Genome-Wide Characterization of the ABI3 Gene Family in Cotton
Abstract
1. Introduction
2. Materials and Methods
2.1. Systematic Identification and Characterization of the ABI3s Gene Family
2.2. Multiple Sequence Alignment Coupled with Molecular Phylogeny Inference
2.3. Chromosomal Localization
2.4. Gene Family Collinearity Analysis
2.5. Gene Structure, Conserved Structural Domains, and Phylogenetic Tree
2.6. Gh_ABI3s Cis-Regulatory Element Analysis and Visualization
2.7. Expression Analysis of Gh_ABI3 in Different Tissues
3. Results
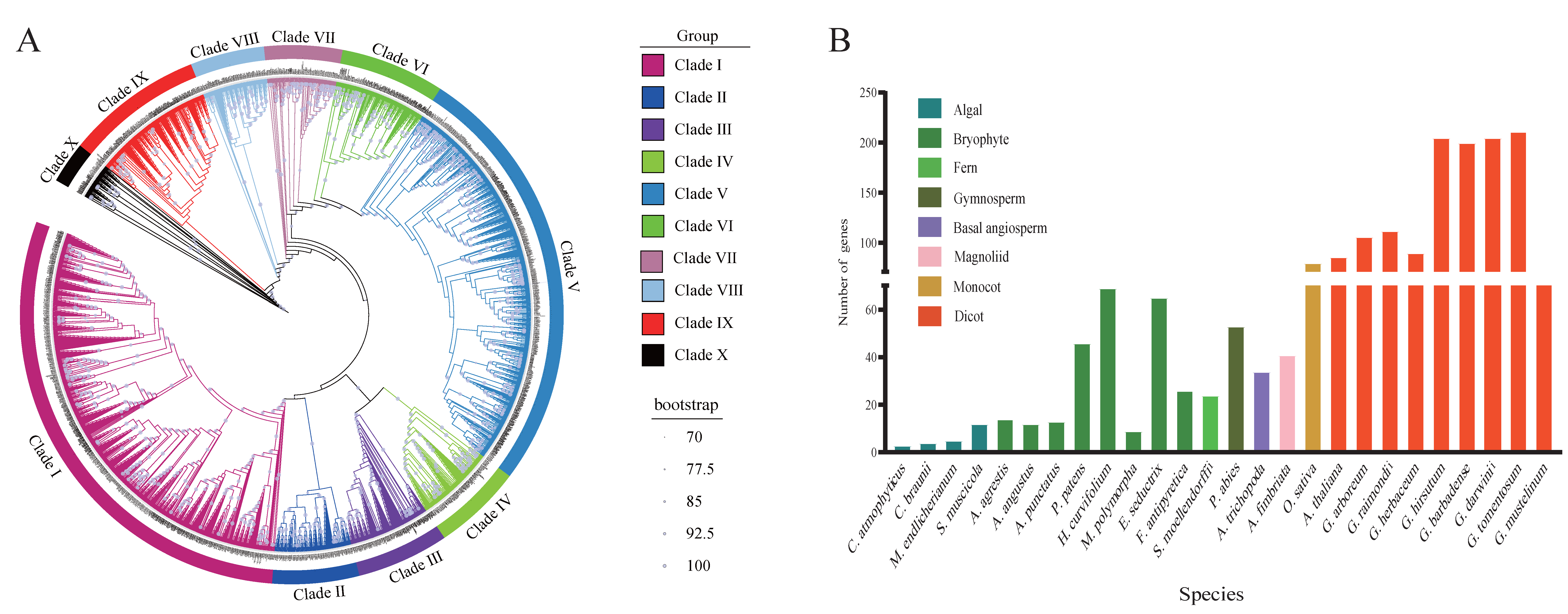
3.1. Comparative Phylogenomics of the ABI3 Gene Family: Identification and Evolutionary Analysis
3.2. ABI3 Chromosome Localization in Eight Cotton Species and Their Physico-Chemical Characteristics
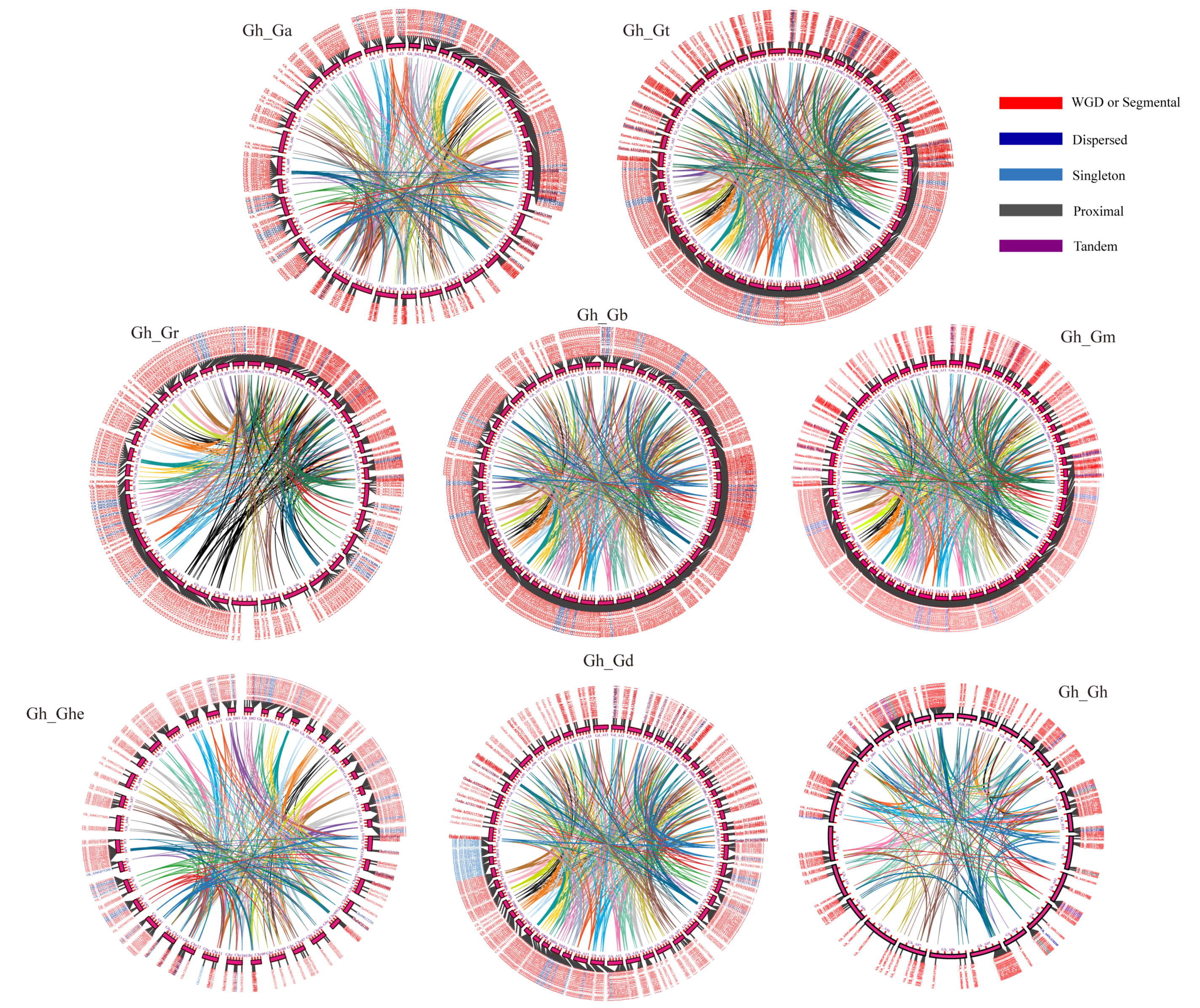
3.3. Evolutionary Analysis of the ABI3 Gene Family Synteny in Cotton
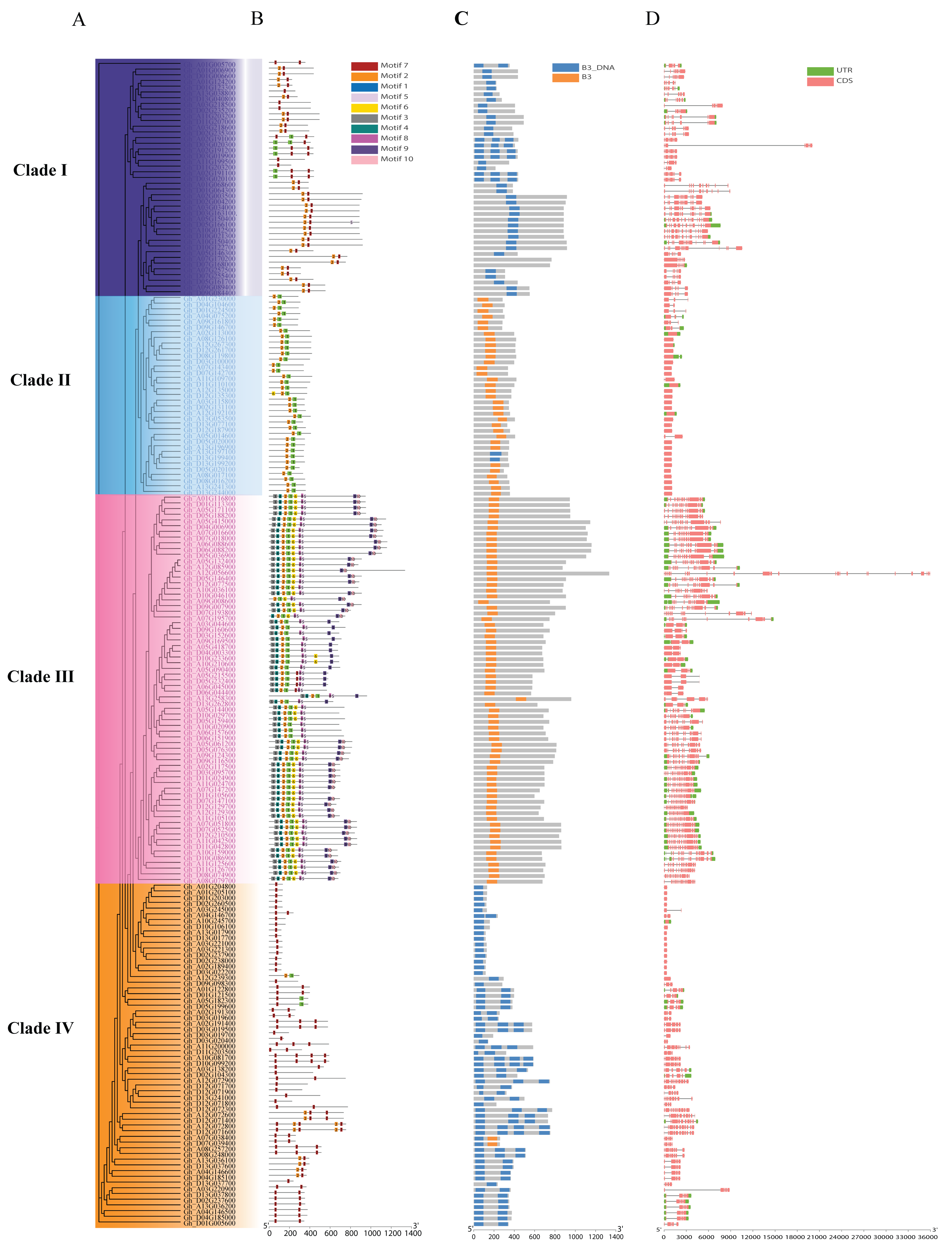
3.4. Comparative Analysis of ABI3 Gene Family Evolution and Structural Features in Upland Cotton
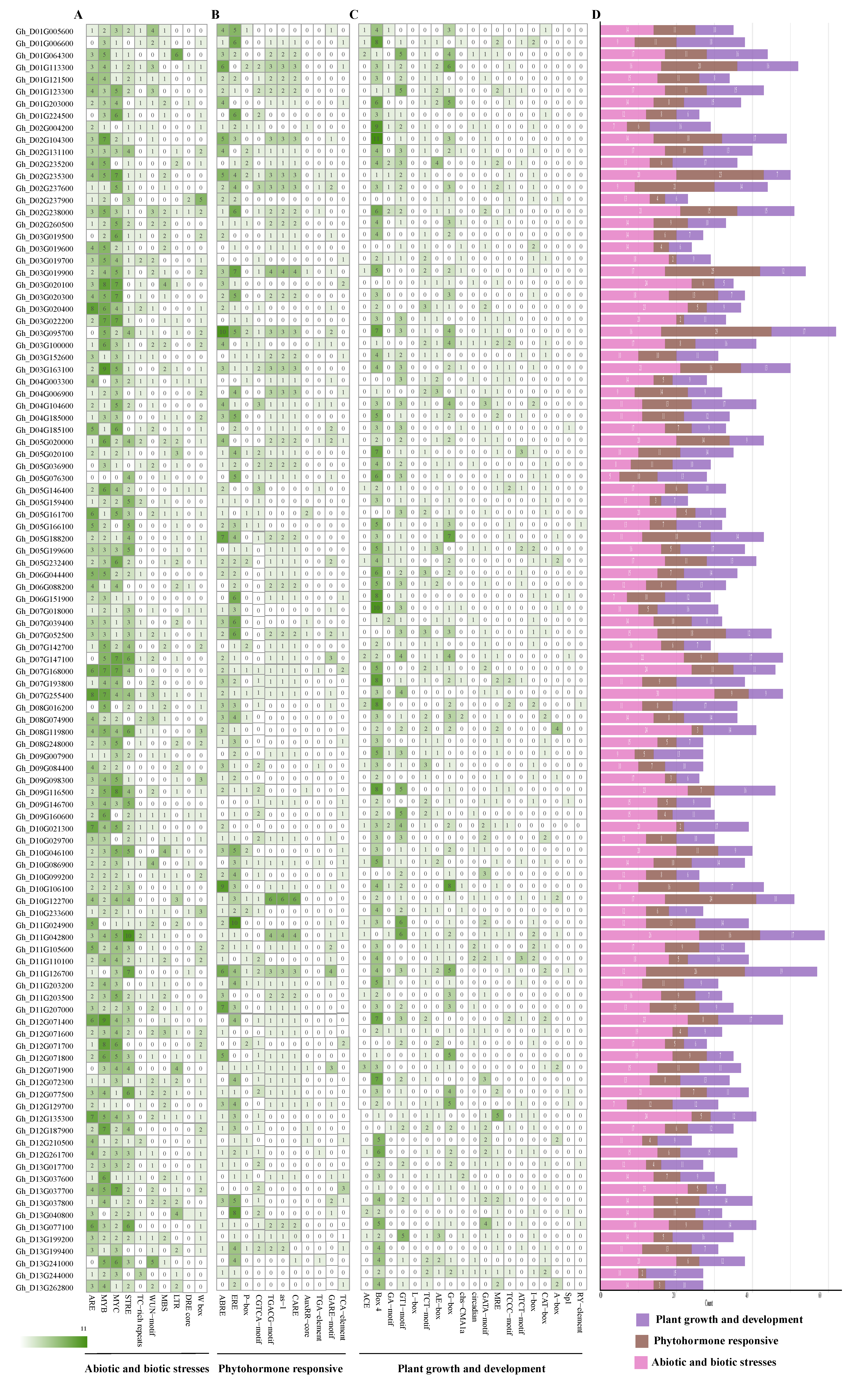
3.5. Promoter Cis-Element Profiling of GhABI3 Genes
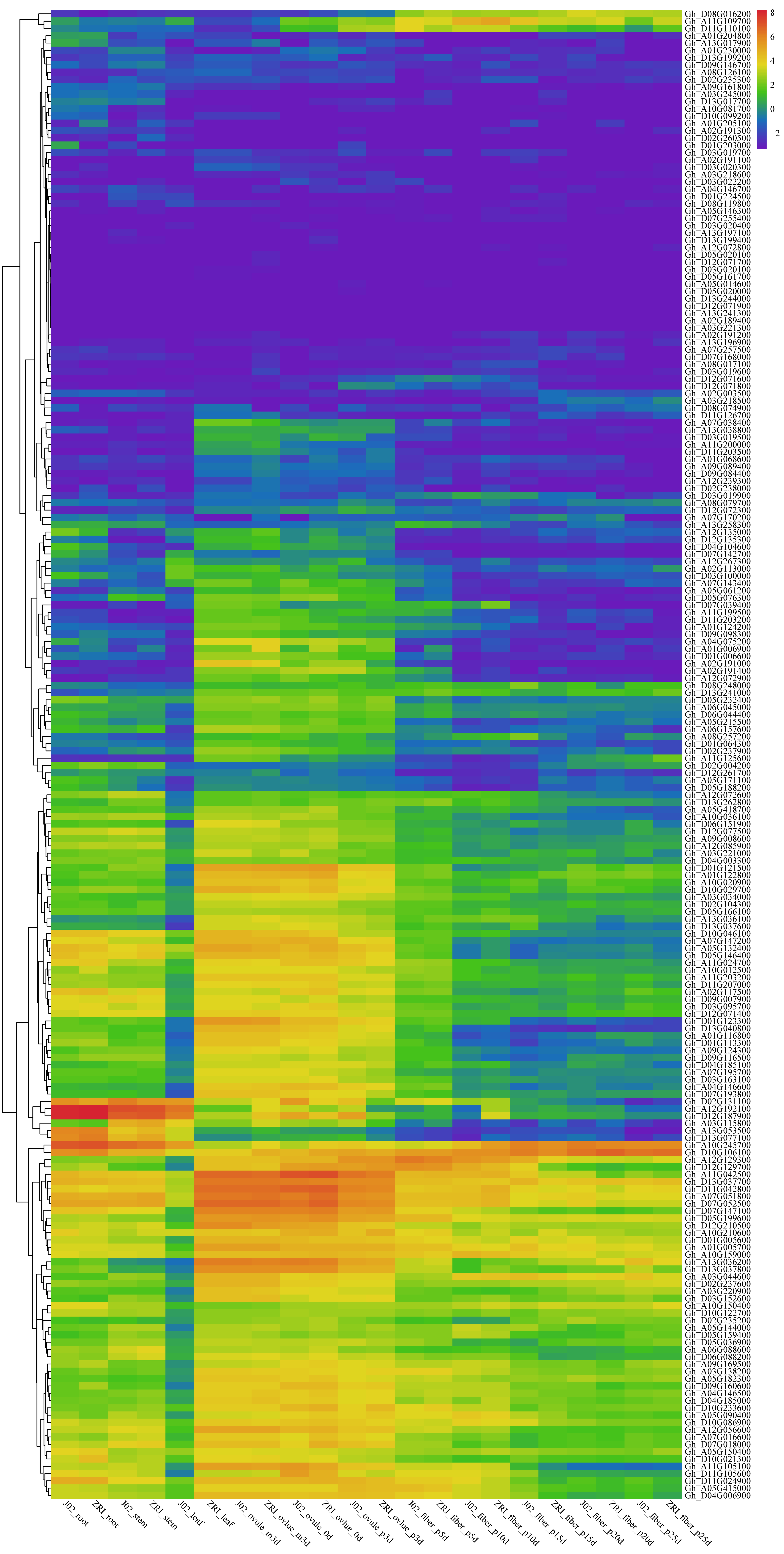
3.6. Tissue-Specific Expression Profiling of GhABI3 Genes in Cotton Fiber Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.; Garreton, V.; Chua, N.H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes. Dev. 2005, 19, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Tao, Z.; He, Y. Embryonic reactivation of FLOWERING LOCUS C by ABSCISIC ACID-INSENSITIVE 3 establishes the vernalization requirement in each Arabidopsis generation. Plant Cell 2022, 34, 2205–2221. