Discovery of Germplasm Resources and Molecular Marker-Assisted Breeding of Oilseed Rape for Anticracking Angle
Abstract
1. Introduction
1.1. Mining Germplasm Resources for Pod Shattering Resistance in Rapeseed
1.2. Mapping QTLs for Pod Shattering Resistance in Rapeseed
2. Materials and Methods
2.1. Plant Materials
- (1)
- Core Test Population: A total of 634 single-plant progenies derived from crosses between the pod shatter-resistant donor parent KL01 and elite parental lines/cultivars, including the NR series, W series, and ‘Yuyou 342’;
- (2)
- Parental Materials: Thirteen commonly used breeding parents, namely KL01, 0911, R10, T8, 187308, and others;
- (3)
- Control Materials: The highly susceptible line 0911 (Shattering Resistance Index, SRI = 0.32) and the resistant source KL01 (SRI = 0.67).
2.2. Field Environmental Conditions and Crop Management
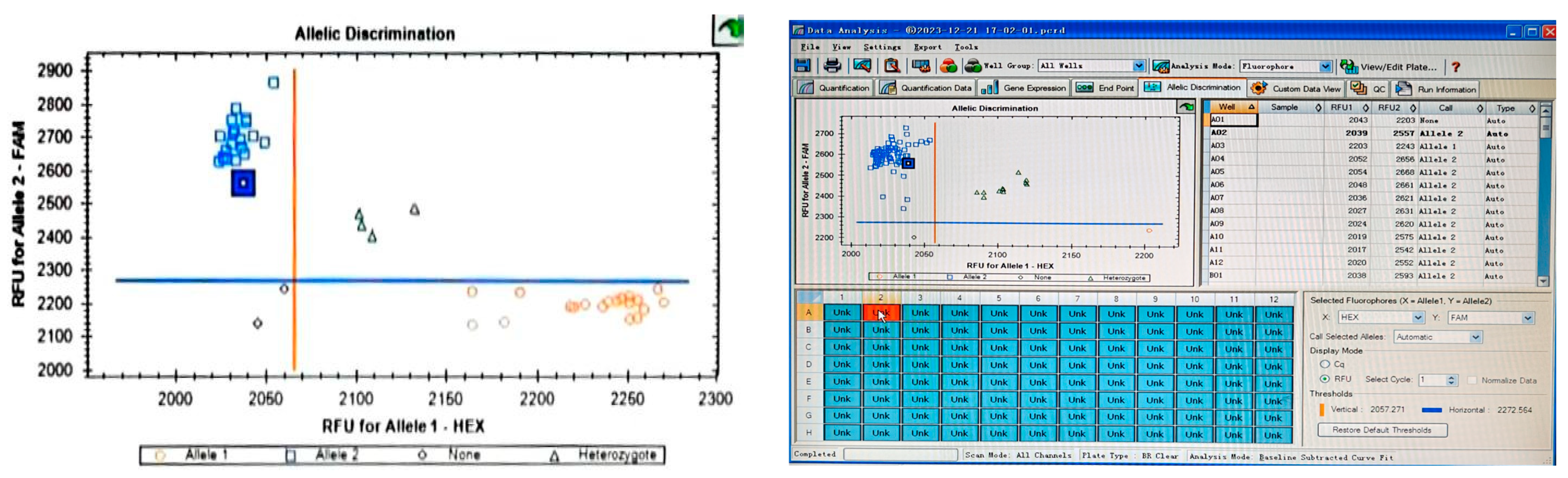
2.3. Molecular Marker Analysis
- S12.68-Fam: 5’-GAAGGTGACCAAGTTCATGCT-3’
- S12.68-Hex: 5’-GAAGGTCGGAGTCAACGGATT-3’
- S12.68-R: 5’-TGGTGGCTTGATGCTCTTC-3’.
