DNA Transfer Between Items Within an Evidence Package
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Mock Casework Items
2.2. Experimental Design
2.3. DNA Processing
2.4. Profile Analyses
2.5. Statistical Analyses
3. Results
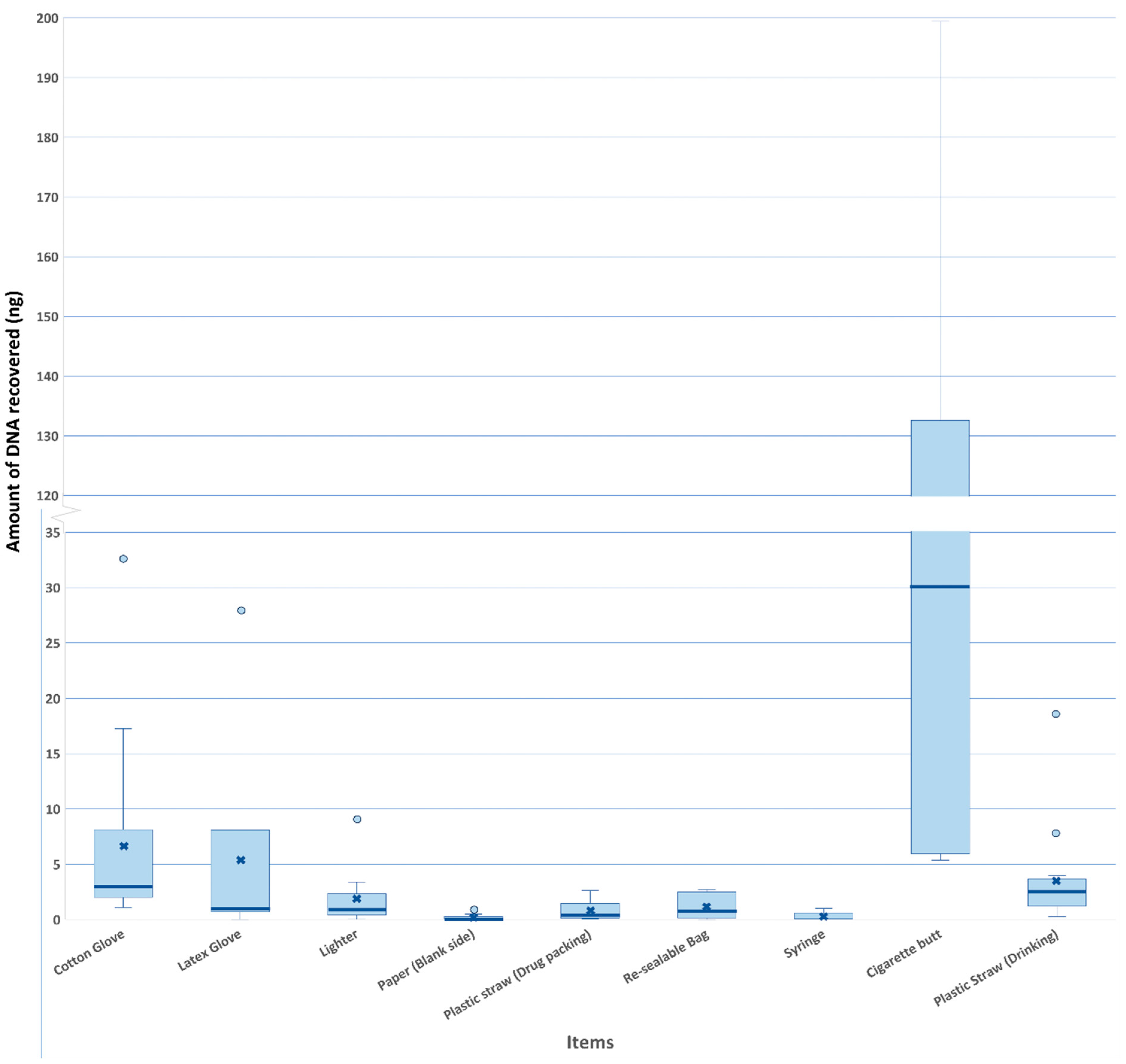
3.1. DNA Recovery from Different Types of Items and Biological Materials
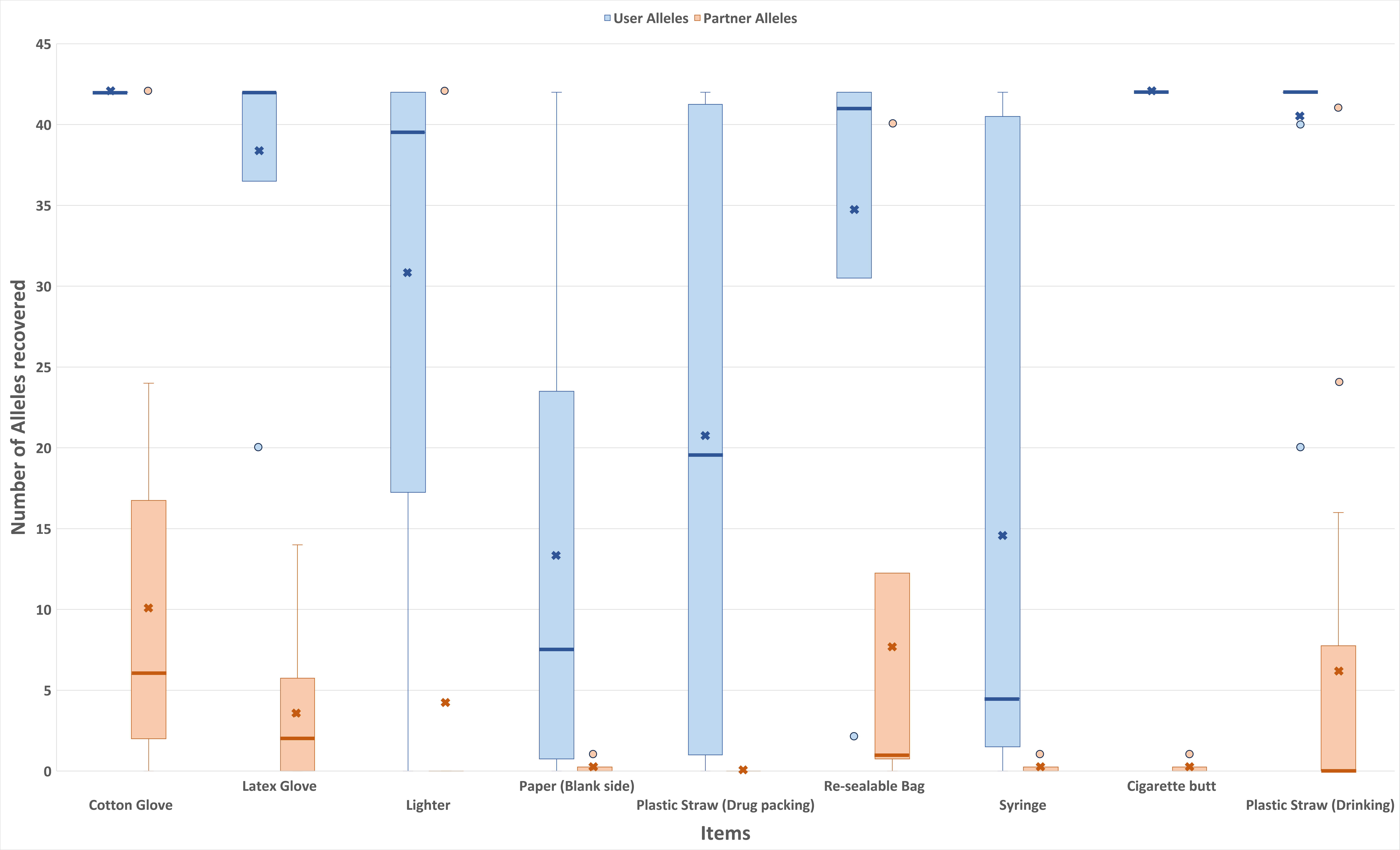
3.2. Recovery of Alleles That Could Be Attributed to the “User” and the “Partner”
3.3. Interpretation of DNA Profiles
4. Discussion
4.1. The Amount of DNA Does Not Correlate with the Number of Transferred Alleles
4.2. DNA Transfer Between Different Types of Items
4.3. DNA Transfer Within the Same Package Could Be a Potential Source of Contamination
4.4. DNA Transfer May Further Interfere with Downstream DNA Profile Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wiegand, P.; Kleiber, M. DNA typing of epithelial cells after strangulation. Int. J. Legal Med. 1997, 110, 181–183. [Google Scholar] [CrossRef]
