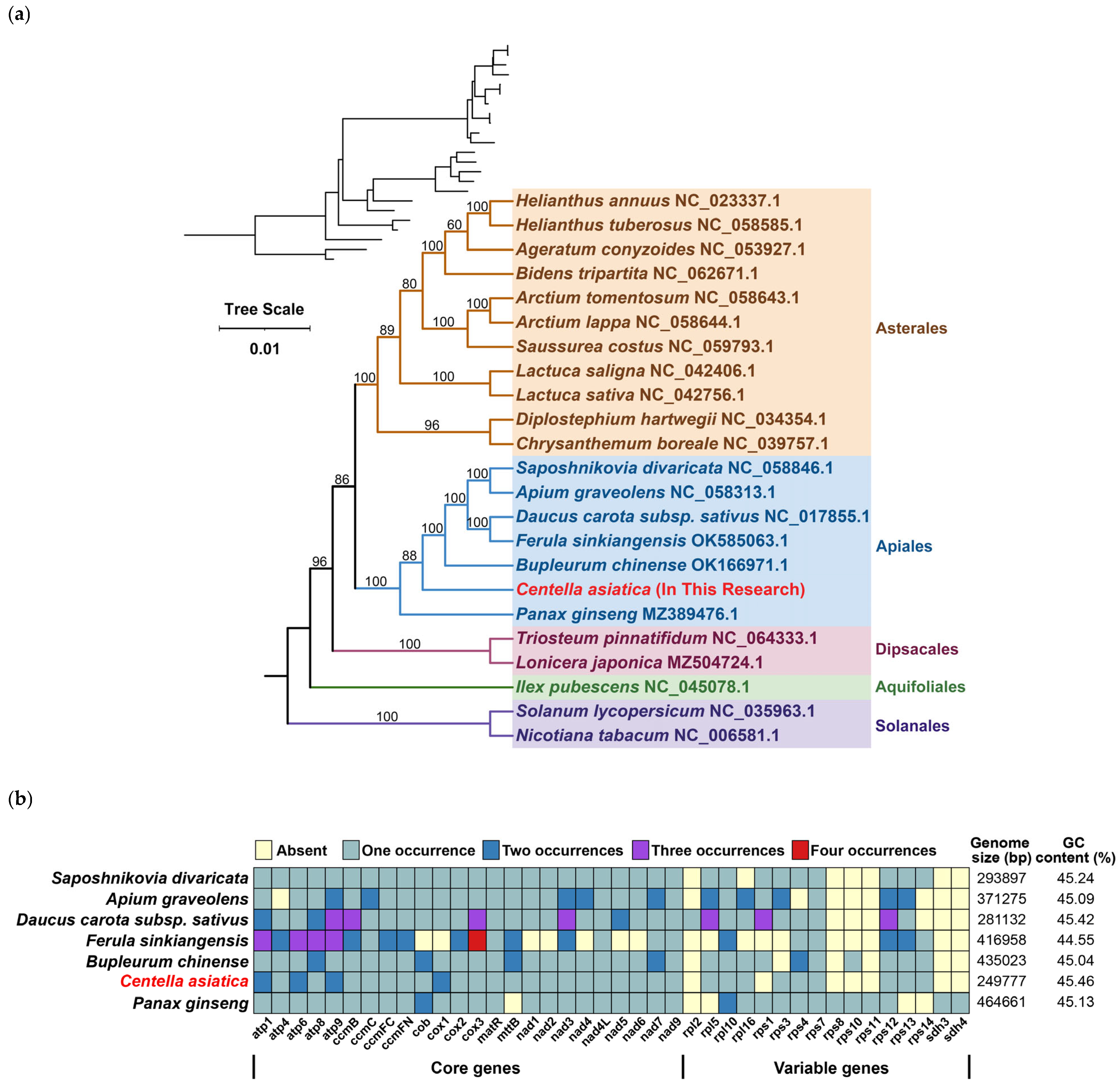
3.5. Phylogenetic Evolution
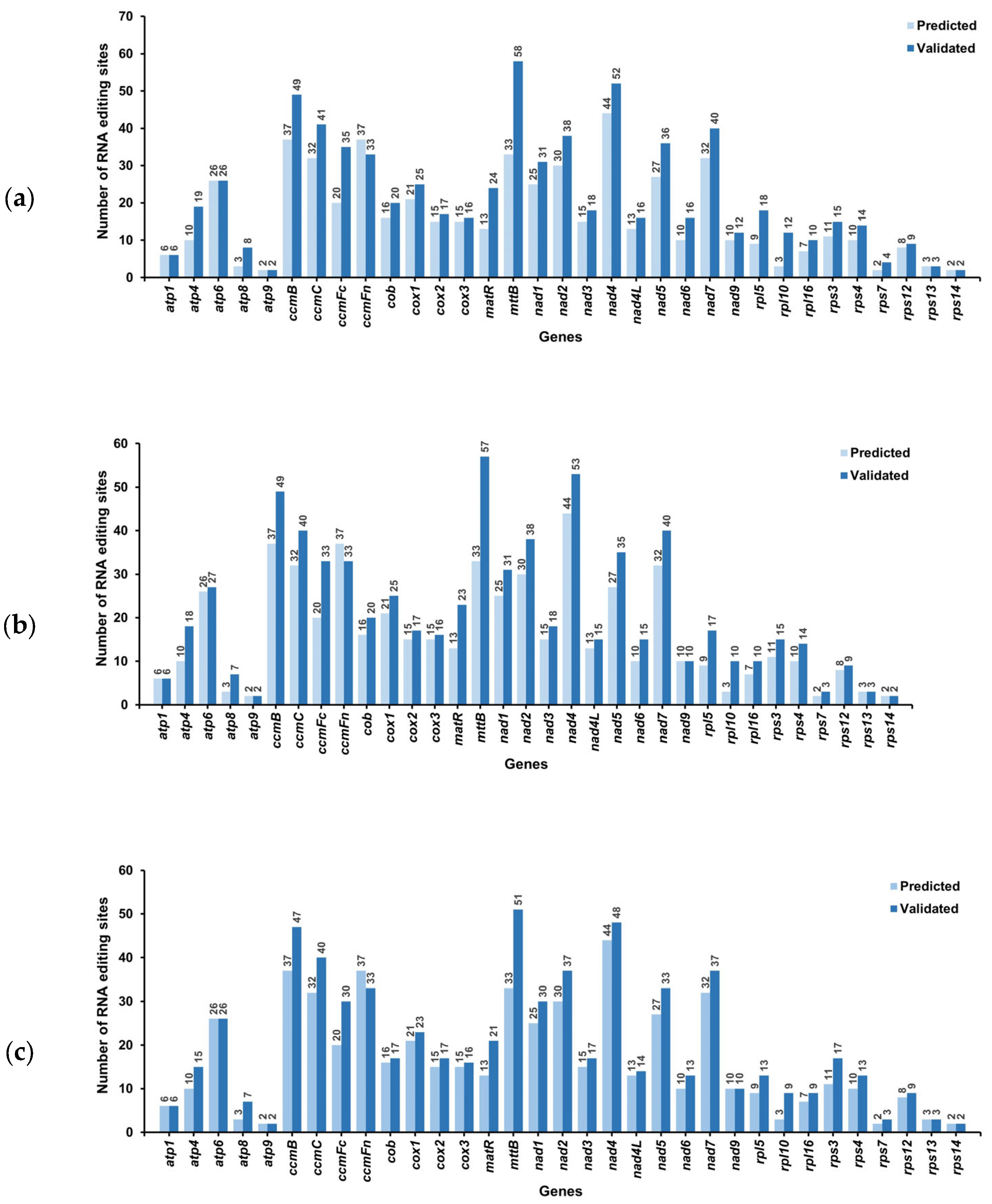
3.6. RNA Editing Analysis in C. asiatica
3.6.1. RNA Editing Prediction
3.6.2. High-Throughput Verification of RNA Editing Sites
4. Discussion
4.1. The Functional Association Between Genome Topology and Repetitive Sequences
4.2. MTPTs and Mitochondrial Functional Autonomy
4.3. Tissue-Specific Regulation of RNA Editing
4.3.1. Tissue-Biased Amino Acid Substitutions
4.3.2. Tissue-Specific Formation of PTC
4.3.3. Tissue-Specific Formation of Start Codons
4.4. Numerous Unpredicted Editing Sites Reveal the Uniqueness of the RNA Editing Machinery in C. asiatica
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, B.; Wu, L.; Wu, Y.; Zhang, C.; Qin, L.; Hayashi, M.; Kudo, M.; Gao, M.; Liu, T. Therapeutic Potential of Centella asiatica and Its Triterpenes: A Review. Front. Pharmacol. 2020, 11, 568032. [Google Scholar] [CrossRef]
- Li, C.; Xie, X.; Li, F.; Tian, E.; Shu, Y.; Chao, Z. The complete chloroplast genome sequence of Centella asiatica (Linnaeus) Urban. Mitochondrial DNA Part B 2020, 5, 2149–2150. [Google Scholar] [CrossRef] [PubMed]
- Kashmira, J.G.; Jagruti, A.P.; Anuradha, K.G. Pharmacological Review on Centella asiatica: A Potential Herbal Cure-all. Indian J. Pharm. Sci. 2010, 72, 546–556. [Google Scholar]
- Abedi, T.F.; Mahin, R.; Reza, D.; Sadegh, A.M.; Tafazoli, M.A.; Neda, S.; Sepideh, E.; Amirhossein, S.; Ahmad, E.S. Ethnobotany, Phytochemistry and Pharmacological Features of Centella asiatica: A Comprehensive Review. Adv. Exp. Med. Biol. 2021, 1308, 451–499. [Google Scholar]
- Yang, L.; Rafael, M.; Bernard, G. 350 my of mitochondrial genome stasis in mosses, an early land plant lineage. Mol. Biol. Evol. 2014, 31, 2586–2591. [Google Scholar] [CrossRef] [PubMed]