- (1)
- Primer stock solutions (100 μM) were diluted to prepare a primer master mix: A total of 15 μL common primer, 6 μL of each allele-specific primer, and ddH2O added to a final volume of 50 μL;
- (2)
- Genomic DNA was extracted from target plants using the CTAB method or other established plant DNA extraction protocols. DNA concentrations were normalized to 10–20 ng/μL;
- (3)
- KASP reactions were assembled in 96-well or 384-well PCR plates according to manufacturer-recommended volumes scaled for the specific plate format (Table 1);
- (4)
- Assembled reactions were subjected to PCR amplification and fluorescence detection in a real-time PCR instrument (capable of detecting FAM, HEX, and ROX channels) (Table 2);
- (5)
- Post-amplification, endpoint fluorescence was measured using platform-specific software. For the Bio-Rad CFX96 Real-Time PCR System employed in this study, genotype calls were generated using the Allelic Discrimination module with the Display Mode set to Relative Fluorescence Units (RFU). Endpoint fluorescence data were analyzed using LGC SNPline Software (v4.1, LGC Genomics, London, UK). Allele calling thresholds were set as ΔRn ≥ 0.3 for cluster separation, with manual verification of ambiguous calls.
2.4. Phenotypic Evaluation
- SRI = ∑[1 − (Number of Shattered Pods/20)]
- Resistance Classification Standard:
- SRI ≤ 0.2: Susceptible (S)
- 0.2 < SRI ≤ 0.4: Moderately Susceptible (MS)
- 0.4 < SRI ≤ 0.6: Low Resistance (LR)
- SRI > 0.6: Moderately Resistant (MR)
- SRI > 0.8: Highly Resistant (HR).
3. Results
3.1. Efficient Introgression and Genetic Stability of Pod Shattering Resistance Allele
- Homozygous Resistant (HR): 73.34% (465 plants)
- Heterozygous (Het): 24.92% (158 plants)
- No Introgression (Susceptible): 0.16% (1 plant).
3.2. Screening of Elite Pod Shatter-Resistant Germplasm and Novel Resistance Mechanisms
3.3. Development of Multi-Trait Elite Breeding Materials
4. Discussion
4.1. Technical Advantages of the Dual-Selection System
4.2. Breakthrough Value of Novel Germplasm and Mechanisms
4.3. Limitations and Future Perspectives
- (1)
- The single-locus specificity of S12.68 necessitates developing multi-gene markers targeting BnSHP1, BnIND, and allied genes to enable resistance pyramiding;
- (2)
- Random-impact assays require multi-location validation to quantify Genotype × Environment (G×E) interactions;
- (3)
- Putative novel loci inferred from 187308 demand linkage analysis or comparative transcriptomics for mechanistic elucidation.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cai, Z.Y. Rapeseed maturity and harvesting. Oil Crop Sci. 1980, 4, 38–39. (In Chinese) [Google Scholar]