- Roberts, D.; Marks, R. The determination of regional and age variations in the rate of desquamation: A comparison of four techniques. J. Investig. Dermatol. 1980, 74, 13–16. [Google Scholar] [CrossRef]
- van Oorschot, R.A.; Jones, M.K. DNA fingerprints from fingerprints. Nature 1997, 387, 767. [Google Scholar] [CrossRef]
- Bragulla, H.H.; Homberger, D.G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009, 214, 516–559. [Google Scholar] [CrossRef]
- Meakin, G.; Jamieson, A. DNA transfer: Review and implications for casework. Forensic Sci. Int. Genet. 2013, 7, 434–443. [Google Scholar] [CrossRef]
- Goray, M.; van Oorschot, R.A.; Mitchell, R.J. DNA transfer within forensic exhibit packaging: Potential for DNA loss and relocation. Forensic Sci. Int. Genet. 2012, 6, 158–166. [Google Scholar] [CrossRef]
- Stella, C.J.; Meakin, G.E.; van Oorschot, R.A.H. DNA transfer in packaging: Attention required. Forensic Sci. Int. Genet. Suppl. Ser. 2022, 8, 303–305. [Google Scholar] [CrossRef]
- Promega Corporation. DNA IQTM Casework Pro Kit for Maxwell® 16 Technical Manual Part# TM332, Revised 11/11.