- Putintseva, Y.A.; Bondar, E.I.; Simonov, E.P.; Sharov, V.V.; Oreshkova, N.V.; Kuzmin, D.A.; Konstantinov, Y.M.; Shmakov, V.N.; Belkov, V.I.; Sadovsky, M.G.; et al. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genom. 2020, 21, 654. [Google Scholar] [CrossRef]
- David, R.S.; Patrick, J.K. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar]
- Yang, L.; Liu, J.; Guo, W.; Zheng, Z.; Xu, Y.; Xia, H.; Xiao, T. Insights into the multi-chromosomal mitochondrial genome structure of the xero-halophytic plant Haloxylon Ammodendron (C.A.Mey.) Bunge ex Fenzl. BMC Genom. 2024, 25, 123. [Google Scholar] [CrossRef]
- Guo, S.; Li, Z.; Li, C.; Liu, Y.; Liang, X.; Qin, Y. Assembly and characterization of the complete mitochondrial genome of Ventilago leiocarpa. Plant Cell Rep. 2024, 43, 77. [Google Scholar] [CrossRef]
- Yang, H.; Ni, Y.; Zhang, X.; Li, J.; Chen, H.; Liu, C. The mitochondrial genomes of Panax notoginseng reveal recombination mediated by repeats associated with DNA replication. Int. J. Biol. Macromol. 2023, 252, 126359. [Google Scholar] [CrossRef]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrok, R.L.; Daniell, H.; Jin, S.; Wu, Z. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zou, Y.; Mower, J.P.; Reeve, W.; Wu, Z. Rethinking the mutation hypotheses of plant organellar DNA. Genom. Commun. 2024, 1, e003. [Google Scholar] [CrossRef]
- Qu, Y.; Zhou, P.; Tong, C.; Bi, C.; Xu, L.A. Assembly and analysis of the Populus deltoides mitochondrial genome: The first report of a multicircular mitochondrial conformation for the genus Populus. J. For. Res. 2022, 34, 717–733. [Google Scholar] [CrossRef]
- Mareike, R.; Helena, T.F.; Stefan, A.R.; Uwe, G.M.; Volker, K. RNA editing: Only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol. Genet. Genom. MGG 2009, 281, 473–481. [Google Scholar]