- Chen, Z.G. Harvesting and storage techniques for rapeseed. Anhui Agric. 1998, 4, 1–9. (In Chinese) [Google Scholar]
- Shen, J.X.; Qi, L.P.; Yang, J.; Wen, J.; Yi, B.; Tu, J.X.; Ma, C.Z.; Fu, T.D. Discovery and preliminary study of a “club-shaped” mutant in Brassica napus L. Oil Crop Sci. 2008, 31, 380–382, (In Chinese with English Abstract). [Google Scholar]
- Wu, C.Y.; Xiao, S.Y.; Jin, M. Comparison and Development Prospects of Rapeseed Harvesting Methods. In Proceedings of the 7th Membership Conference and Annual Symposium of the Oil Crops Professional Committee, Hainan, China, 20 November 2013. (In Chinese). [Google Scholar]
- Tys, J.; Bengtsson, L. Estimation of Rape Silique Resistance to Cracking and Rapeseed Shattering Resistance for Some Selected Varieties and Lines of Spring Rape. In Proceedings of the 8th International Rapeseed Congress, Saskatoon, SK, Canada, 9–11 July 1991; pp. 1848–1858. [Google Scholar]
- He, Y.T.; Li, D.R. Preliminary studies on pod shattering resistance of hybrid Brassica napus. Shaanxi J. Agric. Sci. 1996, 3, 30–31. (In Chinese) [Google Scholar]
- Summers, J.E.; Bruce, D.M.; Vancanneyt, G.; Redig, P.; Werner, C.P.; Morgan, C.; Child, R.D. Pod shatter resistance in the resynthesized Brassica napus line DK142. J. Agric. Sci. 2003, 140, 43–52. [Google Scholar] [CrossRef]
- Sun, C.C.; Wang, W.R.; Li, Y.L.; Qian, X.F. Breeding of ‘Huyou 17’, a new double-low rapeseed cultivar suitable for mechanized harvesting. Oil Crop Sci. 2005, 27, 16–17, (In Chinese with English Abstract). [Google Scholar]
- Wen, Y.C.; Fu, T.D.; Tu, J.X.; Ma, C.; Shen, J.; Zhang, S. Screening and analysis of pod shatter-resistant varieties/lines in Brassica napus. Acta Agron. Sin. 2008, 34, 163–166, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Peng, P.F.; Li, Y.C.; Hu, Q.; Liu, F.L.; Li, Y.D. Identification and varietal screening for pod shattering resistance in Brassica napus. Acta Agric. Boreali-Sin. 2009, 24, 223–226, (In Chinese with English Abstract). [Google Scholar]
- Dong, J.G.; Dong, Z.S.; Meng, Q.; Zhang, B. Screening of pod shatter-resistant germplasm resources in Brassica napus. Acta Agron. Sin. 2014, 40, 2203–2209, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Wang, J.; Wei, Z.F.; Li, D.W.; Wang, L.L.; Zhang, T.P. Screening of pod shatter-resistant germplasm and analysis of related traits in Brassica napus. Resour. Sci. 2017, 36, 51–54. (In Chinese) [Google Scholar]