- Promega Corporation. Maxwell® FSC Instrument Operating Manual TM462 Revised 8/17.
- Applied Biosystems. QuantStudio™ 6/7 Flex Real Time PCR System, Getting Started Guide, Rev A, 2013.
- Applied Biosystems. Quantifiler™ HP and Trio DNA Quantification Kits, User Guide, 2017.
- Applied Biosystems. GlobalFiler™ PCR Amplification Kit, User Guide, July 2016.
- Szkuta, B.; Ballantyne, K.N.; van Oorschot, R.A.H. Transfer and persistence of DNA on the hands and the influence of activities performed. Forensic Sci. Int. Genet. 2017, 28, 10–20. [Google Scholar] [CrossRef]
- Thornbury, D.; Goray, M.; van Oorschot, R.A.H. Indirect DNA transfer without contact from dried biological materials on various surfaces. Forensic Sci. Int. Genet. 2021, 51, 102457. [Google Scholar] [CrossRef]
- Goray, M.; Mitchell, R.J.; van Oorschot, R.A. Investigation of secondary DNA transfer of skin cells under controlled test conditions. Leg. Med. 2010, 12, 117–120. [Google Scholar] [CrossRef]
- Gettings, K.B.; Kiesler, K.M.; Faith, S.A.; Montano, E.; Baker, C.H.; Young, B.A.; Guerrieri, R.A.; Vallone, P.M. Sequence variation of 22 autosomal STR loci detected by next generation sequencing. Forensic Sci. Int. Genet. 2016, 21, 15–21. [Google Scholar] [CrossRef]
- Neste, C.V.; Nieuwerburgh, F.V.; Hoofstat, D.V.; Deforce, D. Forensic STR analysis using massive parallel sequencing. Forensic Sci. Int. Genet. 2012, 6, 810–818. [Google Scholar] [CrossRef]
- Goray, M.; Eken, E.; Mitchell, R.J.; van Oorschot, R.A. Secondary DNA transfer of biological substances under varying test conditions. Forensic Sci. Int. Genet. 2010, 4, 62–67. [Google Scholar] [CrossRef]
- van Oorschot, R.A.H.; Meakin, G.E.; Kokshoorn, B.; Goray, M.; Szkuta, B. DNA Transfer in Forensic Science: Recent Progress towards Meeting Challenges. Genes 2024, 12, 1766. [Google Scholar] [CrossRef]
- Cahill, A.; Volgin, L.; van Oorschot, R.A.H.; Taylor, D.; Goray, M. Where did it go? A study of DNA transfer in a social setting. Forensic Sci. Int. Genet. 2024, 73, 103101. [Google Scholar] [CrossRef]
- Taylor, D.; Volgin, L.; Kokshoorn, B. Accounting for site-to-site DNA transfer on a packaged exhibit in an evaluation given activity level propositions. Forensic Sci. Int. Genet. 2024, 73, 103122. [Google Scholar] [CrossRef]