- He, Z.S.; Zhu, A.; Yang, J.B.; Fan, W.; Li, D.Z. Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing. Int. J. Mol. Sci. 2021, 22, 9842. [Google Scholar] [CrossRef]
- Fang, J.; Jiang, X.; Wang, T.; Deng, Z.; Zhang, A.; Zhang, X. Dynamic landscape of mitochondrial Cytidine-to-Uridine RNA editing in tobacco (Nicotiana tabacum) shows its tissue specificity. Plant Cell Tissue Organ Cult. (PCTOC) 2022, 148, 363–376. [Google Scholar] [CrossRef]
- Li, C.; Liu, H.; Qin, M.; Tan, Y.J.; Ou, X.L.; Chen, X.Y.; Wei, Y.; Zhang, Z.J.; Lei, M. RNA editing events and expression profiles of mitochondrial protein-coding genes in the endemic and endangered medicinal plant, Corydalis saxicola. Front. Plant Sci. 2024, 15, 1332460. [Google Scholar] [CrossRef]
- Tafazoli, A.; Hemmati, M.; Rafigh, M.; Alimardani, M.; Khaghani, F.; Korostyński, M.; Karnes, J.H. Leveraging long-read sequencing technologies for pharmacogenomic testing: Applications, analytical strategies, challenges, and future perspectives. Front. Genet. 2025, 16, 1435416. [Google Scholar] [CrossRef]
- Gao, Y.; Takenaka, K.; Xu, S.M.; Cheng, Y.; Janitz, M. Recent advances in investigation of circRNA/lncRNA-miRNA-mRNA networks through RNA sequencing data analysis. Brief. Funct. Genom. 2025, 24, elaf005. [Google Scholar] [CrossRef]
- Tang, J.; Luo, Z.; Zhang, J.; Chen, L.; Li, L. Multi-Chromosomal Mitochondrial Genome of Medicinal Plant Acorus tatarinowii (Acoraceae): Firstly Reported from Acorales Order. Gene 2023, 892, 147847. [Google Scholar] [CrossRef]
- Fang, B.; Li, J.; Zhao, Q.; Liang, Y.; Yu, J. Assembly of the Complete Mitochondrial Genome of Chinese Plum (Prunus salicina): Characterization of Genome Recombination and RNA Editing Sites. Genes 2021, 12, 1970. [Google Scholar] [CrossRef]