- Zhang, Y.H.; Zhang, Y.J.; Zhou, R.M.; Zhou, Y.T.; Li, A.M. Pod shattering resistance in interspecific hybrids between Brassica napus and Sinapis alba and its correlation with silique traits. Oil Crop Sci. 2017, 39, 18–22, (In Chinese with English Abstract). [Google Scholar]
- Braatz, J.; Harloff, H.J.; Emrani, N.; Elisha, C.; Heepe, L.; Gorb, S.N.; Jung, C. The effect of INDEHISCENT point mutations on silique shatter resistance in oilseed rape (Brassica napus). TAG Theor. Appl. Genet. Theor. Angew. Genet. 2018, 131, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Kadkol, G.P.; Halloran, G.M.; Macmillan, R.H. Inheritance of siliqua strength in Brassica campestris L. I. Studies of F2 and backcross populations. Can. J. Genet. Cytol. 1986, 28, 365–373. [Google Scholar] [CrossRef]
- Mongkolporn, O.; Kadkol, G.P.; Pang, E.C.K.; Taylor, P.W.J. Identification of RAPD markers linked to recessive genes conferring siliqua shatter resistance in Brassica rapa. Plant Breed. 2003, 122, 479–484. [Google Scholar] [CrossRef]
- Peng, P.F. Identification, Genetic Analysis and QTL Mapping of Pod Shattering Resistance in Brassica napus; Chinese Academy of Agricultural Sciences: Beijing, China, 2009; (In Chinese with English Title). [Google Scholar]
- Wen, Y.C. Analysis and QTL Mapping of Pod Shattering Resistance Trait in Brassica napus L.; Huazhong Agricultural University: Wuhan, China, 2012; (In Chinese with English Title). [Google Scholar]
- Hu, Z.Y.; Hua, W.; Huang, S.M.; Yang, H.; Zhan, G.; Wang, X.; Wang, H. Discovery of pod shatter-resistant associated SNPs by deep sequencing of a representative library followed by bulk segregant analysis in rapeseed. PLoS ONE 2012, 7, e40736. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sang, S.F.; Mei, D.S.; Li, Y.C.; Liu, J.; Fu, L.; Wang, J.; Chen, Y.F.; Hu, Q. Genetic analysis of pod shattering resistance in a Brassica napus DH population. Oil Crop Sci. 2014, 36, 437–442, (In Chinese with English Abstract). [Google Scholar]
- Raman, H.; Raman, R.; Kilian, A.; Detering, F.; Carling, J.; Coombes, N.; Wratten, N. Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS ONE 2014, 9, e101673. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, J.; Wang, H.; Zhou, R.; Mei, D.; Hu, Q. Multigenic control of pod shattering resistance in Chinese rapeseed germplasm revealed by genome-wide association and linkage analyses. Plant Sci. 2016, 7, 1058. [Google Scholar] [CrossRef] [PubMed]
- Braatz, J.; Harloff, H.J.; Jung, C. EMS-induced point mutations in ALCATRAZ homoeologs increase silique shatter resistance of oilseed rape (Brassica napus). Euphytica 2018, 214, 29. [Google Scholar] [CrossRef]