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Wang, F.; Zheng, Q.; Niza, V.; Downie, A.B.; Perry, S.E. Direct and indirect targets of the arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. 2020, 103, 1679–1694. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Wen, X.; Zhang, W.; Huang, L.; Peng, Y.; Jin, L.; Han, H.; Zhang, L.; Li, W.; Guo, H. Arabidopsis EIN2 represses ABA responses during germination and early seedling growth by inactivating HLS1 protein independently of the canonical ethylene pathway. Plant J. 2023, 115, 1514–1527. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Sun, W. Arabidopsis seed-specific vacuolar aquaporins are involved in maintaining seed longevity under the control of ABSCISIC ACID INSENSITIVE 3. J. Exp. Bot. 2015, 66, 4781–4794. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Devic, M.; Lepiniec, L.; Zhou, D.X. Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol. J. 2015, 13, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Datta, S.; Mitra, S.; Nag Chaudhuri, R. ABSCISIC ACID INSENSITIVE 3 promotes auxin signalling by regulating SHY2 expression to control primary root growth in response to dehydration stress. J. Exp. Bot. 2024, 75, 5111–5129. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, C.; Ji, Z.; Lu, J.; Zhang, L.; Li, C.; Huang, J.; Yang, G.; Yan, K.; Zhang, S.; et al. Regulation of drought tolerance in Arabidopsis involves the PLATZ4-mediated transcriptional repression of plasma membrane aquaporin PIP2;8. Plant J. 2023, 115, 434–451. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xie, Y.; Gao, Q.; Pan, Y.; Tang, X.; Liu, Y.; Li, W.; Guo, H. Distinct regulation of mRNA decay pathways by ABA enhances Nitrate Reductase 1/2-derived siRNAs production and stress adaptation. Mol. Plant 2025, 18, 853–871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Wang, C.; Xiao, J.; Huang, M.; Zhuo, L.; Zhang, D. Enhancement of salt tolerance of alfalfa: Physiological and molecular responses of transgenic alfalfa plants expressing Syntrichia caninervis-derived ScABI3. Plant Physiol. Biochem. 2024, 207, 108335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ekwealor, J.T.B.; Mishler, B.D.; Silva, A.T.; Yu, L.; Jones, A.K.; Nelson, A.D.L.; Oliver, M.J. Syntrichia ruralis: Emerging model moss genome reveals a conserved and previously unknown regulator of desiccation in flowering plants. New Phytol. 2024, 243, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, J.; Zhang, Z.; Wu, W.; Lin, X.; Gao, M.; Yang, Y.; Zhao, P.; Xu, S.; Yang, C.; et al. Deciphering the Transcriptional Regulatory Network Governing Starch and Storage Protein Biosynthesis in Wheat for Breeding Improvement. Adv. Sci. 2024, 11, e2401383. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.J.; Piati, G.L.; Leite, R.C.; Zanella, M.S.; Osorio, C.R.; Lima, S.F. Nitrogen and mepiquat chloride can affect fiber quality and cotton yield. Rev. Bras. Eng. Agríc. Ambient. 2020, 24, 238–243. [Google Scholar] [CrossRef]