- Wu, D.; Fu, W.; Fan, G.; Huang, D.; Wu, K.; Zhan, Y.; Tu, X.; He, J. Characteristics and Comparative Analysis of the Special-Structure (Non-Single-Circle) Mitochondrial Genome of Capsicum pubescens Ruiz & Pav. Genes 2024, 15, 152. [Google Scholar] [PubMed]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; Depamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Heng, L.; Richard, D. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar]
- Michael, T.; Pascal, L.; Tommaso, P.; Ulbricht, J.E.S.; Axel, F.; Ralph, B.; Stephan, G. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.J.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, research0082.1. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [PubMed]
- Sebastian, B.; Thomas, T.; Thomas, M.; Uwe, S.; Martin, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ivica, L.; Peer, B. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Laiho, A.; Venäläinen, M.S.; McGlinchey, A.J.; Wang, N.; Elo, L.L. Systematic evaluation of differential splicing tools for RNA-seq studies. Brief. Bioinform. 2019, 21, 2052–2065. [Google Scholar] [CrossRef] [PubMed]
- Flati, T.; Gioiosa, S.; Spallanzani, N.; Tagliaferri, I.; Diroma, M.A.; Pesole, G.; Chillemi, G.; Picardi, E.; Castrignanò, T. HPC-REDItools: A novel HPC-aware tool for improved large scale RNA-editing analysis. BMC Bioinform. 2020, 21, 353. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, Q. Complete mitochondrial genome of Syzygium samarangense reveals genomic recombination, gene transfer, and RNA editing events. Front. Plant Sci. 2024, 14, 1301164. [Google Scholar] [CrossRef]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef]
- Gui, L.; Zhang, Z.; Song, L.; Feng, C.; Yu, H.; Pan, L.; Fu, J.; Liang, W.; Huang, Q.; Sappah, A.H.E.; et al. Mitogenome of Uncaria rhynchophylla: Genome structure, characterization, and phylogenetic relationships. BMC Genom. 2025, 26, 199. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber, L.F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, J.Y.; Li, D.Z. Remarkable mitochondrial genome heterogeneity in Meniocus linifolius (Brassicaceae). Plant Cell Rep. 2024, 43, 36. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Liang, W.; Yu, H.; Zhang, K.; Li, L.; Feng, S.; Song, L.; Yang, C.; Wan, L.; Zeng, D.; et al. Assembly and Comparative Analysis of the Complete Mitochondrial Genomes of Smilax glabra and Smilax zeylanica. Genes 2025, 16, 450. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, Z.; Hao, Y.; Li, M.; Yu, H.; Zhang, X.; Mi, H.; Cheng, L.; Zhao, Y. Decoding the complete organelle genomic architecture of Stewartia gemmata: An early-diverging species in Theaceae. BMC Genom. 2024, 25, 114. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.W.; Guo, W.; Mower, J.P.; Palmer, J.D.; Purugganan, M. High and variable rates of repeat-mediated mitochondrial genome rearrangement in a genus of plants. Mol. Biol. Evol. 2018, 35, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Xu, Y.; Zhang, Z.; Wei, Y.; Hu, Y.; Zheng, C.; Qu, X. Assembly and comparative analysis of the first complete mitochondrial genome of a traditional Chinese medicine Angelica biserrata (Shan et Yuan) Yuan et Shan. Int. J. Biol. Macromol. 2023, 257, 128571. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhang, X.; Li, Z.; Song, Y.; Sun, Z. Assembly and comparative analysis of the complete mitochondrial genome of Bupleurum chinense DC. BMC Genom. 2022, 23, 664. [Google Scholar] [CrossRef]
- Sloan, D.B.; Wu, Z. History of plastid DNA insertions reveals weak deletion and at mutation biases in angiosperm mitochondrial genomes. Genome Biol. Evol. 2014, 6, 3210–3221. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Härtel, B.; Brennicke, A. RNA Editing in Plants and Its Evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef]