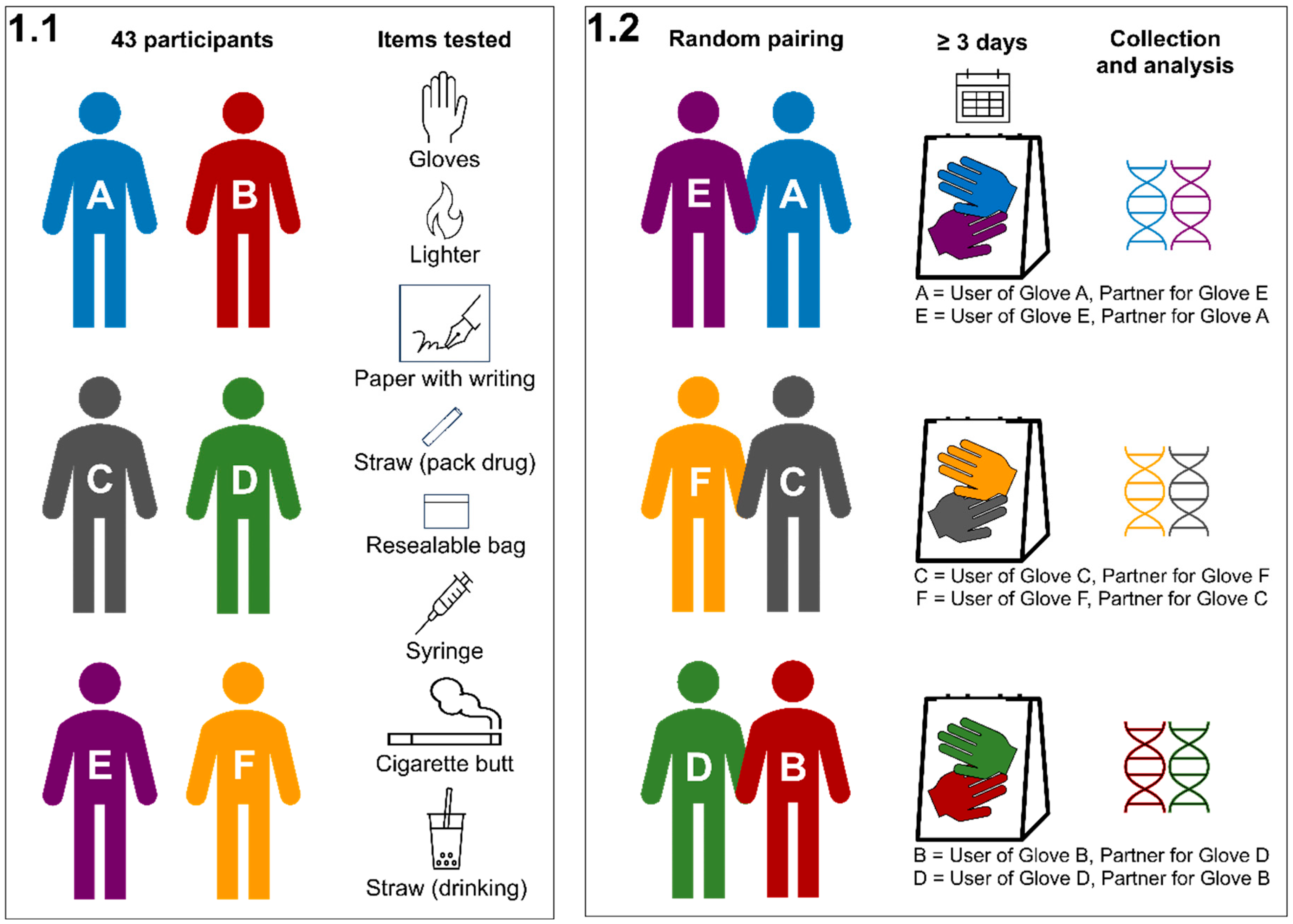
| Item | N | Way of Use | Note |
|---|---|---|---|
| Group A | |||
| Cotton glove | 16 | Worn for whole day for ≥2 days | Interior of glove facing out when placed in envelope |
| Latex glove | 6 | Worn for ≥1 h | Interior of glove facing out when placed in envelope |
| Lighter | 10 | Held for 1 min and rolled the wheel at least 3 times every hour for 6 h | |
| Paper | 10 | Wrote 3 sentences on it (landscape) and folded it twice, with each fold bringing shorter ends to each other, unfolded, and pressed down on the table (blank side in contact with the table) to mimic a debt collector pressing the paper onto the door of the debtor’s house before fixing it with adhesive tape. | Folded form when placed in envelope |
| Plastic straw for drug packing | 10 | Cut and heat sealed | |
| Re-sealable bag | 6 | Touched ≥ 5 times within a day | |
| Syringe | 10 | Opened cap and plunged 3 times, then capped back | |
| Group B | |||
| Cigarette butt | 10 | Held as when smoking | Provided by participants after smoking |
| Plastic straw for drinking | 16 | Used to drink beverage | |
| Total | 94 |
| Group | Item Type | Count (% of Total Number of Each Item Type) |
|---|---|---|
| A | Cotton glove (n = 16) | 14 (88%) |
| Latex glove (n = 6) | 4 (67%) | |
| Lighter (n = 10) | 1 (10%) | |
| Paper (n = 10) | 2 (20%) | |
| Plastic straw for drug packing (n = 10) | 0 (0%) | |
| Resealable bag (n = 6) | 5 (83%) | |
| Syringe (n = 10) | 2 (20%) | |
| B | Cigarette butt (n = 10) | 2 (20%) |
| Plastic straw for drinking (n = 16) | 7 (44%) |
| Category of Comparison | Item (A) | Item with Significant Difference with Item (A) |
|---|---|---|
| Amount of DNA | Cotton glove | Paper |
| Plastic straw for drug packing | ||
| Syringe | ||
| Cigarette butt | Lighter | |
| Paper | ||
| Plastic straw for drug packing | ||
| Resealable bag | ||
| Syringe | ||
| Plastic straw for drinking | Paper | |
| Syringe | ||
| Number of alleles attributed to “user” | Cotton glove | Paper |
| Plastic straw for drug packing | ||
| Syringe | ||
| Cigarette butt | Paper | |
| Plastic straw for drug packing | ||
| Syringe | ||
| Plastic straw for drinking | Paper | |
| Plastic straw for drug packing | ||