- Kim, H.J.; Triplett, B.A. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001, 127, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Zhao, P.; Zhang, Y. A Pivotal Role of Hormones in Regulating Cotton Fiber Development. Front. Plant Sci. 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tu, L.; Ye, Z.; Wang, M.; Gao, W.; Zhang, X. A cotton fiber-preferential promoter, PGbEXPA2, is regulated by GA and ABA in Arabidopsis. Plant Cell Rep. 2015, 34, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, J.; Huang, Y.; Wang, S.; Wei, L.; Liu, D.; Weng, Y.; Xiang, J.; Zhu, Q.; Yang, Z.; et al. CottonMD: A multi-omics database for cotton biological study. Nucleic Acids Res. 2023, 51, D1446–D1456. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. Corrigendum to: IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, X.; Wang, L.; Nazir, M.F.; Fu, G.; Peng, Z.; Chen, B.; Xing, A.; Zhu, M.; Ma, X.; et al. Exploring the regulatory role of non-coding RNAs in fiber development and direct regulation of GhKCR2 in the fatty acid metabolic pathway in upland cotton. Int. J. Biol. Macromol. 2024, 266, 131345. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, S.; Dia, S.; Sun, G.; Liu, X.; Wang, X.; Pan, Z.; Jia, Y.; Wang, L.; Pang, B.; et al. Alien genomic introgressions enhanced fiber strength in upland cotton (Gossypium hirsutum L.). Ind. Crop Prod. 2021, 159, 113028. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Qanmber, G.; Liu, J.; Yu, D.; Liu, Z.; Lu, L.; Mo, H.; Ma, S.; Wang, Z.; Yang, Z. Genome-Wide Identification and Characterization of the PERK Gene Family in Gossypium hirsutum Reveals Gene Duplication and Functional Divergence. Int. J. Mol. Sci. 2019, 20, 1750. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.Y.; Chen, B.J.; Pei, X.X.; Wang, X.; Wang, X.; Nazir, M.F.; Wang, J.; Zhang, X.; Xing, A.; Pan, Z.; et al. Genome-wide analysis of the serine carboxypeptidase-like protein family reveals Ga09G1039 is involved in fiber elongation in cotton. Plant Physiol. Biochem. 2023, 201, 107759. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.J.; Xiang, Y. REVERSAL OF RDO5 1, a Homolog of Rice Seed Dormancy4, Interacts with bHLH57 and Controls ABA Biosynthesis and Seed Dormancy in Arabidopsis. Plant Cell 2020, 32, 1933–1948. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Ray, A.; Mandal, D.; Nag Chaudhuri, R. ABI3 mediated repression of RAV1 gene expression promotes efficient dehydration stress response in Arabidopsis thaliana. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194582. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Lee, H.; Lee, J.; Kim, H.; Ryu, H. ABSCISIC ACID-INSENSITIVE 3 is involved in brassinosteroid-mediated regulation of flowering in plants. Plant Physiol. Biochem. 