- Tao, Z.; Huang, Y.; Zhang, L.; Wang, X.; Liu, G.; Wang, H. BnLATE, a Cys2/His2-Type Zinc-Finger Protein, Enhances Silique Shattering Resistance by Negatively Regulating Lignin Accumulation in the Silique Walls of Brassica napus. PLoS ONE 2017, 12, e0168046. [Google Scholar] [CrossRef] [PubMed]

| Reagent | 96-Well Plate Volume | 384-Well Plate Volume |
|---|---|---|
| 2× KASP V3 Master Mix (LGC) | 5 μL | 2.5 μL |
| Genomic DNA Template | 2 μL | 1 μL |
| Primer Master Mix | 0.14 μL | 0.07 μL |
| ddH2O | 2.86 μL | 1.43 μL |
| Total Volume | 10 μL | 5 μL |
| Step | Temperature | Time | Notes |
|---|---|---|---|
| 1 | 94 °C | 15 min | Initial Denaturation |
| 2 | 94 °C | 20 s | Denaturation |
| 3 | 61 °C | 60 s | Annealing/Extension (touchdown: −0.6 °C per cycle) |
| 4 | Go to Step 2 | --- | Repeat for 9 cycles (total touchdown) |
| 5 | 94 °C | 20 s | Denaturation |
| 6 | 55 °C | 60 s | Annealing/Extension |
| 7 | Go to Step 5 | --- | Repeat for 26 cycles |
| 8 | 37 °C | 60 s | Fluorescence Measurement |
| Material Source | Homozygous Susceptible | Homozygous Resistant | Heterozygous | No Target Allele Detected | Total |
|---|---|---|---|---|---|
| 0911 | 1 | 1 | |||
| KL01 | 1 | 1 | |||
| R10 | 1 | 1 | |||
| T8 | 1 | 1 | |||
| KL01/NR17 | 23 | 2 | 3 | 28 | |
| KL01/NR20 | 22 | 2 | 24 | ||
| KL01/NR21 | 3 | 2 | 5 | ||
| KL01/NR22 | 26 | 26 | |||
| KL01/NR23 | 24 | 3 | 27 | ||
| KL01/NR24 | 53 | 53 | |||
| KL01/NR25 | 12 | 15 | 27 | ||
| KL01/NR26 | 26 | 1 | 27 | ||
| KL01/NR27 | 6 | 6 | |||
| NR17/KL01 | 11 | 2 | 13 | ||
| NR20/KL01 | 1 | 8 | 9 | ||
| NR21/KL01 | 24 | 24 | |||
| NR22/KL01 | 25 | 25 | |||
| NR23/KL01 | 23 | 3 | 26 | ||
| NR24/KL01 | 26 | 1 | 27 | ||
| NR25/KL01 | 12 | 13 | 25 | ||
| NR26/KL01 | 15 | 15 | |||
| NR27/KL01 | 26 | 1 | 27 | ||
| W1/KL01 | 11 | 15 | 1 | 27 | |
| W2/KL01 | 1 | 1 | |||
| W3/KL01 | 11 | 10 | 1 | 22 | |
| W4/KL01 | 11 | 5 | 2 | 18 | |
| W5/KL01 | 7 | 19 | 26 | ||
| W6/KL01 | 2 | 2 | |||
| W7/KL01 | 12 | 14 | 26 | ||
| W8/KL01 | 26 | 1 | 27 | ||
| W9/KL01 | 1 | 1 | |||
| W10/KL01 | 27 | 27 | |||
| Zao 5/KL01 | 1 | 15 | 16 | ||
| Yuyou 342/KL01 | 27 | 27 | |||
| Total | 4 | 466 | 159 | 10 | 638 |
| Material Source | Homozygous Susceptible | Homozygous Resistant | Heterozygous |
|---|---|---|---|
| KL01 | • | ||
| 0911 | • | ||
| R10 | • | ||
| ZY-13 | • | ||
| T8 | • | ||
| 0911A | • | ||
| 18Z23 | • | ||
| 186037 | • | ||
| 187308 | • | ||
| 187338 | • | ||
| 10032 | • | ||
| 10021 | • | ||