| Syringe | ||
| Number of alleles attributed to “partner” | Cotton glove | Lighter |
| Paper | ||
| Plastic straw for drug packing | ||
| Syringe | ||
| Cigarette butt |
| Interpretation | Count of Sample |
|---|---|
| Reportable | 72 (76.6%) |
| • Attributed to the “user” | 62 |
| • Attributed to the “partner” | 1 |
| • Mix of the “user” and the “partner” | 8 |
| • Mix of the “user” and foreign person | 1 |
| Not reportable | 21 (22.3%) |
| Uninterpretable | 1 (1.1%) |
| Item Type | Attributed to the “User” | Attributed to the “User” and the “Partner” | Attributed to the “Partner” | Attributed to the “User” and Foreign Person |
|---|---|---|---|---|
| Cotton glove (n = 16) | 11 (69%) | 4 (25%) | ||
| Latex glove (n = 6) | 6 (100%) | |||
| Lighter (n = 10) | 8 (80%) | 1 (10%) | ||
| Paper (n = 10) | 3 (30%) | |||
| Plastic straw for drug packing (n = 10) | 4 (40%) | 1 (10%) | ||
| Resealable bag (n = 6) | 4 (67%) | 1 (17%) | ||
| Syringe (n = 10) | 3 (30%) | |||
| Cigarette butt (n = 10) | 10 (100%) | |||
| Plastic straw for drinking (n = 16) | 13 (81%) | 3 (19%) |
| Number of Items | Expected Profile If No DNA Transfer | Observed Profile |
|---|---|---|
| 1 | Not reportable single-source profile of the “user” but less than 16 alleles | “partner” being the major contributor |
| 3 | Essentially a single-source profile of the “user” | “user” and “partner” being approximately equal co-contributors |
| 5 | Essentially a single-source profile of the “user” | “user” being the major contributor and “partner” being the minor contributor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.S.; Syn, C.K.-C. DNA Transfer Between Items Within an Evidence Package. Genes 2025, 16, 894. https://doi.org/10.3390/genes16080894
Lee YS, Syn CK-C. DNA Transfer Between Items Within an Evidence Package. Genes. 2025; 16(8):894. https://doi.org/10.3390/genes16080894
Chicago/Turabian StyleLee, Yong Sheng, and Christopher Kiu-Choong Syn. 2025. "DNA Transfer Between Items Within an Evidence Package" Genes 16, no. 8: 894. https://doi.org/10.3390/genes16080894
APA StyleLee, Y. S., & Syn, C. K.-C. (2025). DNA Transfer Between Items Within an Evidence Package. Genes, 16(8), 894. https://doi.org/10.3390/genes16080894