2019, 139, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Manan, S.; Zhao, J. Role of Glycine max ABSCISIC ACID INSENSITIVE 3 (GmABI3) in lipid biosynthesis and stress tolerance in soybean. Funct. Plant Biol. 2021, 48, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, P.; Chen, S.; Sun, L.; Mao, J.; Tan, S.; Xiang, C. The ABI3-ERF1 module mediates ABA-auxin crosstalk to regulate lateral root emergence. Cell Rep. 2023, 42, 112809. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Gampala, S.S.; Ritchie, G.L.; Payton, P.; Burke, J.J.; Rock, C.D. Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol. J. 2014, 12, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.; Sun, Y.; Peng, X.; Wu, G.; Bao, F.; He, Y.; Zhou, H.; Lin, H. ABSCISIC ACID INSENSITIVE3 Is Involved in Cold Response and Freezing Tolerance Regulation in Physcomitrella patens. Front. Plant Sci. 2017, 8, 1599. [Google Scholar] [CrossRef] [PubMed]
- Delmas, F.; Sankaranarayanan, S.; Deb, S.; Widdup, E.; Bournonville, C.; Bollier, N.; Northey, J.G.; McCourt, P.; Samuel, M.A. ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA 2013, 110, E3888–E3894. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xia, L.; Deng, J.; Sun, S.; Yue, D.; You, J.; Wang, M.; Jin, S.; Zhu, L.; Lindsey, K.; et al. Evolution and subfunctionalization of CIPK6 homologous genes in regulating cotton drought resistance. Nat. Commun. 2024, 15, 5733. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Z.; You, C.; Qi, Z.; You, J.; Grover, C.E.; Long, Y.; Huang, X.; Lu, S.; Wang, Y.; et al. Convergence and divergence of diploid and tetraploid cotton genomes. Nat. Genet. 2024, 56, 2562–2573. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, E. Do not panic: An intron-centric guide to alternative splicing. Plant Cell 2023, 35, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Sakharkar, M.K.; Chow, V.T.; Kangueane, P. Distributions of exons and introns in the human genome. Silico Biol. 2004, 4, 387–393. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.; Luo, S.; Ma, W.; Li, N.; Zhang, W.; Tikunov, Y.; Xuan, S.; Zhao, J.; Wang, Y.; et al. Discovery of a DFR gene that controls anthocyanin accumulation in the spiny Solanum group: Roles of a natural promoter variant and alternative splicing. Plant J. 2022, 111, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Oudelaar, A.M.; Higgs, D.R. The relationship between genome structure and function. Nat. Rev. Genet. 2021, 22, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Monzo, C.; Liu, T.; Conesa, A. Transcriptomics in the era of long-read sequencing. Nat. Rev. Genet. 2025, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Glinos, D.A.; Garborcauskas, G.; Hoffman, P.; Ehsan, N.; Jiang, L.; Gokden, A.; Dai, X.; Aguet, F.; Brown, K.L.; Garimella, K.; et al. Transcriptome variation in human tissues revealed by long-read sequencing. Nature 2022, 608, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, Y.; Li, Y.; Zhang, R.; Yang, J.; Ma, H.; Min, L.; Zhang, X. Reversing anther thermotolerance by manipulating the cis-elements in the promoter of a high-temperature upregulated gene Casein Kinase I in upland cotton. Sci. China Life Sci. 2025, 68, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, Z.R.; Li, F.G. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol. J. 2019, 17, 1706–1722. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, G.; Yang, Y.; Mahmood, T.; Liu, X.; Xie, Z.; Zhao, Z.; Dong, Y.; Tian, Y.; Farooq, J.; Sharif, I.; et al. Genome-Wide Characterization of the ABI3 Gene Family in Cotton. Genes 2025, 16, 854. https://doi.org/10.3390/genes16080854
Fu G, Yang Y, Mahmood T, Liu X, Xie Z, Zhao Z, Dong Y, Tian Y, Farooq J, Sharif I, et al. Genome-Wide Characterization of the ABI3 Gene Family in Cotton. Genes. 2025; 16(8):854. https://doi.org/10.3390/genes16080854
Chicago/Turabian StyleFu, Guoyong, Yanlong Yang, Tahir Mahmood, Xinxin Liu, Zongming Xie, Zengqiang Zhao, Yongmei Dong, Yousheng Tian, Jehanzeb Farooq, Iram Sharif, and et al. 2025. "Genome-Wide Characterization of the ABI3 Gene Family in Cotton" Genes 16, no. 8: 854. https://doi.org/10.3390/genes16080854
APA StyleFu, G., Yang, Y., Mahmood, T., Liu, X., Xie, Z., Zhao, Z., Dong, Y., Tian, Y., Farooq, J., Sharif, I., & Li, Y. (2025). Genome-Wide Characterization of the ABI3 Gene Family in Cotton. Genes, 16(8), 854. https://doi.org/10.3390/genes16080854