| 20M99 | • | ||
| NR209-1 | • | ||
| NR215-6 | • | ||
| NR232-2 | • | ||
| NR236-1 | • | ||
| NR280-6 | • | ||
| NR301-2 | • | ||
| NR318-6 | • |
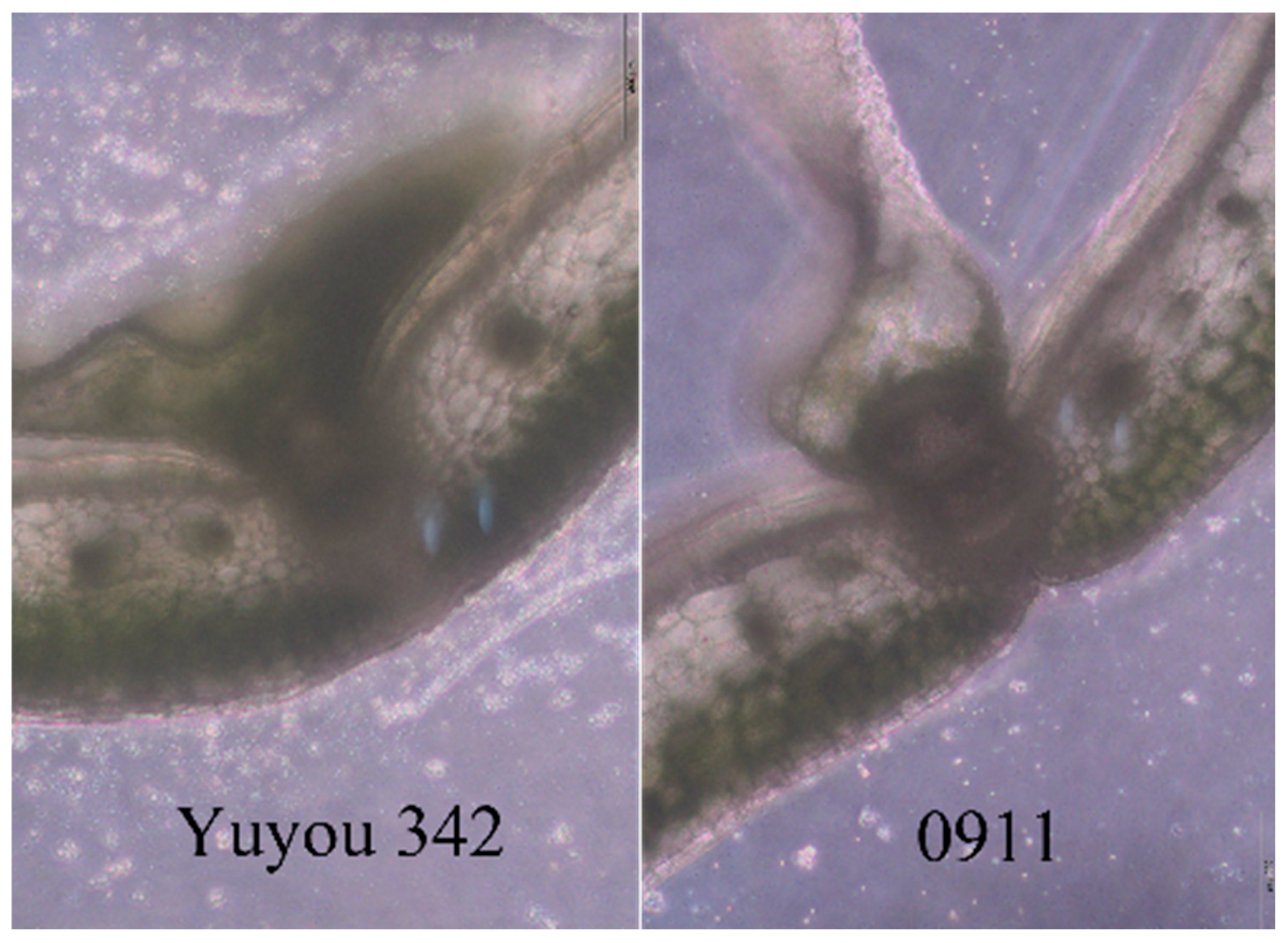
| Material | Shattered Pods (Stage 1) | Shattered Pods (Stage 2) | Shattered Pods (Stage 3) | Shattered Pods (Stage 4) | Shattered Pods (Stage 5) | SRI | Resistance Category |
|---|---|---|---|---|---|---|---|
| KL01 | 2 | 3 | 2 | 2 | 1 | 0.67 | MR |
| 0911 | 15 | 0 | 4 | 0 | 1 | 0.12 | S |
| Yuyou 342 | 4 | 1 | 1 | 3 | 5 | 0.62 | MR |
| 10032 | 20 | 0 | 0 | 0 | 0 | 0 | S |
| 187308 | 0 | 3 | 2 | 1 | 2 | 0.78 | MR |
| Qingyou3 | 3 | 6 | 5 | 4 | 0 | 0.38 | MS |
| W10/KL01 | 0 | 0 | 0 | 1 | 1 | 0.97 | HR |
| 187338/KL01 | 0 | 3 | 1 | 2 | 3 | 0.76 | MR |
| Yuyou 342/KL01 | 0 | 0 | 0 | 1 | 1 | 0.97 | HR |
| NR21/KL01 | 0 | 0 | 0 | 0 | 0 | 1 | HR |
| Material ID | Pod Shattering Genotype | SRI | Oil Content (%) | Resistance Category | Plant Height (cm) | Breeding Application |
|---|---|---|---|---|---|---|
| NR215-6 | Homozygous Resistant | 0.92 | 46.8 | HR | 162 | Elite parent for improving high-yielding cultivars |
| NR232-2 | Homozygous Resistant | 0.88 | 47.2 | HR | 155 | Cultivars optimized for mechanized harvesting |
| NR236-1 | Homozygous Resistant | 0.85 | 45.9 | HR | 168 | Multi-disease resistant breeding lines |
| NR280-6 | Homozygous Resistant | 0.91 | 44.7 | HR | 158 | Novel varieties for mechanized systems |
| NR301-2 | Homozygous Resistant | 0.94 | 48.7 | HR | 172 | Ultra-high oil content breeding |
| NR318-6 | Homozygous Resistant | 0.89 | 47.5 | HR | 161 | Broadly adapted cultivars for diverse agroecosystems |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Li, Z.; Liu, R.; Huang, T. Discovery of Germplasm Resources and Molecular Marker-Assisted Breeding of Oilseed Rape for Anticracking Angle. Genes 2025, 16, 831. https://doi.org/10.3390/genes16070831
Zhu C, Li Z, Liu R, Huang T. Discovery of Germplasm Resources and Molecular Marker-Assisted Breeding of Oilseed Rape for Anticracking Angle. Genes. 2025; 16(7):831. https://doi.org/10.3390/genes16070831
Chicago/Turabian StyleZhu, Cheng, Zhi Li, Ruiwen Liu, and Taocui Huang. 2025. "Discovery of Germplasm Resources and Molecular Marker-Assisted Breeding of Oilseed Rape for Anticracking Angle" Genes 16, no. 7: 831. https://doi.org/10.3390/genes16070831
APA StyleZhu, C., Li, Z., Liu, R., & Huang, T. (2025). Discovery of Germplasm Resources and Molecular Marker-Assisted Breeding of Oilseed Rape for Anticracking Angle. Genes, 16(7), 831. https://doi.org/10.3390/genes16070831