- Gallagher, L.J.; Betz, S.K.; Chase, C.D. Mitochondrial RNA editing truncates a chimeric open reading frame associated with S male-sterility in maize. Curr. Genet. 2002, 42, 179–184. [Google Scholar] [CrossRef]
- Sota, F.; Ian, S. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011, 191, 37–47. [Google Scholar] [CrossRef] [PubMed]
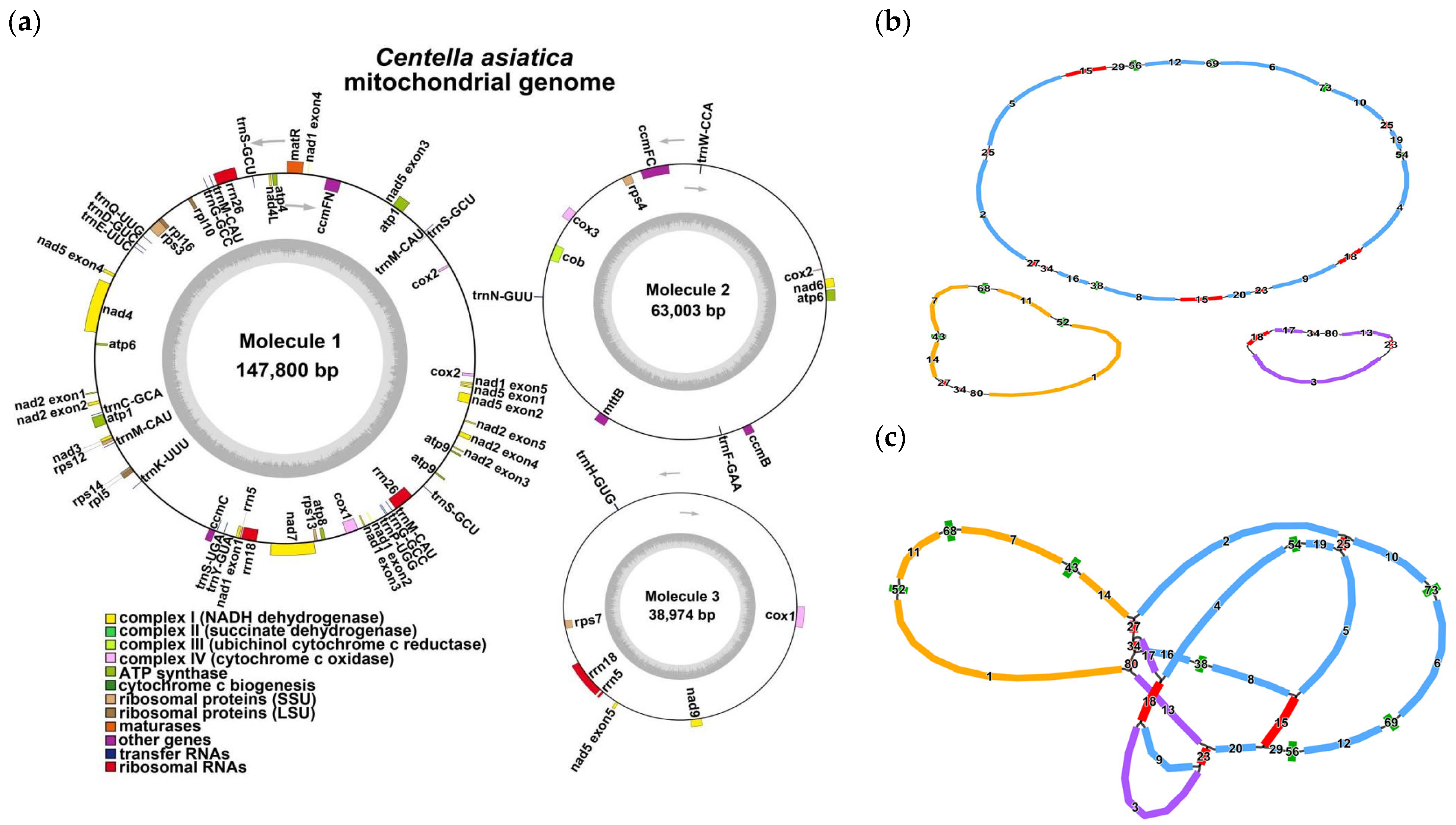


| NCBI Accession Number | Contigs | Type | Length | GC Content |
|---|---|---|---|---|
| PV739434-PV739436 | Molecule 1–3 | Branched | 249,777 bp | 45.46% |
| PV739434 | Molecule 1 | circular | 147,800 bp | 46.15% |
| PV739434 | Molecule 2 | circular | 63,003 bp | 44.38% |
| PV739434 | Molecule 3 | circular | 38,974 bp | 44.65% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Liang, W.; Qin, Y.; Li, Y.; Wei, S.; Huang, Q.; El-Sappah, A.H.; Tan, G.; Wei, Y.; Gui, L.; et al. Mitochondrial Genome and RNA Editing Tissue Specificity of Centella asiatica. Genes 2025, 16, 953. https://doi.org/10.3390/genes16080953
Yang C, Liang W, Qin Y, Li Y, Wei S, Huang Q, El-Sappah AH, Tan G, Wei Y, Gui L, et al. Mitochondrial Genome and RNA Editing Tissue Specificity of Centella asiatica. Genes. 2025; 16(8):953. https://doi.org/10.3390/genes16080953
Chicago/Turabian StyleYang, Cuihong, Wenjing Liang, Ya Qin, Yuqiong Li, Shugen Wei, Qiulan Huang, Ahmed H. El-Sappah, Guiyu Tan, Ying Wei, Lingjian Gui, and et al. 2025. "Mitochondrial Genome and RNA Editing Tissue Specificity of Centella asiatica" Genes 16, no. 8: 953. https://doi.org/10.3390/genes16080953
APA StyleYang, C., Liang, W., Qin, Y., Li, Y., Wei, S., Huang, Q., El-Sappah, A. H., Tan, G., Wei, Y., Gui, L., & Wan, L. (2025). Mitochondrial Genome and RNA Editing Tissue Specificity of Centella asiatica. Genes, 16(8), 953. https://doi.org/10.3390/genes16080953







